Copyright
©The Author(s) 2021.
World J Clin Cases. Dec 16, 2021; 9(35): 10884-10898
Published online Dec 16, 2021. doi: 10.12998/wjcc.v9.i35.10884
Published online Dec 16, 2021. doi: 10.12998/wjcc.v9.i35.10884
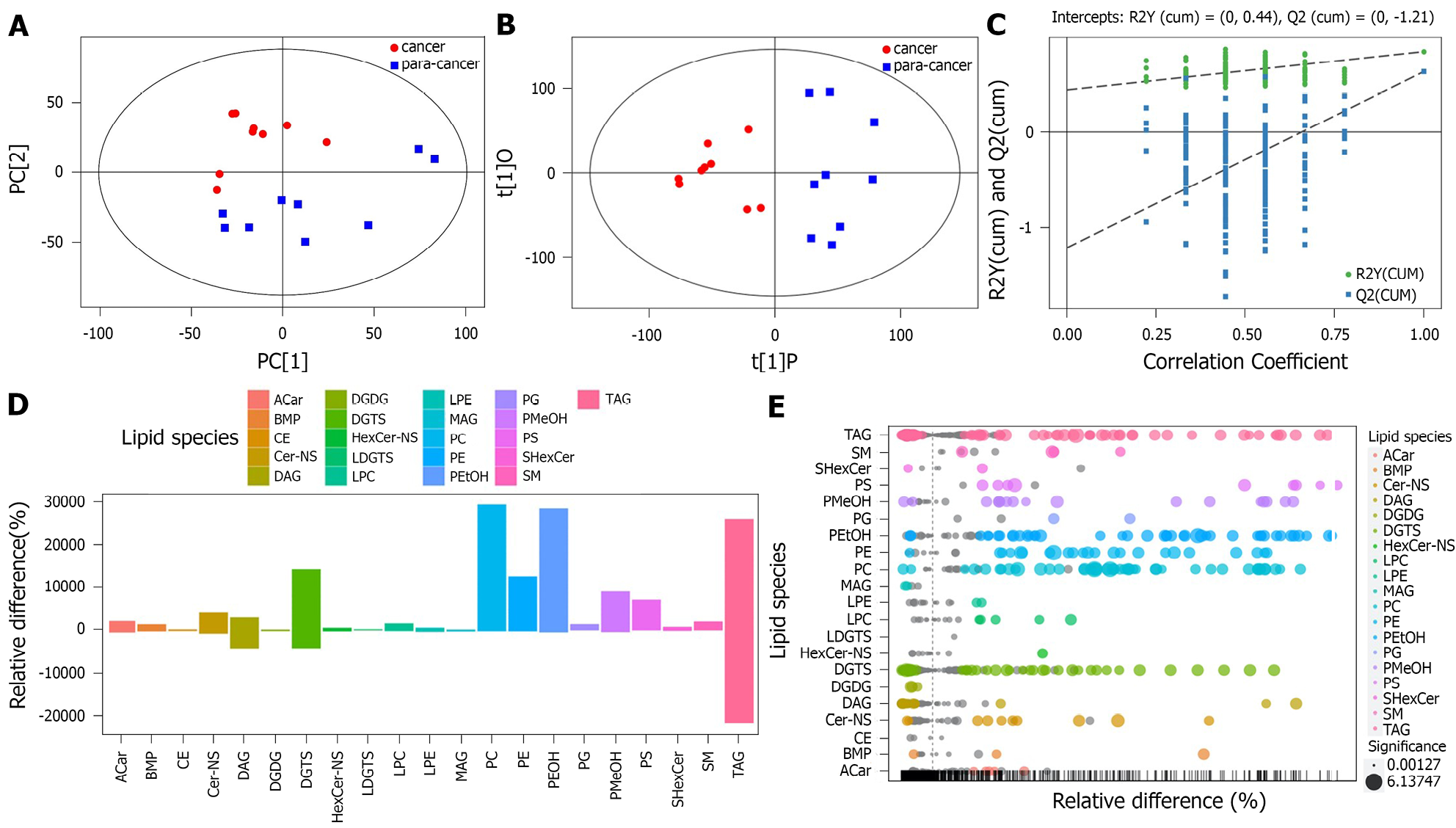
Figure 1 Identification of differential lipid metabolites.
