Copyright
©The Author(s) 2023.
World J Clin Cases. Jul 6, 2023; 11(19): 4579-4600
Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4579
Published online Jul 6, 2023. doi: 10.12998/wjcc.v11.i19.4579
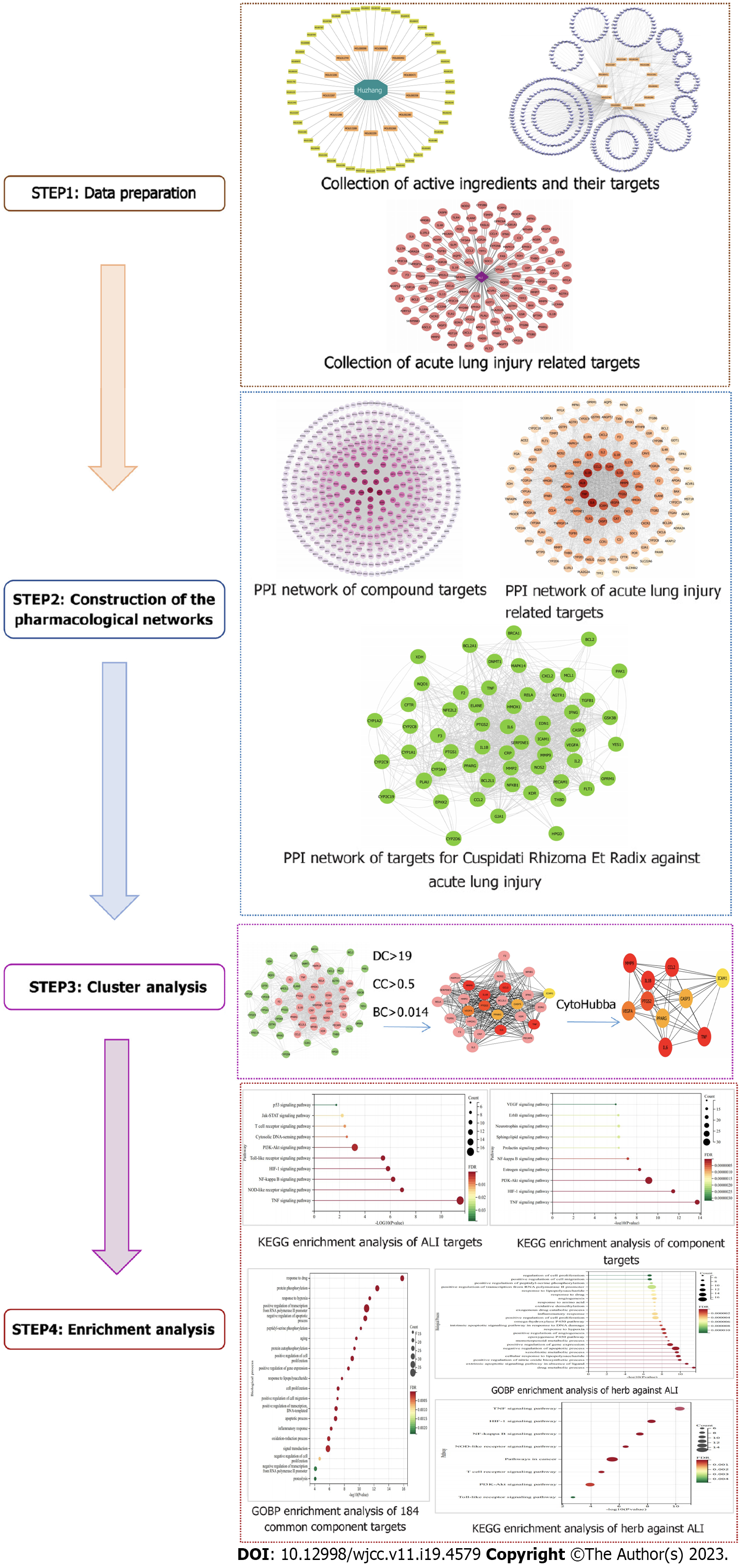
Figure 1 Schematic representation of network pharmacology study of Polygoni Cuspidati Rhizoma et Radix in the treatment of acute lung injury.
PCRR: Polygoni Cuspidati Rhizoma et Radix; ALI: Acute lung injury; PPI: Protein–protein interaction; KEGG: Kyoto Encyclopedia of Genes and Genomes.
