Copyright
©The Author(s) 2024.
World J Clin Cases. Jun 26, 2024; 12(18): 3505-3514
Published online Jun 26, 2024. doi: 10.12998/wjcc.v12.i18.3505
Published online Jun 26, 2024. doi: 10.12998/wjcc.v12.i18.3505
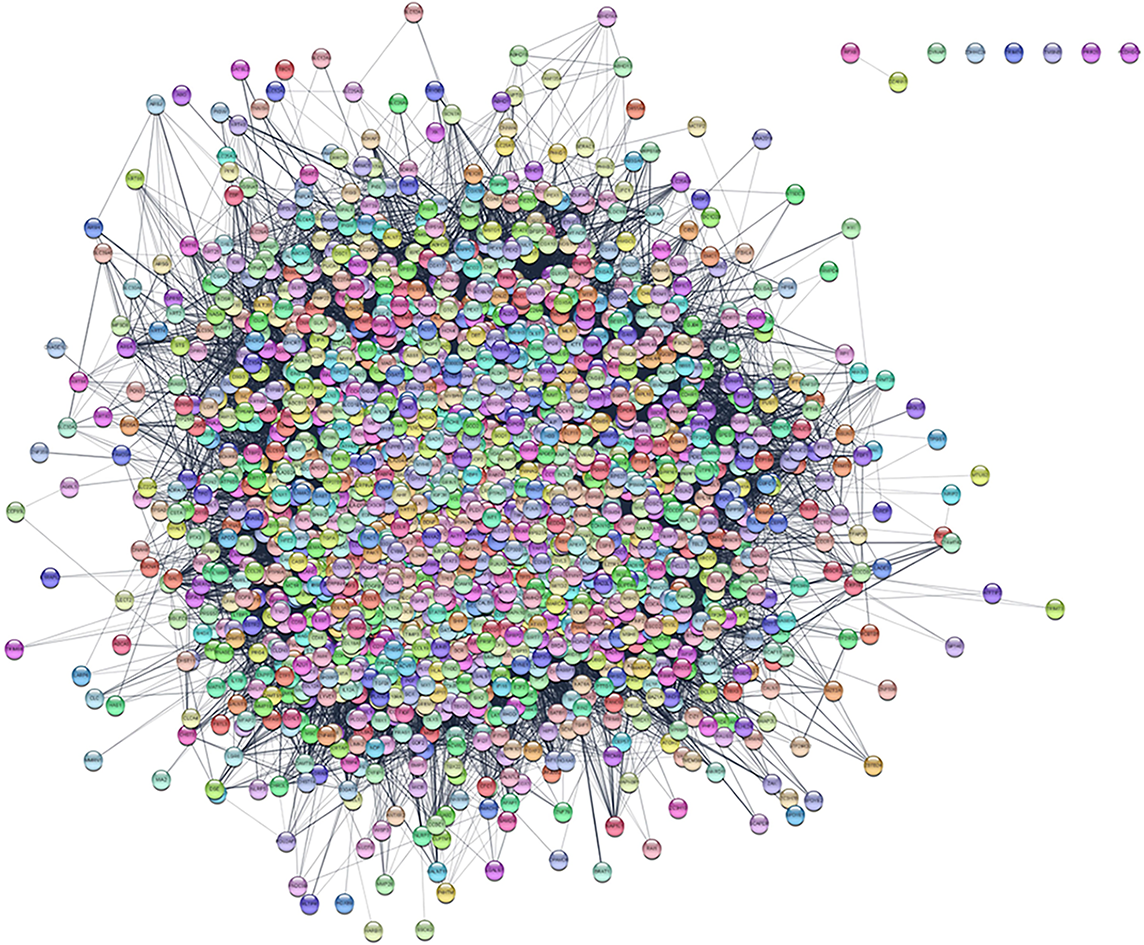
Figure 1 The protein-protein interaction network of hypertrophic scar-related genes.
