Copyright
©The Author(s) 2023.
World J Clin Cases. Oct 26, 2023; 11(30): 7318-7328
Published online Oct 26, 2023. doi: 10.12998/wjcc.v11.i30.7318
Published online Oct 26, 2023. doi: 10.12998/wjcc.v11.i30.7318
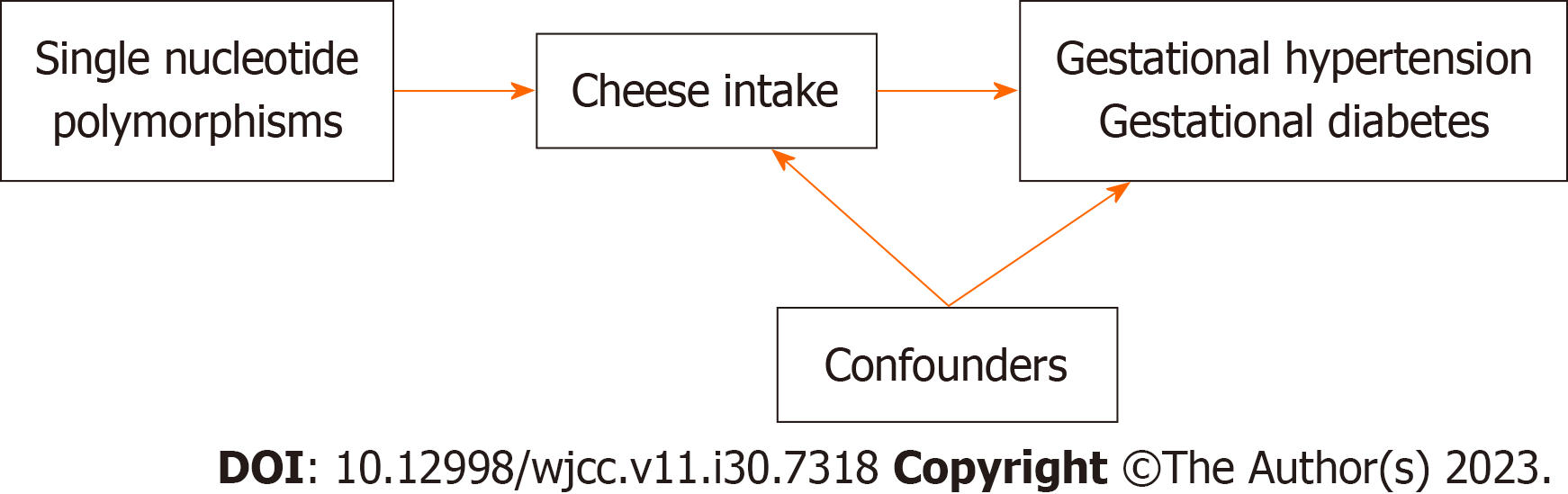
Figure 1 Flow chart of the design of the present study.
The selected genetic variant (single nucleotide polymorphisms) was strongly associated with cheese intake. The selected genetic variants were strongly associated with confounding factors, but not associated with gestational hypertension gestational diabetes.