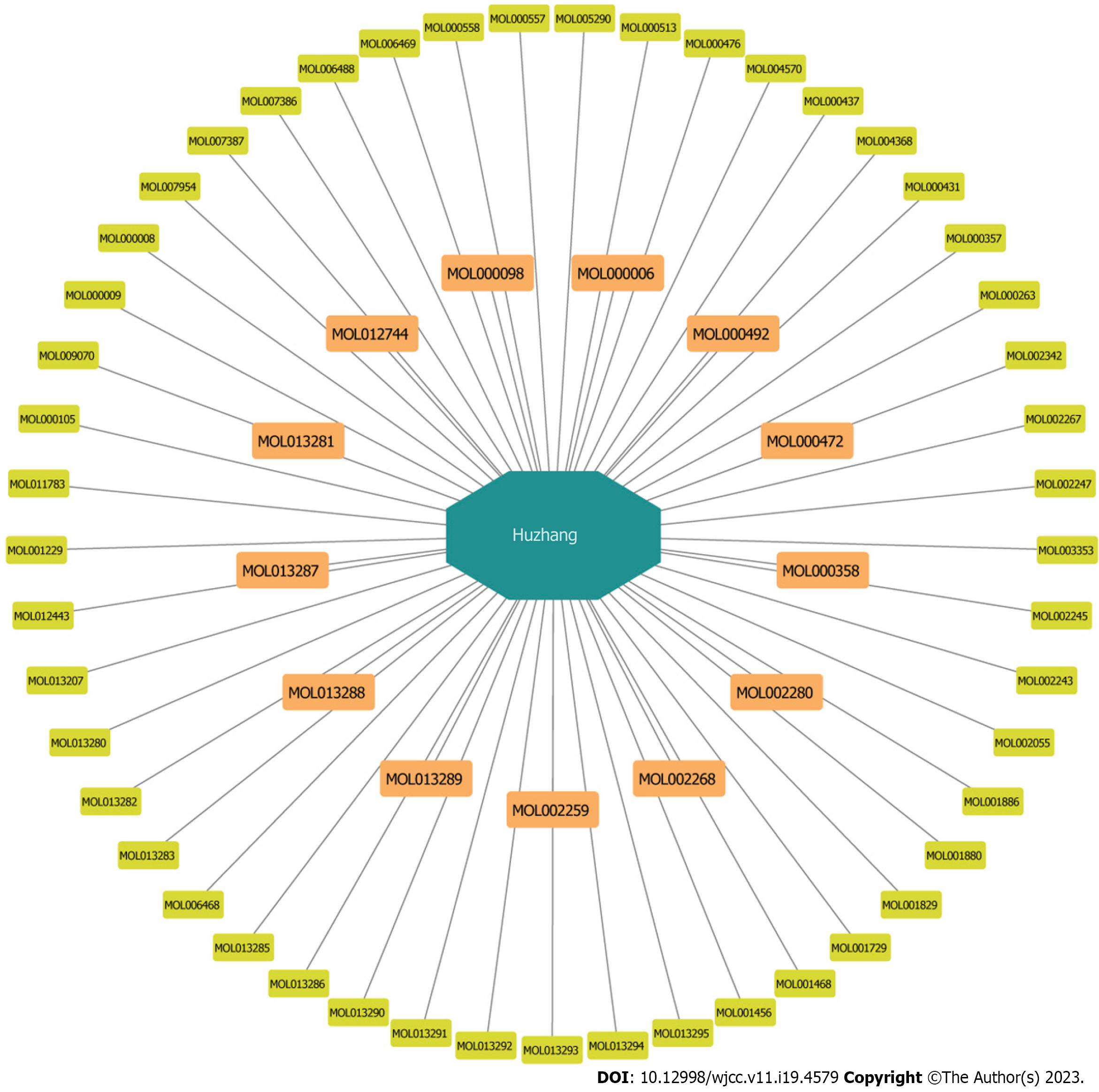
Figure 2 The active ingredients of Polygoni Cuspidati Rhizoma et Radix, orange round rectangles represent the 13 active ingredients selected finally.
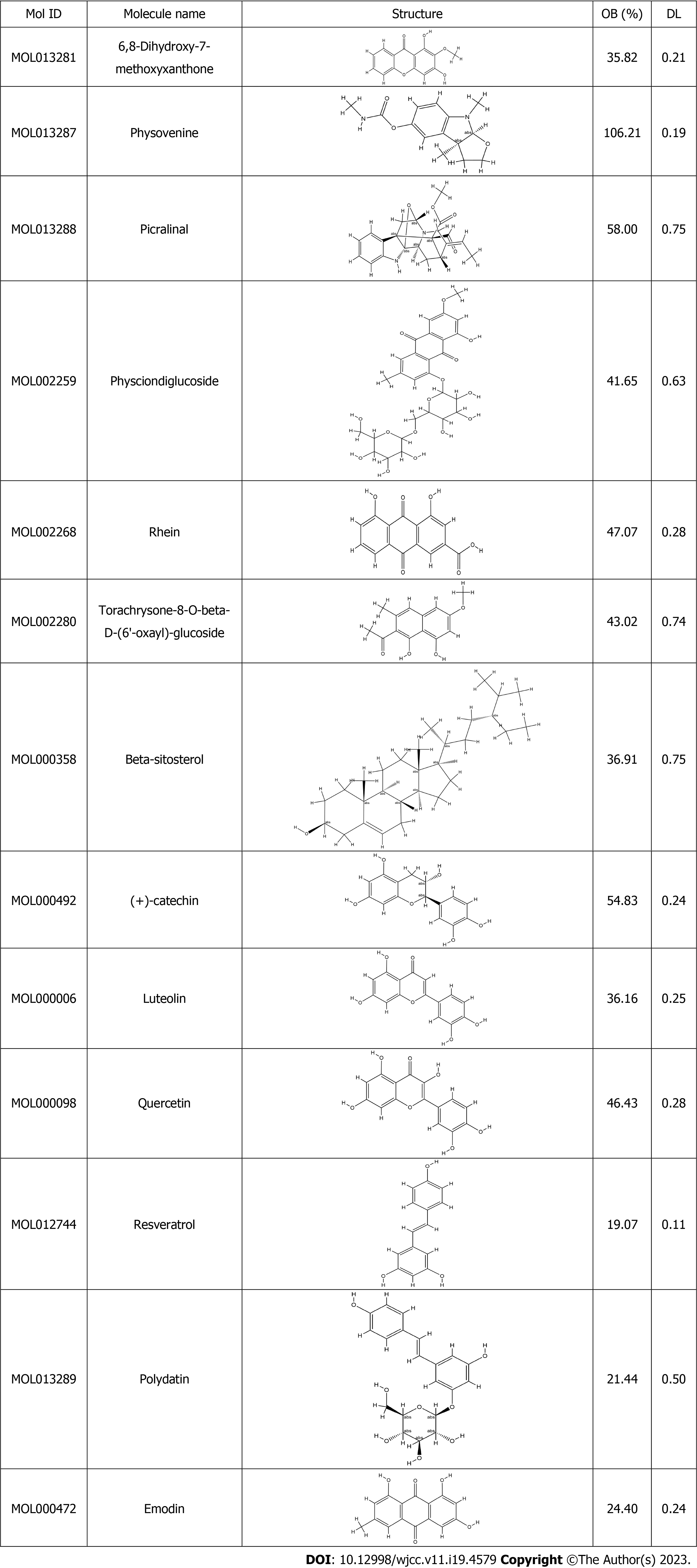
Figure 3 Thirteen active compounds from Polygoni Cuspidati Rhizoma et Radix.
OB: Oral bioavailability; DL: Drug likeness.