A: Score scatter plot of the PCA model for the cancer group vs. the paracarcinoma tissue group; B: Score scatter plot of the orthogonal projections to latent structures-discriminant analysis (OPLS-DA) model for the cancer group vs. the paracancer group; C: Permutation test of the OPLS-DA model for the cancer group vs. the para-cancer group; D: Bar plot for the cancer group vs. the paracancer group. TAG: triacylglycerols; SM: sphingomyelin; SHeXcer: sulfatide; PS: phosphatidylserines; PMeOH: phosphatidylmethanol; PG: phosphatidylglycerols; PEtOH: phosphatidylethanol; PE: phosphatidylethanolamines; PC: phosphatidylcholines; MAG: myelin-associated glycoprotein; LPE: lyso-PE; LPC: lyso-PC; LDGTS: lyso-DGTS; HexCer: hexosylceramide; ACar: acaraben; DGTS: diacylglyceryl trimethylhomoserine; DGDG: digalactosyl diacylglycerols; DAG: diacylglycerols; Cer: ceramide; CE: cholesteryl esters; BMP: bis(monoacylglycerol)phosphate.
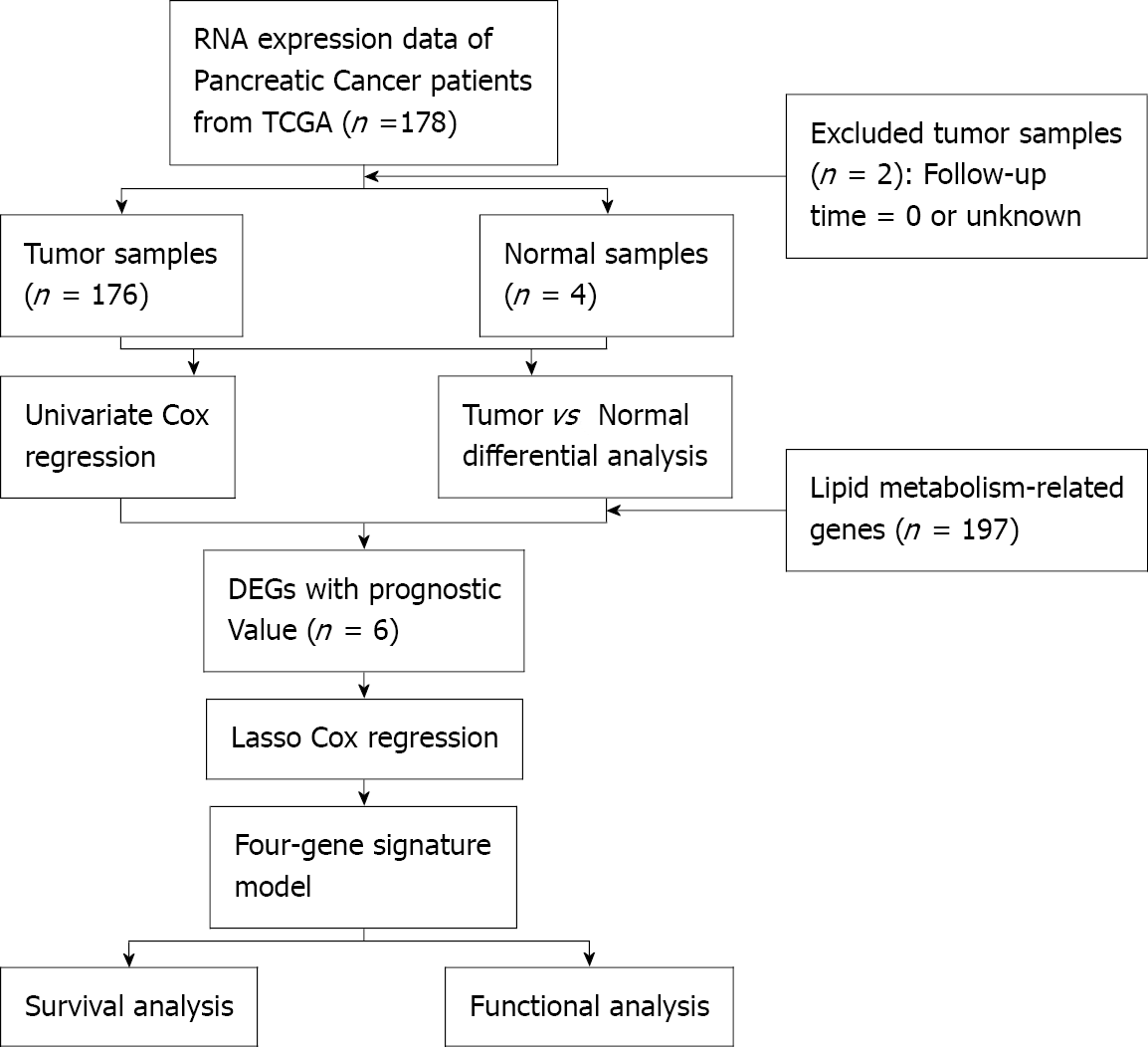
Figure 2 Flow chart of data collection and analysis.