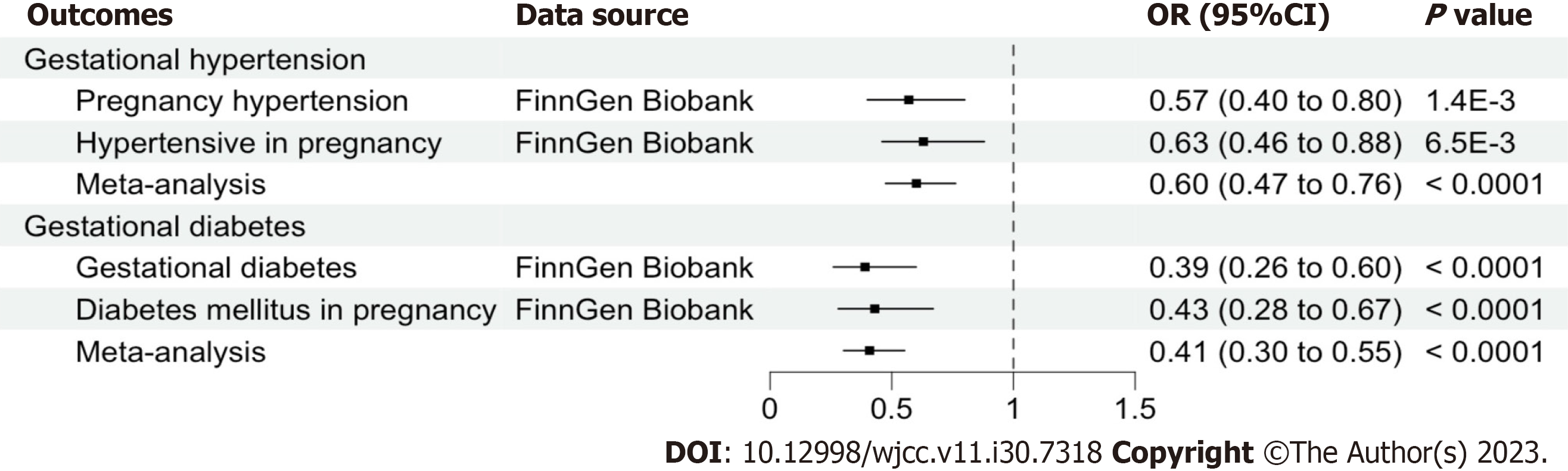
Figure 2 Forest plot showing the main Mendelian randomization estimates for the association of cheese intake with gestational hypertension and gestational diabetes by a meta-analysis of fixed-effect model.
OR: Odds ratio; CI: Confidence interval.
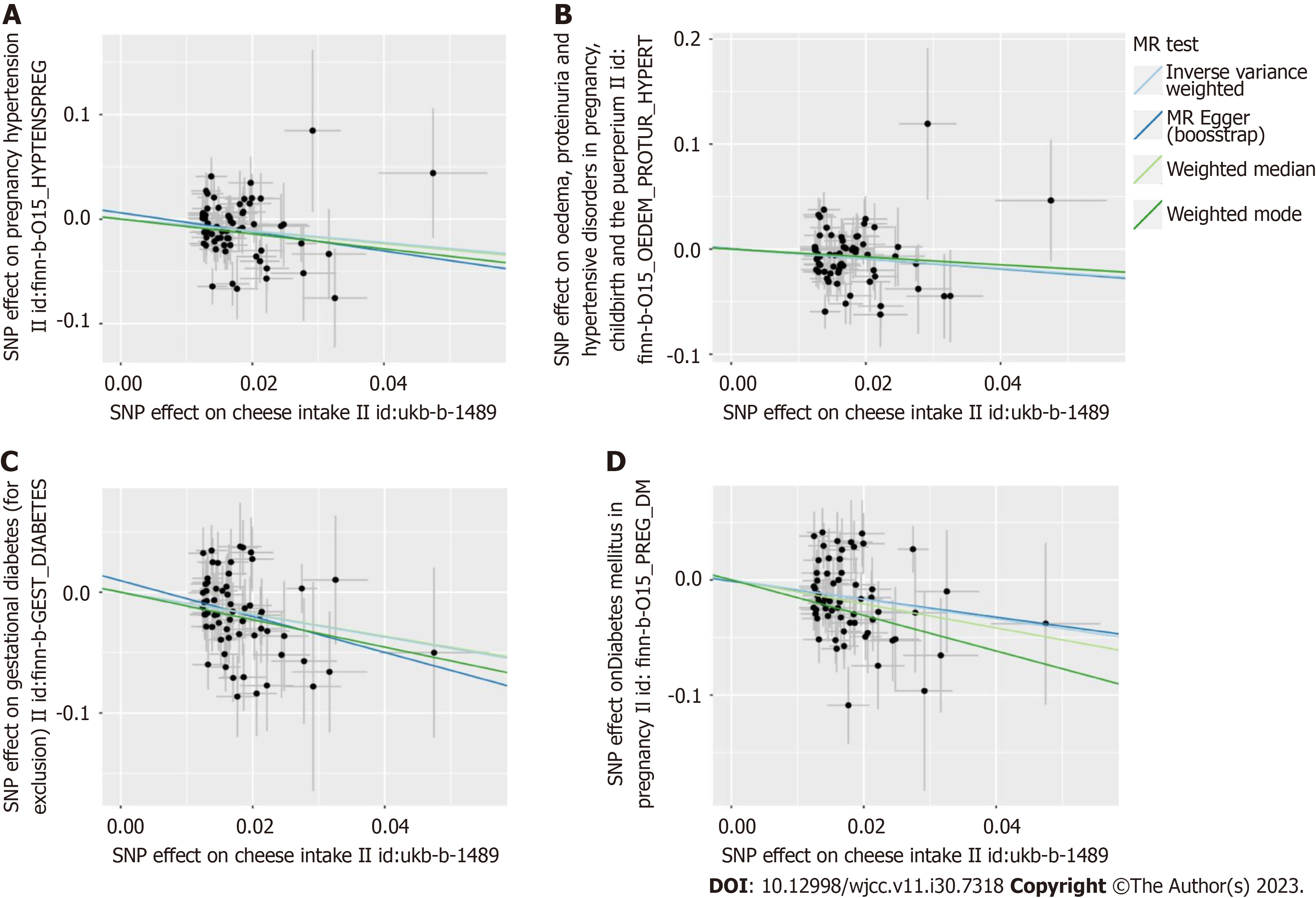
Figure 3 Scatter plot to visualize causal effect of cheese intake on gestational hypertension and gestational diabetes risk using four Mendelian randomization methods.
The slope of the straight line indicates the magnitude of the causal association. The plot presents the effect sizes of the single nucleotide polymorphisms (SNP)-cheese intake association (X-axis, standard deviation units) and the SNP-gestational hypertension and gestational diabetes association [Y-axis, log (odds ratio)] with 