Figure 4 Construction of the protein-protein interaction network of durg active compounds and associated targets.
Orange represents active compounds of durg. Blue represents gene symbols of targets
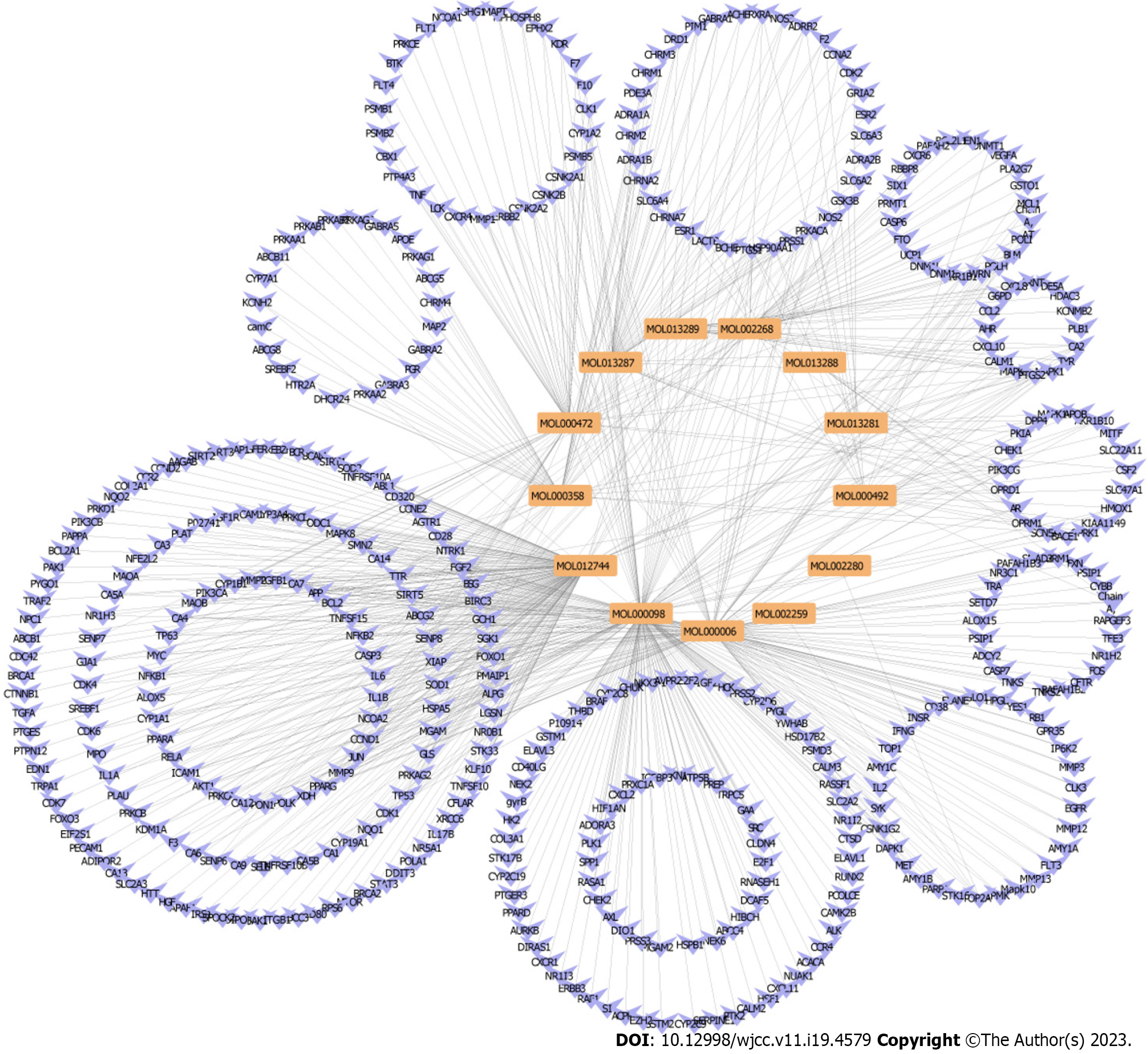
Figure 5 The putative target of active Polygoni Cuspidati Rhizoma et Radix ingredients.