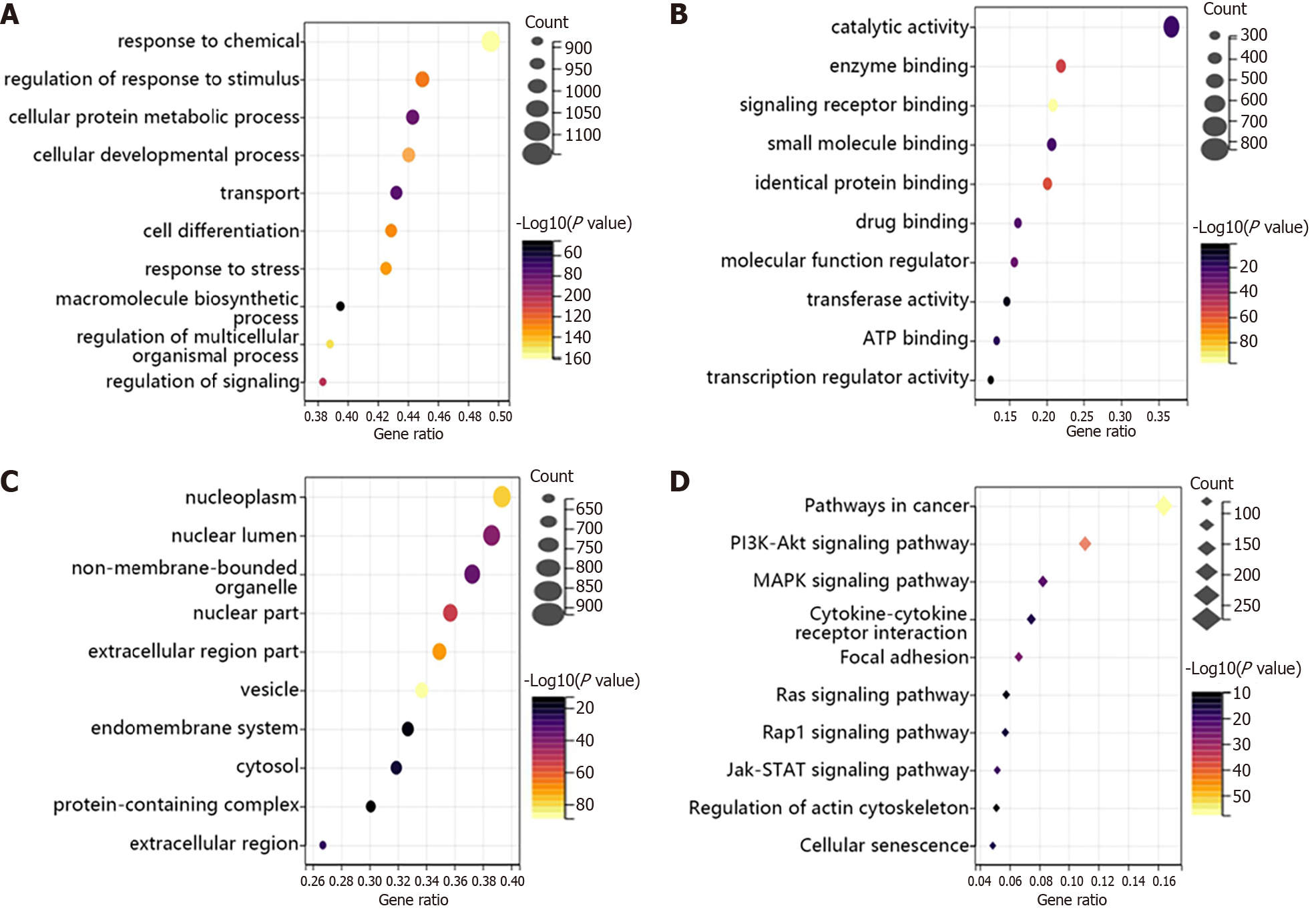
Figure 2 The enrichment analysis of hypertrophic scar-related genes.
A-C: The Gene Ontology analysis of hypertrophic scar (HTS)-related genes in biological process (A), molecular function (B), and cell composition (C) terms; D: Kyoto Encyclopaedia of Genes and Genome pathway enrichment of the candidate targets of HTS.