95% confidence intervals. The regression slopes of the lines correspond to causal estimates using the four Mendelian randomization methods. A: The Scatter plot visualize causal effect of cheese intake on gestational hypertension risk in the FinnGen Biobank database; B: The Scatter plot visualize causal effect of cheese intake on gestational hypertension risk in the FinnGen Biobank database; C: The Scatter plot visualize causal effect of cheese intake on gestational diabetes risk in the FinnGen Biobank database; D: The Scatter plot visualize causal effect of cheese intake on gestational diabetes risk in the FinnGen Biobank database. SNPs: Single nucleotide polymorphisms; MR: Mendelian randomization.
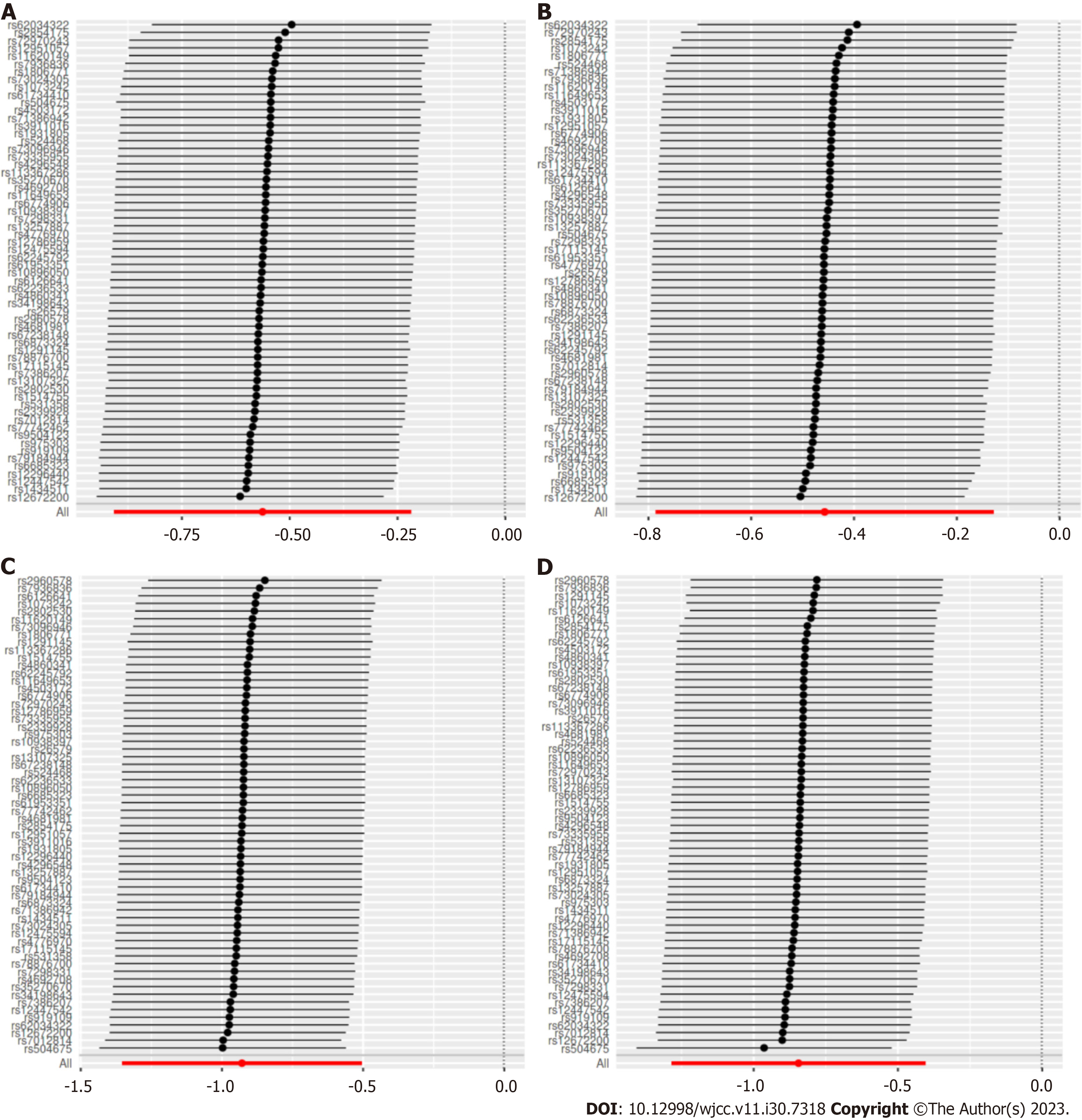
Figure 4 Leave-one-out of each single nucleotide polymorphisms associated of cheese intake with gestational hypertension and gestational diabetes risk.
Each black point represents result of the inverse variance weighted (IVW) Mendelian randomization method applied to estimate the causal effect of cheese intake on gestational hypertension and gestational diabetes excluding particular single nucleotide polymorphism (SNP) from the analysis. Each red point depicts the IVW estimate using all SNPs. A: Mendelian randomization (MR) leave-one-out sensitivity analysis of gestational hypertension risk in FinnGen Biobank database; B: MR leave-one-out sensitivity analysis of gestational hypertension in FinnGen Biobank database; C: MR leave-one-out sensitivity analysis of gestational diabetes in FinnGen Biobank database; D: MR leave-one-out sensitivity analysis of gestational diabetes in FinnGen Biobank database.