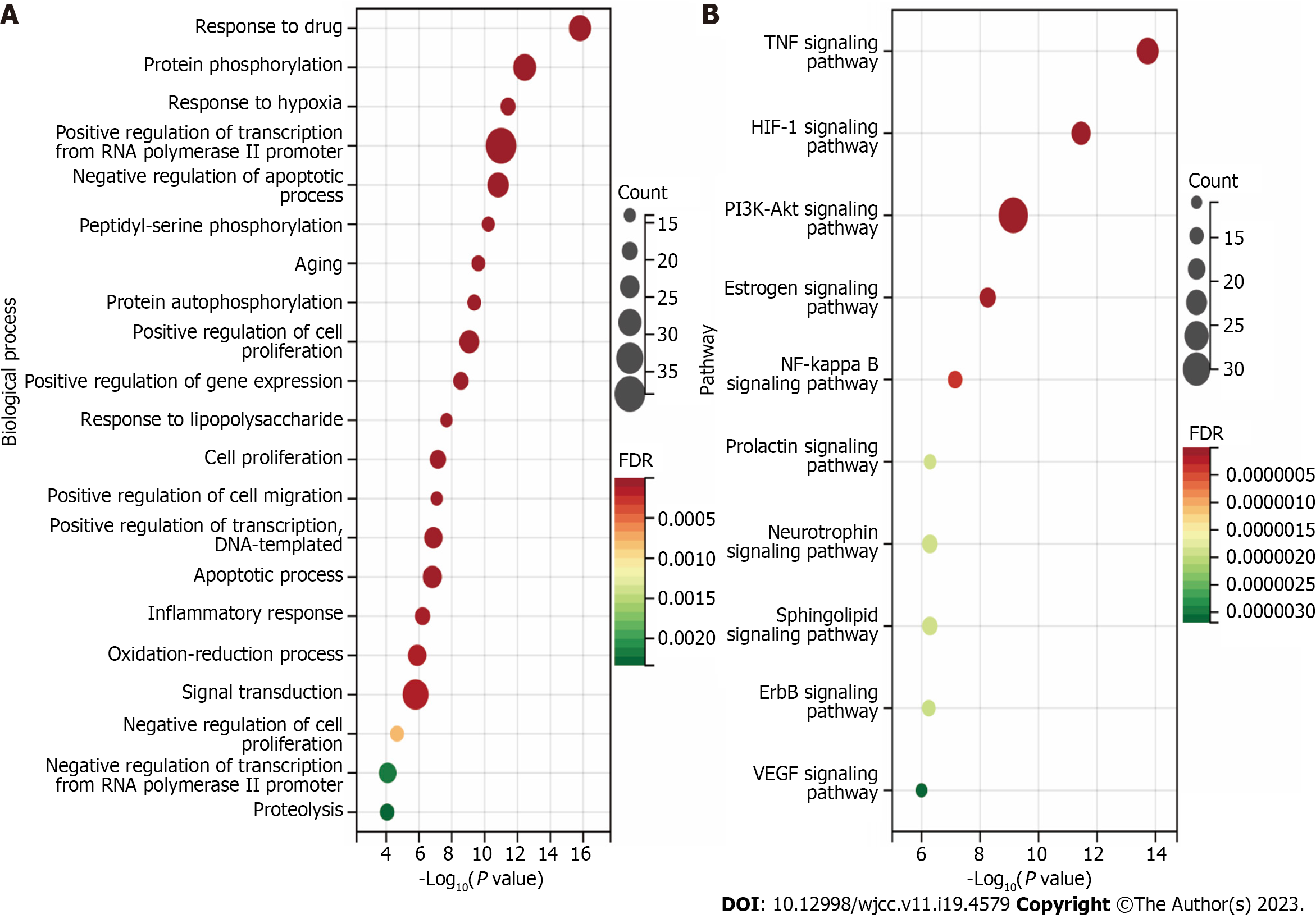
A: Biological processes analysis for targets of active ingredients of Polygoni Cuspidati Rhizoma et Radix; B: Kyoto Encyclopedia of Genes and Genomes pathways through enrichment method to indicate the top 10 signaling pathways of active ingredients of Polygoni Cuspidati Rhizoma et Radix. HIF-1: Hypoxia-inducible factor-1; TNF: Tumor necrosis factor; FDR: False discovery rate.
