Copyright
©The Author(s) 2018.
World J Clin Cases. Nov 6, 2018; 6(13): 577-588
Published online Nov 6, 2018. doi: 10.12998/wjcc.v6.i13.577
Published online Nov 6, 2018. doi: 10.12998/wjcc.v6.i13.577
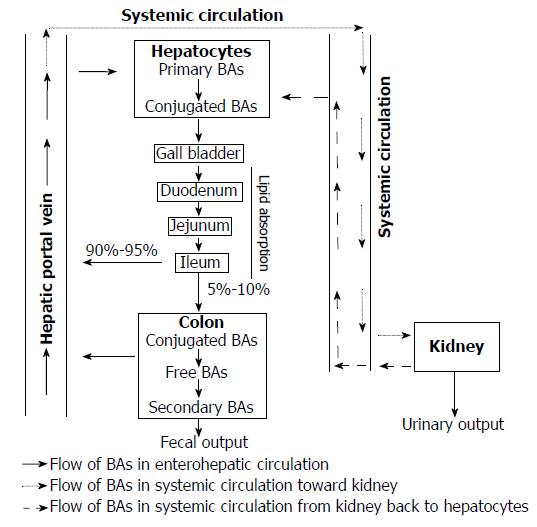
Figure 1 Bile acid circulation.
Primary bile acids (BAs) are mainly synthesized in liver from cholesterols. After that, they are conjugated with glycine or taurine, excreted in the bile, and stored in the gallbladder. After a meal, conjugated BAs are stimulated to release into the intestinal tract for facilitating the digestion of dietary lipids and fat-soluble vitamins. Then, BAs are efficiently reabsorbed in the ileum and most of them (90%-95%) is transported back to the liver via the hepatic portal vein to be cleared, re-secreted in the bile, and ready for new circulation. This is called enterohepatic circulation. In addition to enterohepatic circulation, about 10% of the total BA pool reaches the systemic circulation to the kidney to be filtrated by the renal glomeruli and then return to the liver for subsequent circulation. A small amount of BAs (5%-10%), which escapes from ileum re-absorption, flows to the large intestines, where some of it is de-conjugated by bacterial bile salt hydrolases to become free BA and converted to secondary BAs. They are then reabsorbed into colonocytes to return to the liver for detoxification and then re-cycling. Only a small amount of these BAs (about 5%-10%) are lost via feces. BAs: Bile acids.
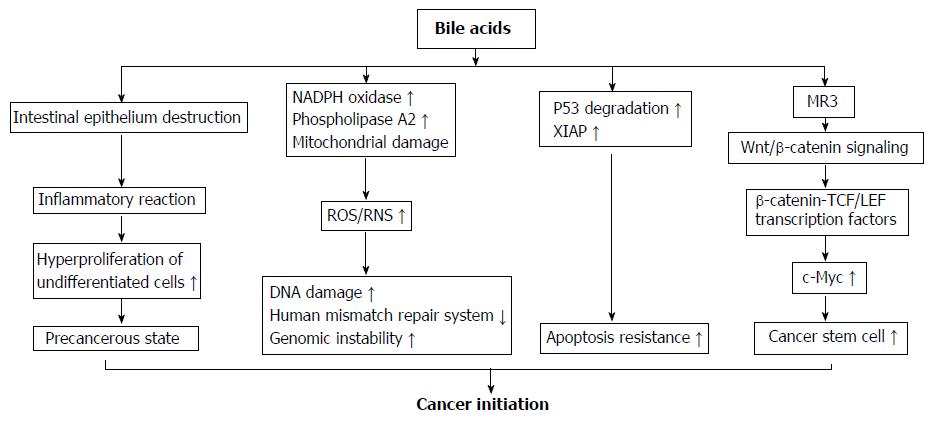
Figure 2 Mechanisms of bile acids initiating colorectal cancer development.
(1) High concentrations of bile acids (BAs) could cause a focal destruction of intestinal epithelium, subsequently stimulate repair mechanisms involving inflammatory reactions and hyper-proliferation of undifferentiated cells. These processes could cause a cell transition into a precancerous state and are considered as an early priming step in colorectal tumorigenesis; (2) BAs strongly induce reactive oxygenic species and reactive nitrogen species production via its stimulatory effect on nicotinamide adenine dinucleotide phosphate oxidase and phospholipase A2, and mitochondrial damage. These reactive species cause damage on DNA, and disrupt the base excision repair pathways; (3) After chronic exposure to BAs at physiological concentration, colon epithelial cells become resistant to apoptosis. This is because BAs induce the degradation of tumor suppressor p53, and up-regulate the expression of X-linked inhibitor of apoptosis protein protein. The cells with genomic errors coupled with apoptosis resistance ability rapidly get much further mutation and ultimately become cancer cells; (4) BAs induce colonic epithelial cells becoming CSCs through muscarinic cholinergic receptor receptor and Wnt/β-catenin signaling. This pathway leads to nuclear translocation of β-catenin to form a complex with T cell factor/lymphoid enhancer factor family transcription factors that acts as a co-activator to express c-Myc, a gene regulating cell stemness. NADPH oxidase: Nicotinamide adenine dinucleotide phosphate oxidase; MR3: Muscarinic cholinergic receptor 3; ROS: Reactive oxygenic species; RNS: Reactive nitrogen species; XIAP: X-linked inhibitor of apoptosis protein; TCF/LEF: T cell factor/lymphoid enhancer factor.