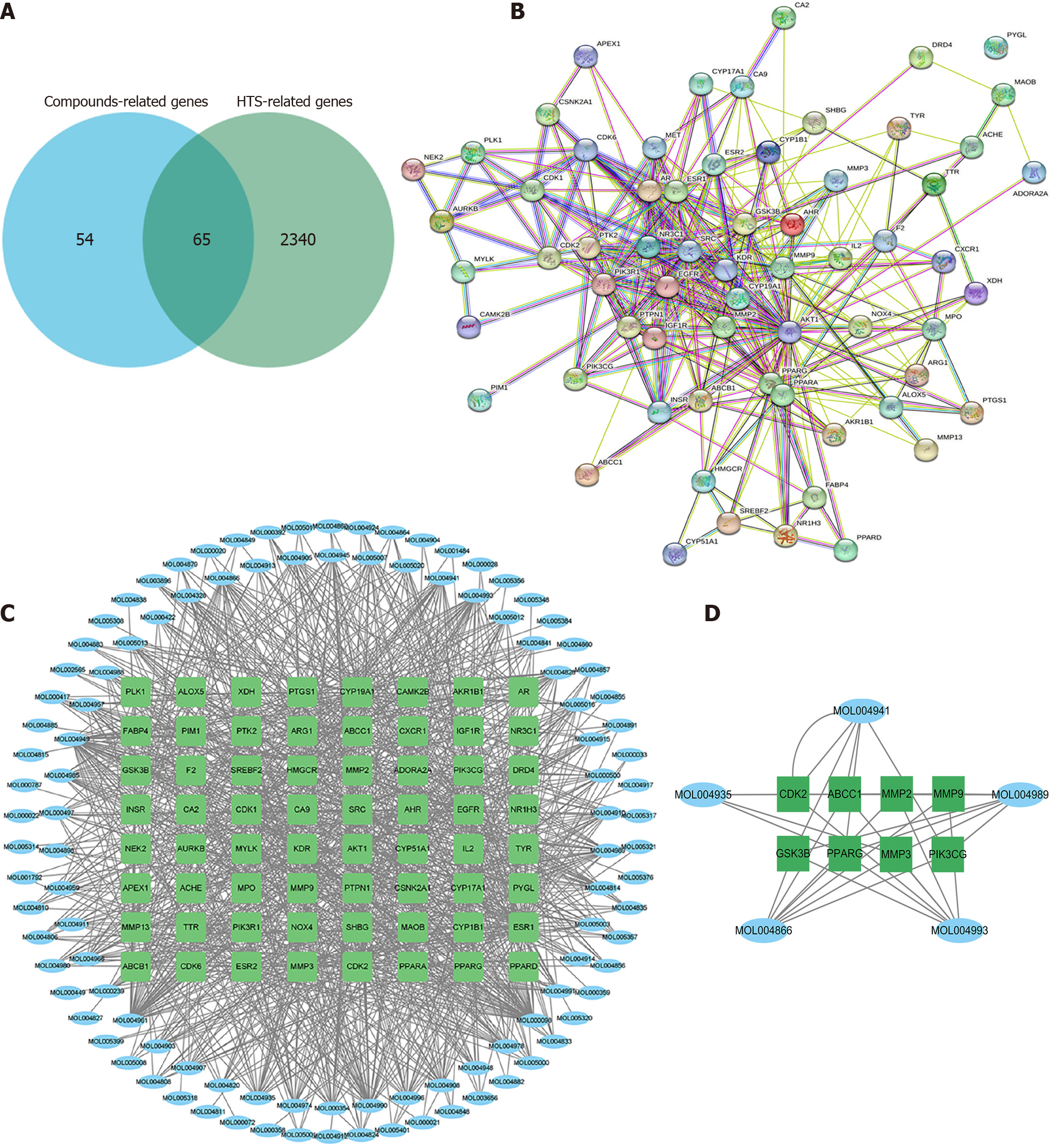
Figure 3 The compound-target network analysis.