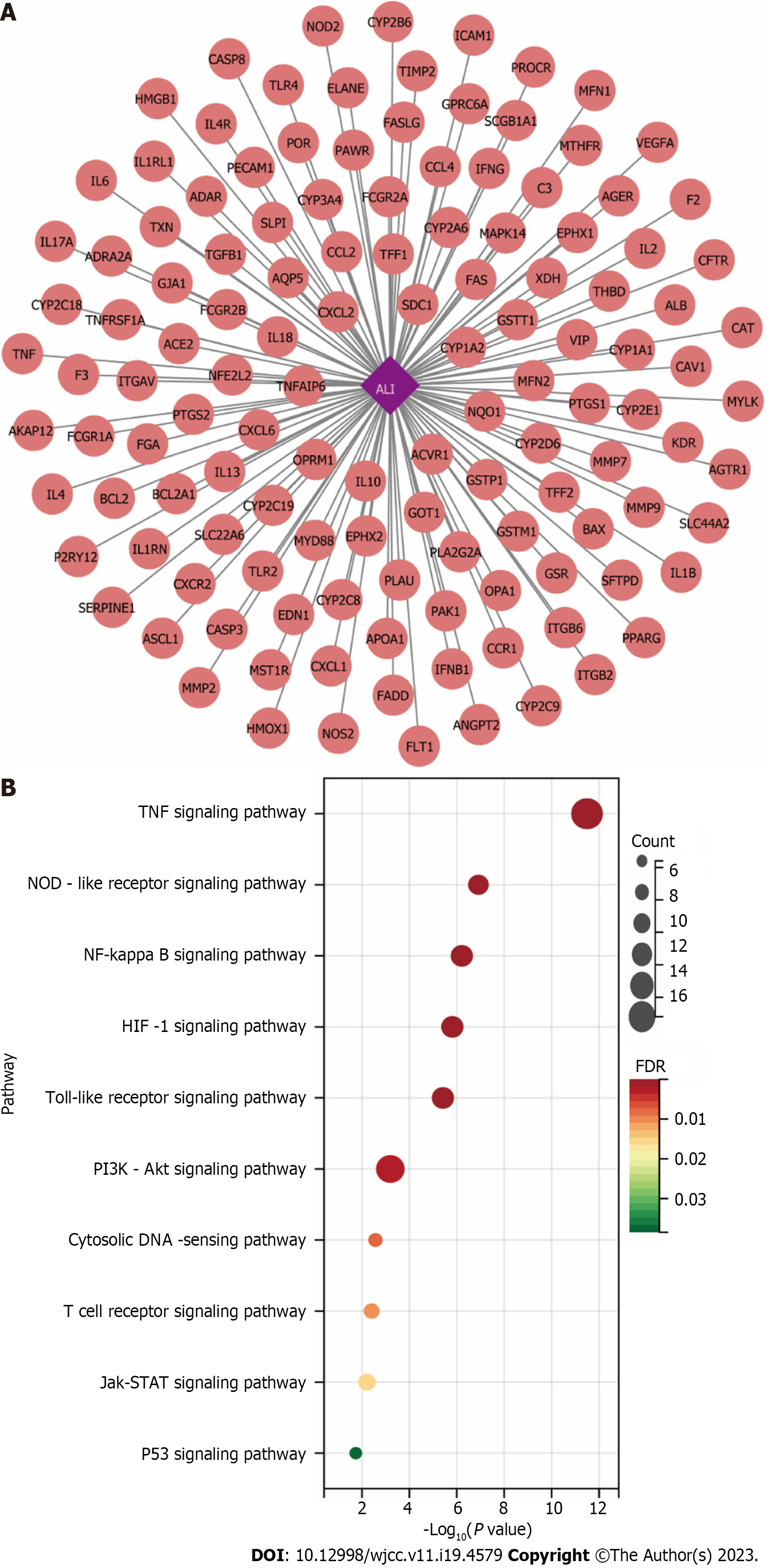
Figure 6 Identification of acute lung injury-Related targets.
A: The related targets of acute lung injury; B: Kyoto Encyclopedia of Genes and Genomes pathways through enrichment method to indicate the top 10 signaling pathways of the related targets of acute lung injury. HIF-1: Hypoxia-inducible factor-1; TNF: Tumor necrosis factor; ALI: Acute lung injury.