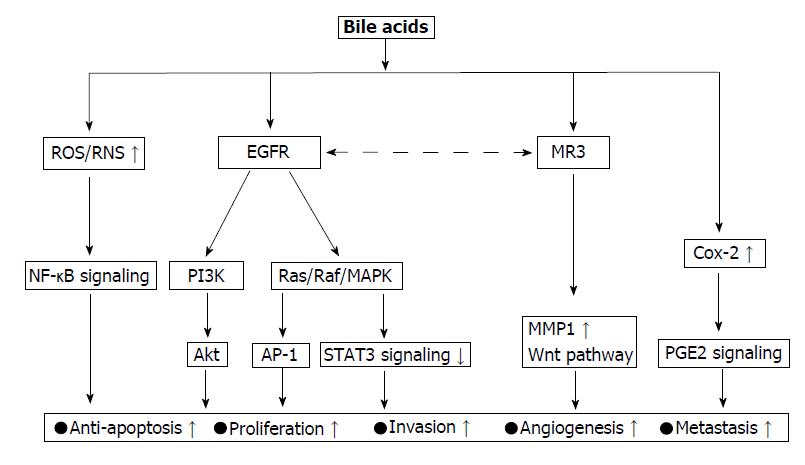
Figure 3 Oncogenic signaling network activated by bile acids.
The epidermal growth factor receptor (EGFR) pathway is central signaling for bile acid (BA) action associated with colorectal cancer (CRC) progression. Over-activation of this pathway in tumor cells is associated with tumor cell proliferation, survival, angiogenesis, invasion, and metastasis. PI3K and Ras/Raf/MAPK are two dominant downstream signaling cascades of EGFR activation. Muscarinic cholinergic receptor 3 (MR3) is another actor that mainly contribute to CRC initiation and progression mediated by BAs. This action of CR3 requires cross-talk with EGFR transactivation. MR3 activation stimulates MMP1 expression, which promotes CRC invasion and mediates for BA-induced CSCs in the colonic epithelial population via Wnt pathway. NF-κB is also one of major signaling activated by BAs. This signaling is activated by oxidative stress and as downstream of PI3K signaling. It protects cells against apoptosis and enhances cell proliferation. Cox-2/PGE2 is one of the signaling events of BAs action, BAs increase the synthesis of PGE2 via Cox-2 enzyme induction. This signaling increases the expression of Bcl-2, that then suppresses p53-induced apoptosis thereby promoting tumorigenesis. Additionally, PGE2 signaling is known to stimulate cAMP production, which can stimulate tumor growth by suppressing apoptosis. ROS: Reactive oxygen species; RNS: Reactive nitrogen species, NF-κB: Nuclear factor kappa-light-chain-enhancer of activated B cells; PI3K: Phosphatidylinositol-4,5-bisphosphate 3-kinase; EGFR: Epidermal growth factor receptor; MAPK: Mitogen activated protein kinase; AP-1: Activator protein 1; STAT3: Signal transducer and activator of transcription 3; Cox-2: Cyclooxygenase 2; PGE2: Prostaglandin E2; MR3: Muscarinic cholinergic receptor 3.
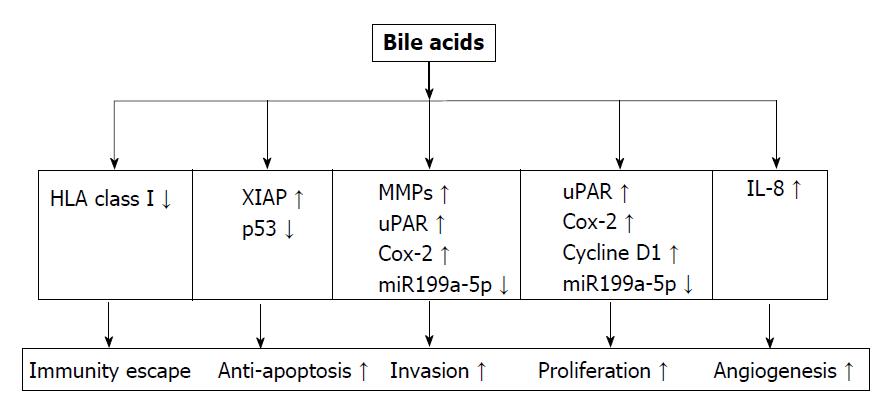
Figure 4 Bile acids regulate gene expression towards tumor development.
Bile acids (BAs) decrease the expression of human leukocyte antigen class I antigens on the surface of colorectal cancer cells, helping cancer cells escape from immune surveillance. They enhance the apoptosis resistance ability of cancer cells via up-regulating XIAP expression, and degrading p53 protein. BAs stimulates cancer proliferation and invasion via their effect on a series of oncogenes including MMPs, uPAR, Cox-2, Cycline D1, and miR199a-5p. By inducing IL-8 expression, BAs enhance angiogenesis activity for tumor growth and metastasis. HLA: Human leukocyte antigen; MMP: Matrix metalloproteinase; UPAR: Urokinase receptor; Cox: Cyclooxygenase; IL-8: Interleukin-8; VEGF: Vascular endothelial growth factor, XIAP: X-linked inhibitor of apoptosis protein.
- Citation: Nguyen TT, Ung TT, Kim NH, Jung YD. Role of bile acids in colon carcinogenesis. World J Clin Cases 2018; 6(13): 577-588
- URL: https://www.wjgnet.com/2307-8960/full/v6/i13/577.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v6.i13.577