A: Venn diagram of compounds-related genes and hypertrophic scar-related genes; B: The protein-protein interaction network for the 65 genes; C: The interaction between 65 genes and compounds of WuFuYin; D: The key subnetwork from A by MCODE. HTS: Hypertrophic scar.
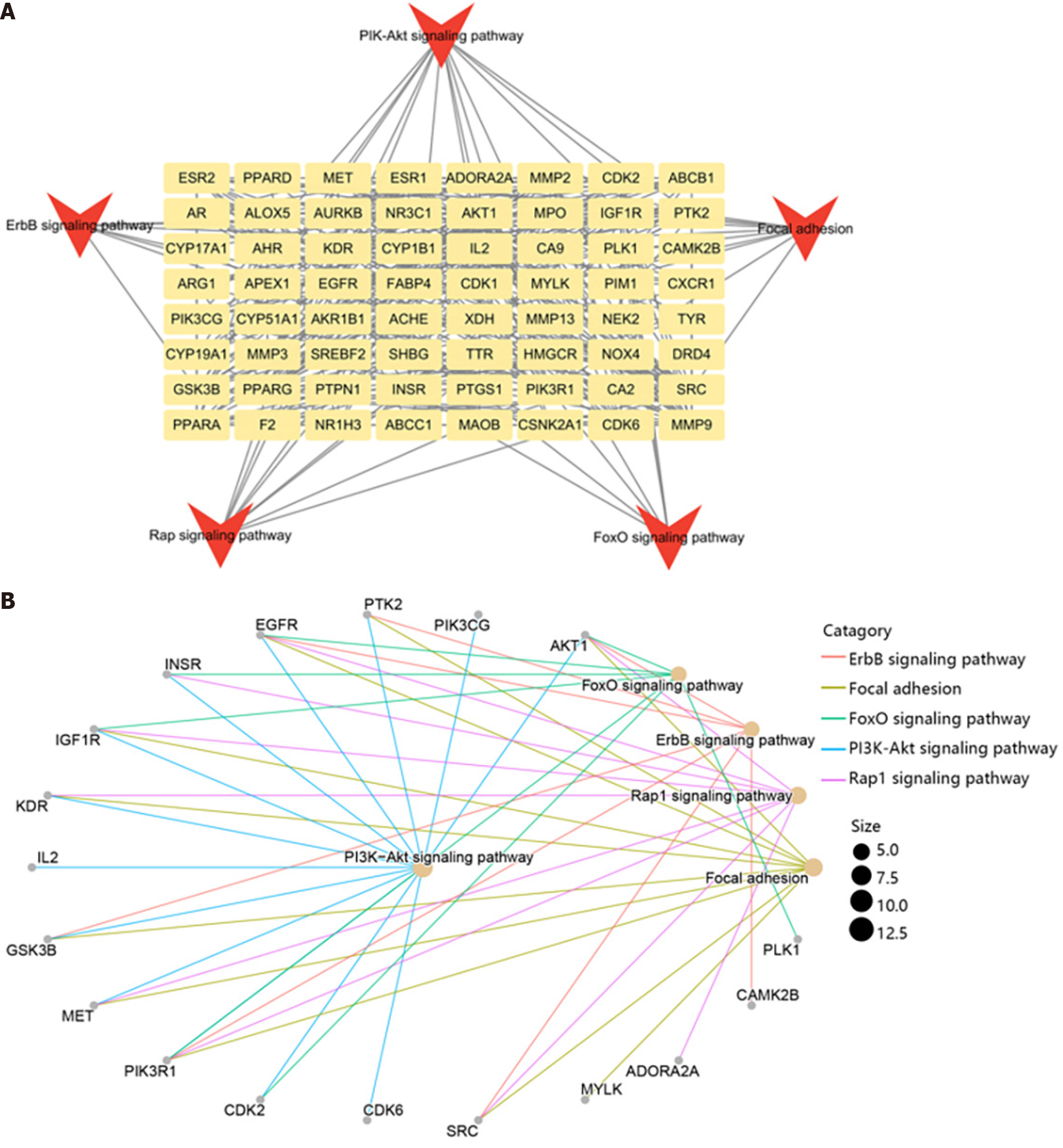
Figure 4 The genes enriched in key pathways.