TCGA: The Cancer Genome Atlas; DEGs: differentially expressed genes.
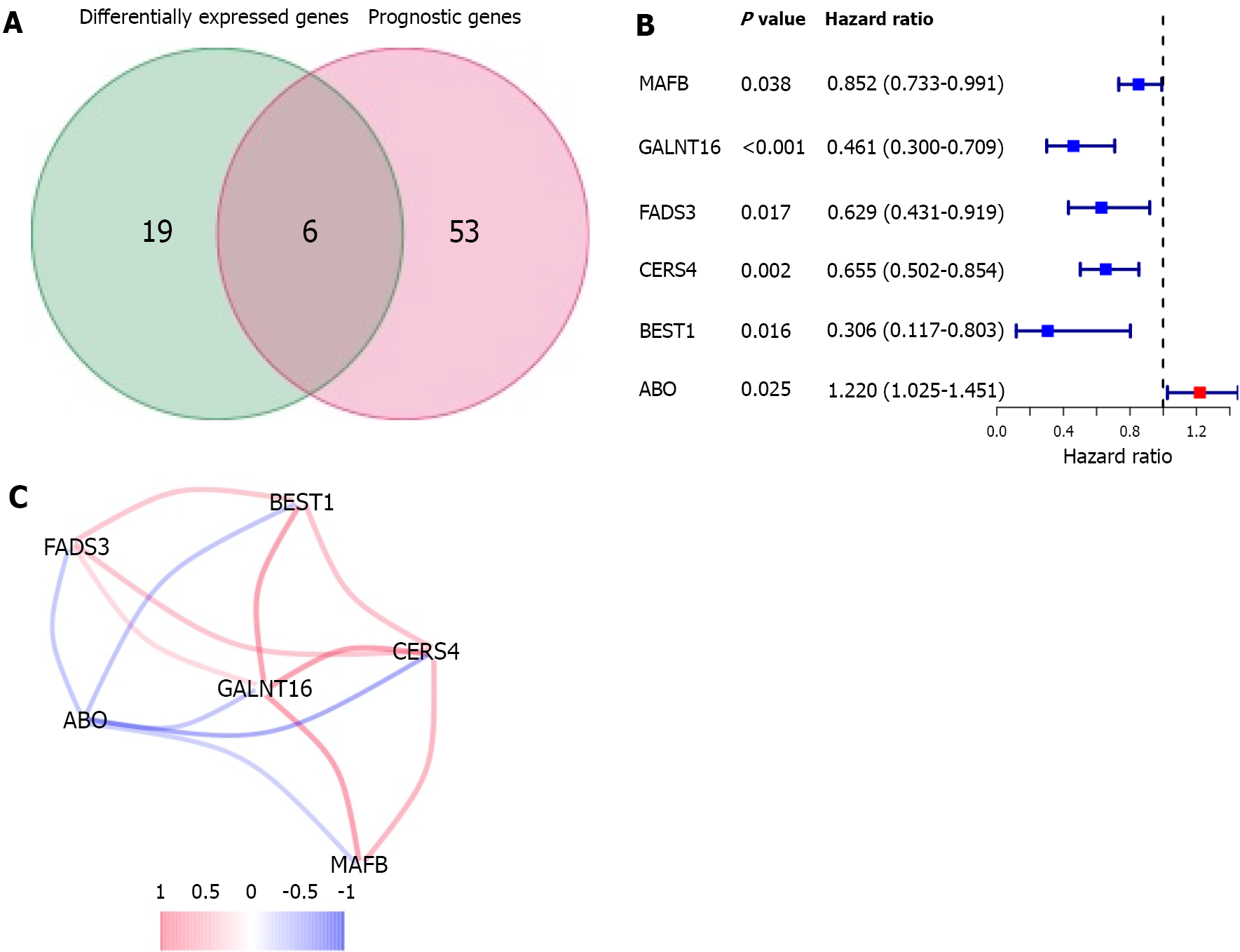
Figure 3 Candidate genes related to lipid metabolism in The Cancer Genome Atlas.
A: Venn diagrams were used to identify differentially expressed genes associated with overall survival between tumor tissues and adjacent normal tissues; B: The forest plot shows the relationship between gene expression and overall survival by univariate Cox regression analysis; C: Correlation coefficients in the correlation network of candidate genes are shown in different colors.