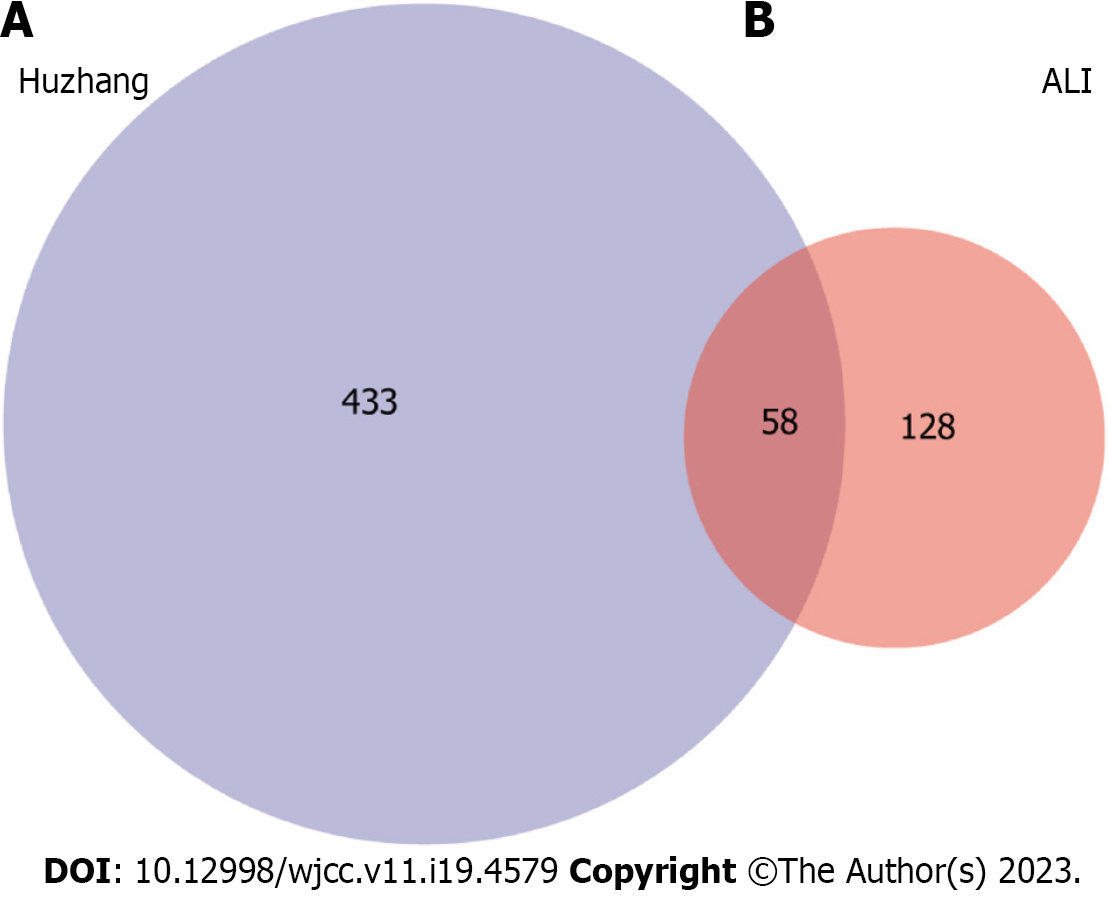
Figure 7 Venn diagram summarizing differentially expressed targets of Polygoni Cuspidati Rhizoma et Radix and acute lung injury.
A: Indicates gene targets of Polygoni Cuspidati Rhizoma et Radix; B: Indicates gene targets related to acute lung