A: The network of 65 genes and main signal pathways; B: The top 5 pathways related to the genes.
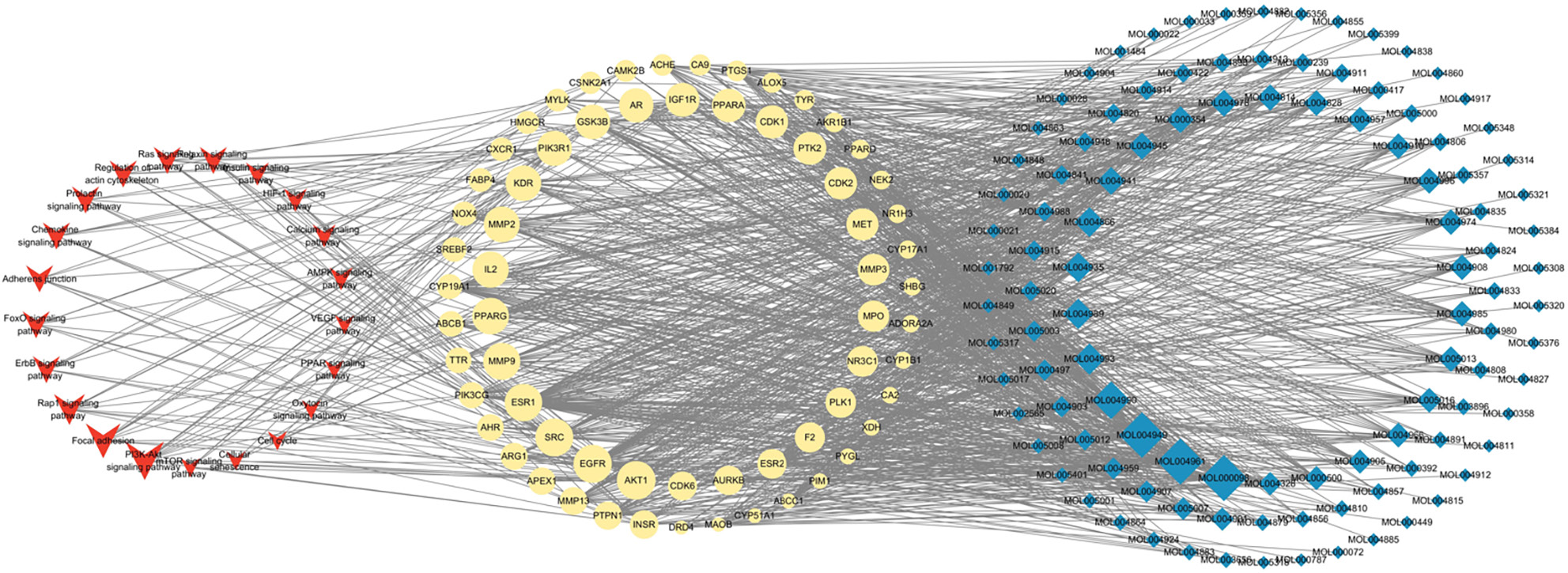
Figure 5 The topological network analysis among compounds, genes and pathways.
The red V was pathways; the yellow ellipse was genes; the blue diamond was compounds. The size of the red V, yellow ellipse and blue diamond were gradually and respectively changed according to the related gene numbers, degree and EC value, with larger size indicating a higher value.