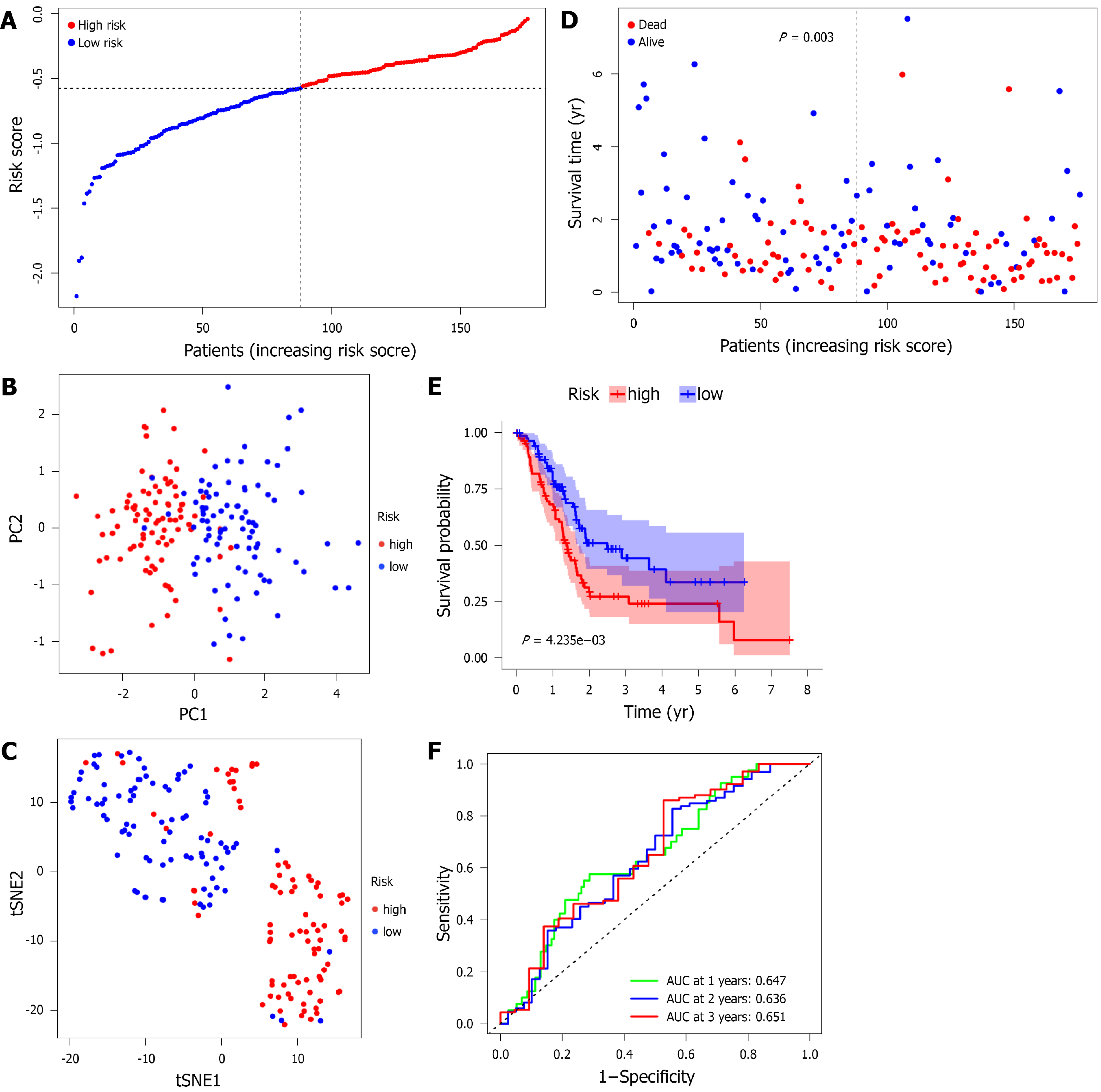
Figure 4 Prognostic analysis of 4-gene signature models in The Cancer Genome Atlas.
A: Distribution of risk score values in The Cancer Genome Atlas (TCGA); B: Principal component analysis plot of the pancreatic adenocarcinoma (PAAD) cohort from TCGA; C: t-distributed stochastic neighbor embedding analysis of the PAAD cohort from TCGA; D: Distribution of overall survival status, survival time, and risk scores in the cohort from TCGA; E: The Kaplan-Meier curve presents the overall survival of patients in the high-risk and low-risk groups in the PAAD cohort from TCGA; F: The prognostic performance of the risk score in the PAAD cohort from TCGA was verified by the area under the curve of the time-dependent receiver operating characteristic curve. PC: Principal component; tSNE: t-distributed stochastic neighbor embedding.