injury. The common part of A and B indicates the 58 overlapping genes between the disease and drug. ALI: Acute lung injury.
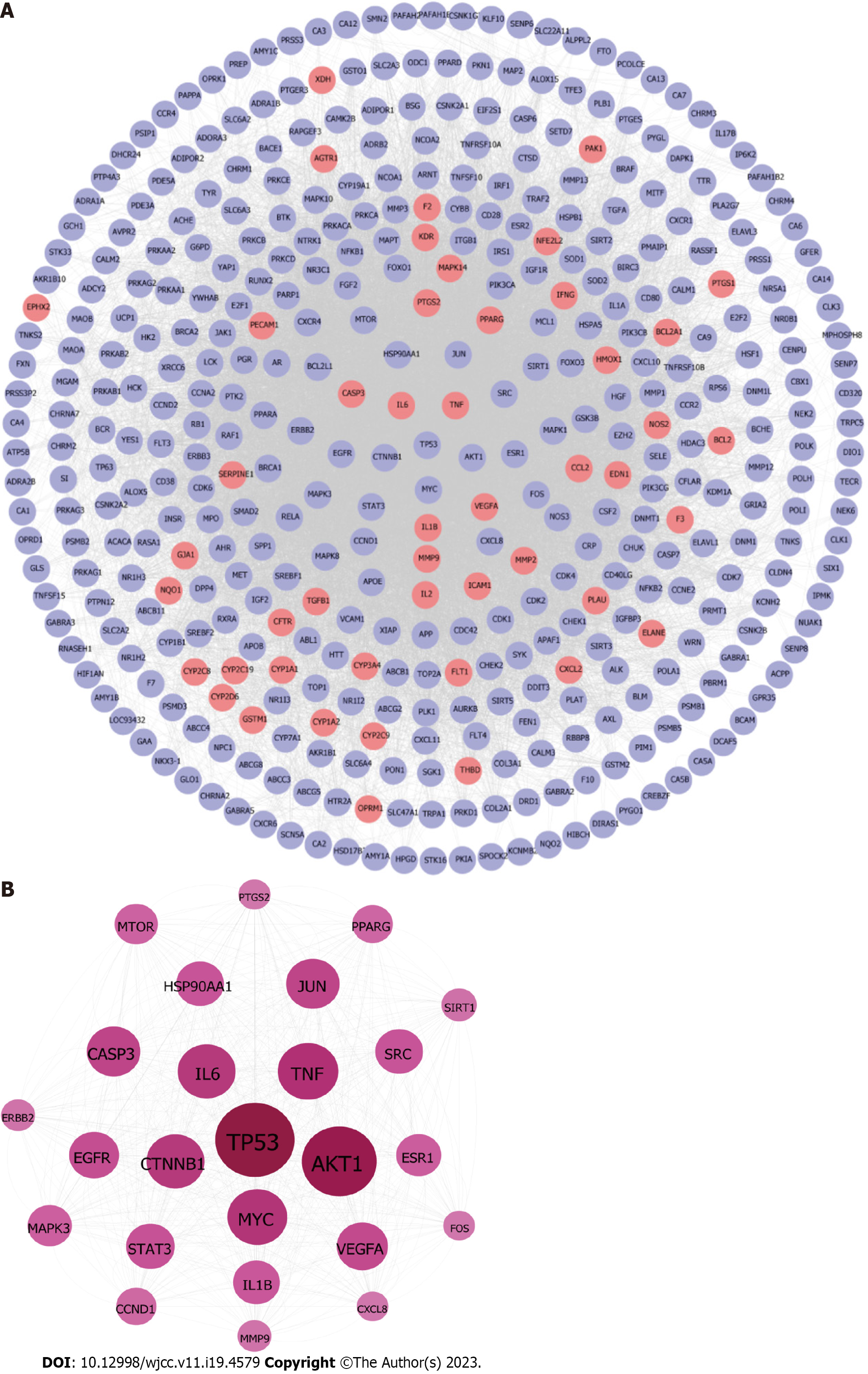
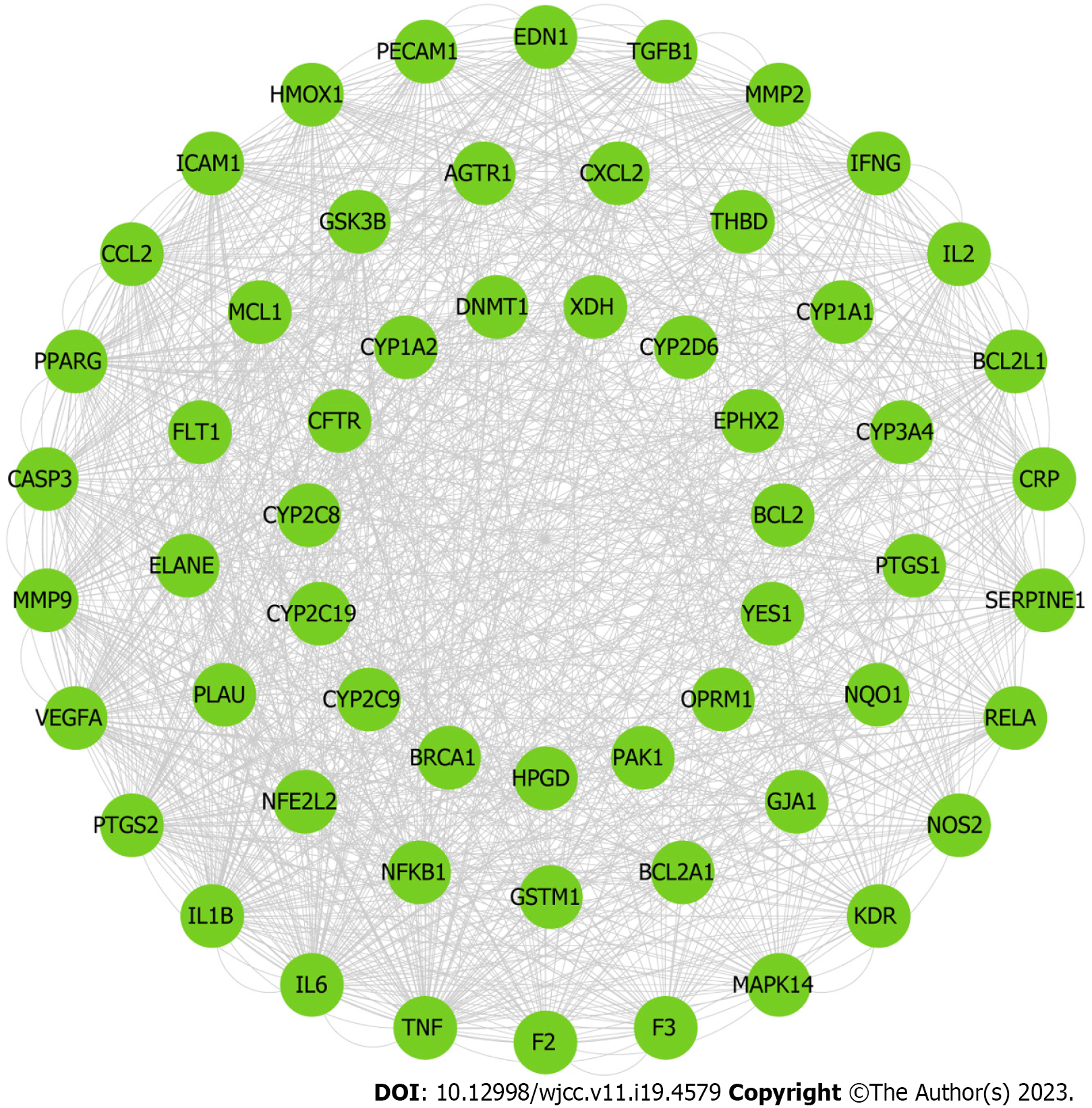
Figure 8 Construction of protein-protein interaction network of compound targets.