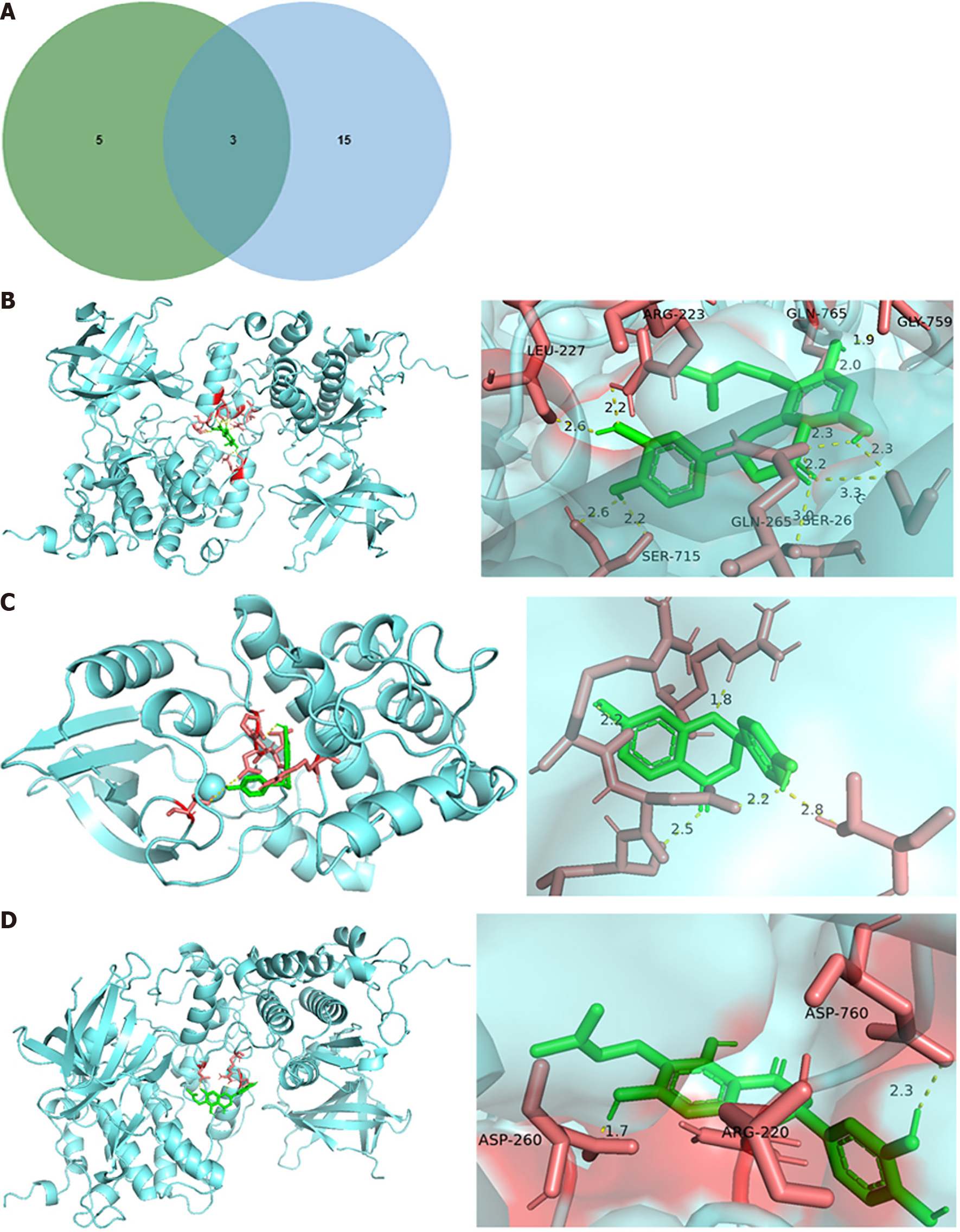
Figure 6 The molecular docking of hub genes.
A: The hub genes were screened by Venn. The green indicated the gene played vital roles in the interaction between genes and compounds. The blue indicated the genes involved in the top 5 pathways; B-D: Molecular docking diagrams of hypertrophic scar-related targets with main compounds of WuFuYin.
- Citation: Zhang SY, Guo SX, Chen LL, Zhu JY, Hou MS, Lu JK, Shen XX. Exploring the potential mechanism of WuFuYin against hypertrophic scar using network pharmacology and molecular docking. World J Clin Cases 2024; 12(18): 3505-3514
- URL: https://www.wjgnet.com/2307-8960/full/v12/i18/3505.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i18.3505