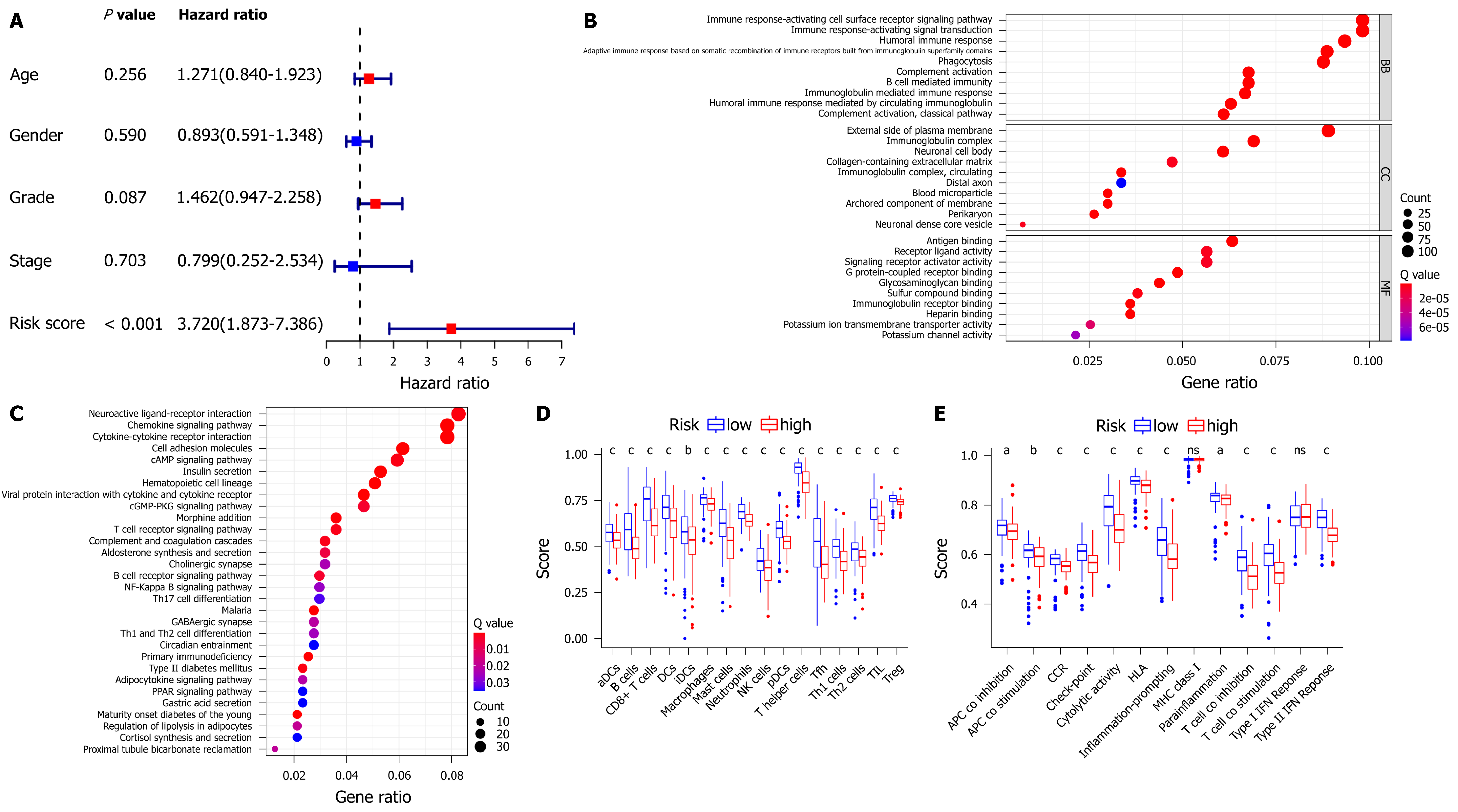
Figure 5 Construction of the predictive prognostic model and preliminary exploration of the potential mechanism.
A: Results of univariate Cox regression analyses of overall survival in The Cancer Genome Atlas cohort; B: Gene Ontology enrichment; C: Kyoto Encyclopedia of Genes and Genomes pathways; D: The scores of 16 immune cells; E: The scores of 13 immune-related functions. ns was P ≥ 0.05; aP < 0.05; bP < 0.01; cP < 0.001.
- Citation: Xu H, Sun J, Zhou L, Du QC, Zhu HY, Chen Y, Wang XY. Development of a lipid metabolism-related gene model to predict prognosis in patients with pancreatic cancer. World J Clin Cases 2021; 9(35): 10884-10898
- URL: https://www.wjgnet.com/2307-8960/full/v9/i35/10884.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i35.10884