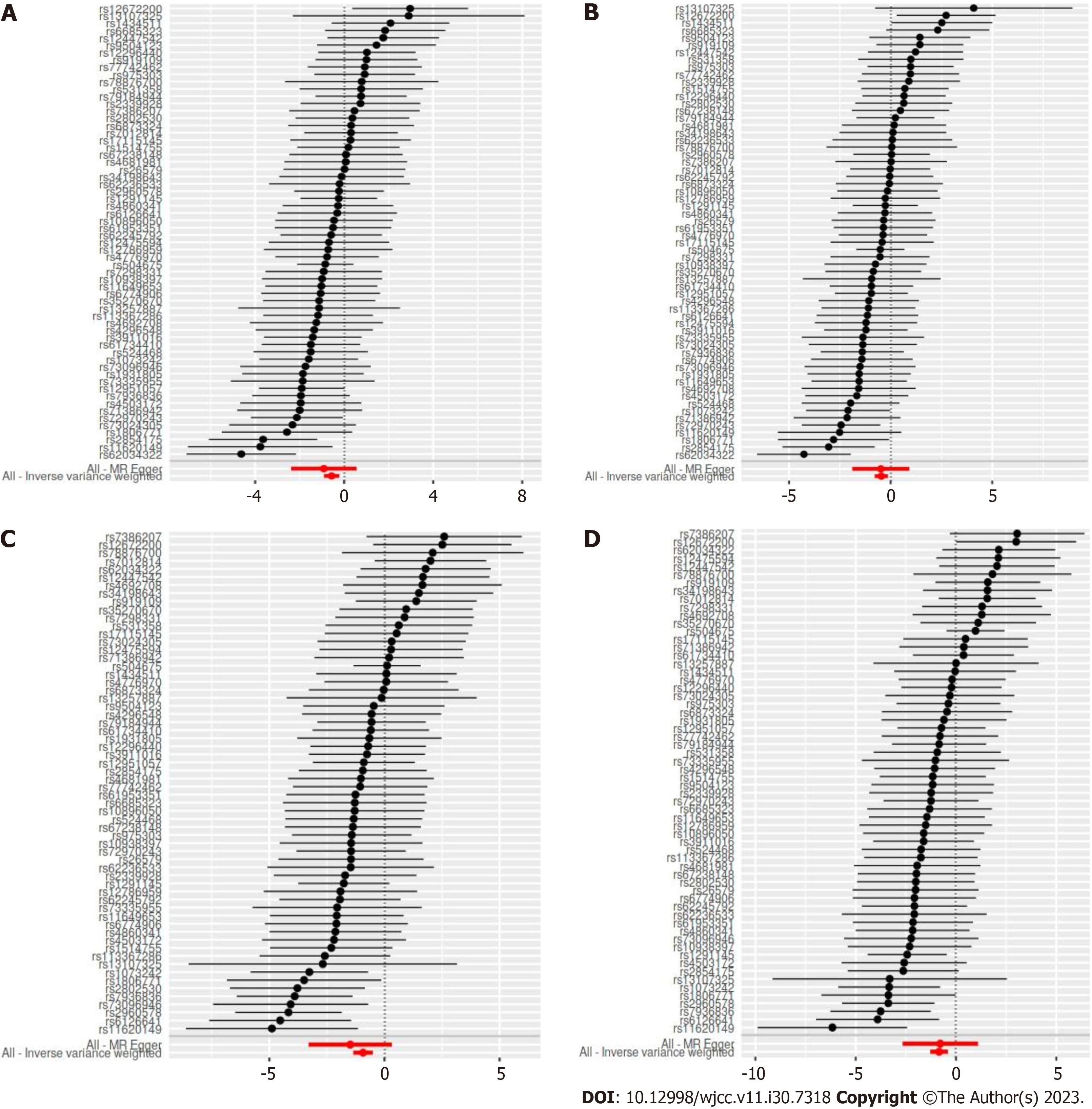
Figure 5 Forest plot showing the association of each single nucleotide polymorphisms with gestational hypertension and gestational diabetes.
Each black point represents result of the inverse variance weighted (IVW) Mendelian randomization (MR) method applied to estimate the causal effect of each single nucleotide polymorphism (SNP) on gestational hypertension and gestational diabetes. Each red point depicts the IVW and MR egger estimate using all SNPs. A: Forest plot of gestational hypertension in FinnGen Biobank database; B: Forest plot of gestational hypertension in FinnGen Biobank database; C: Forest plot of gestational diabetes in FinnGen Biobank database; D: Forest plot of gestational diabetes in FinnGen Biobank database.
- Citation: Zhong T, Huang YQ, Wang GM. Causal relationship association of cheese intake with gestational hypertension and diabetes result from a Mendelian randomization study. World J Clin Cases 2023; 11(30): 7318-7328
- URL: https://www.wjgnet.com/2307-8960/full/v11/i30/7318.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i30.7318