A: The protein-protein interaction networks of compound targets. The purple nodes represent compound targets, and the pink nodes stand for common targets of disease and drugs; B The significant Polygoni Cuspidati Rhizoma et Radix-related targets after topology analysis.
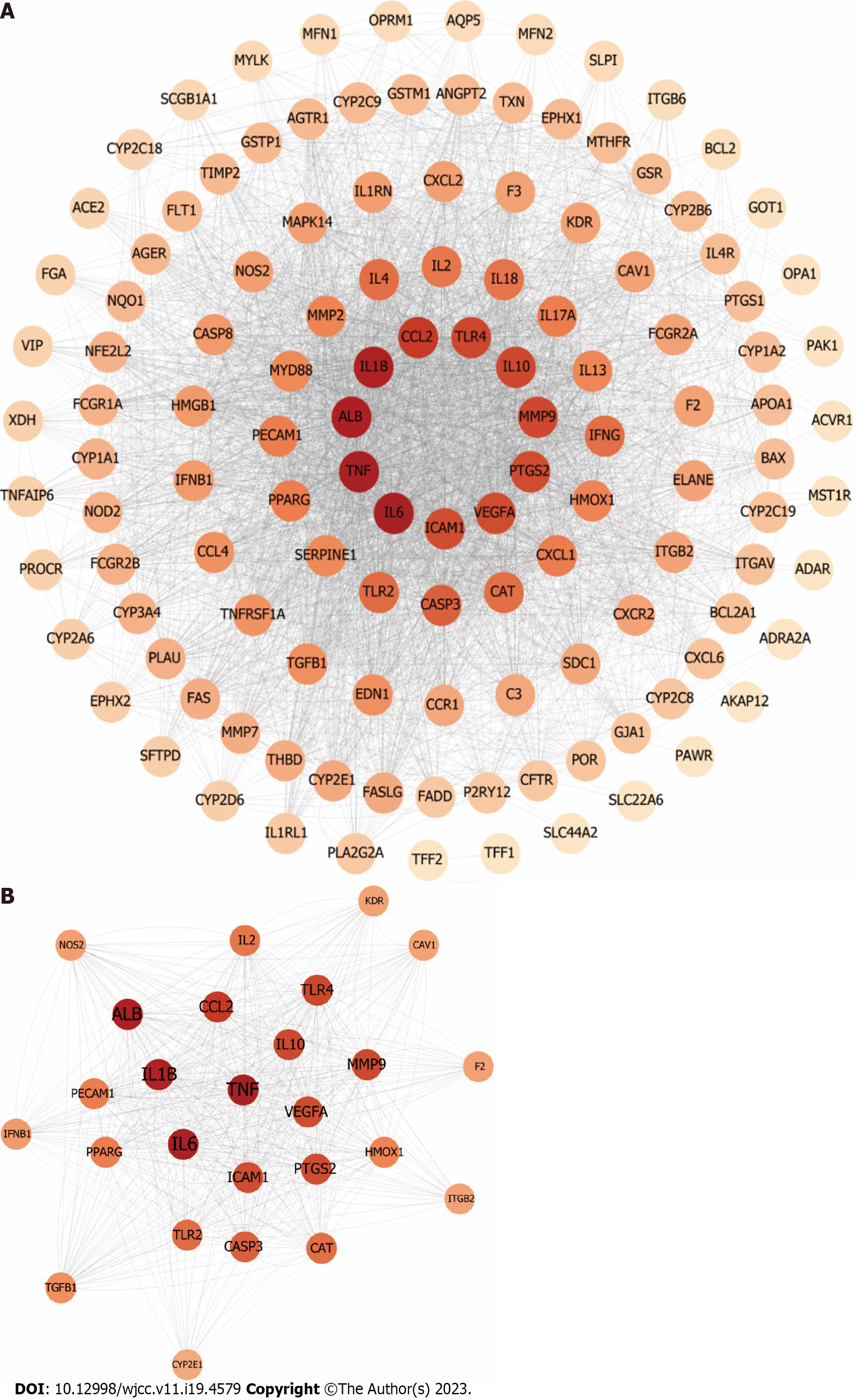
Figure 9 Identification of candidate protein targets associated with acute lung injury.
A: The protein-protein interaction networks of acute lung injury-related targets. The node color changes from light to dark reflect the degree value changes from low to high in the network; B: The significant Polygoni Cuspidati Rhizoma et Radix-related targets after topology analysis.
Figure 10 Protein–protein interaction Network graph of 58 Hub Nodes based on their direct interactions for Polygoni Cuspidati Rhizoma et Radix in acute lung injury.
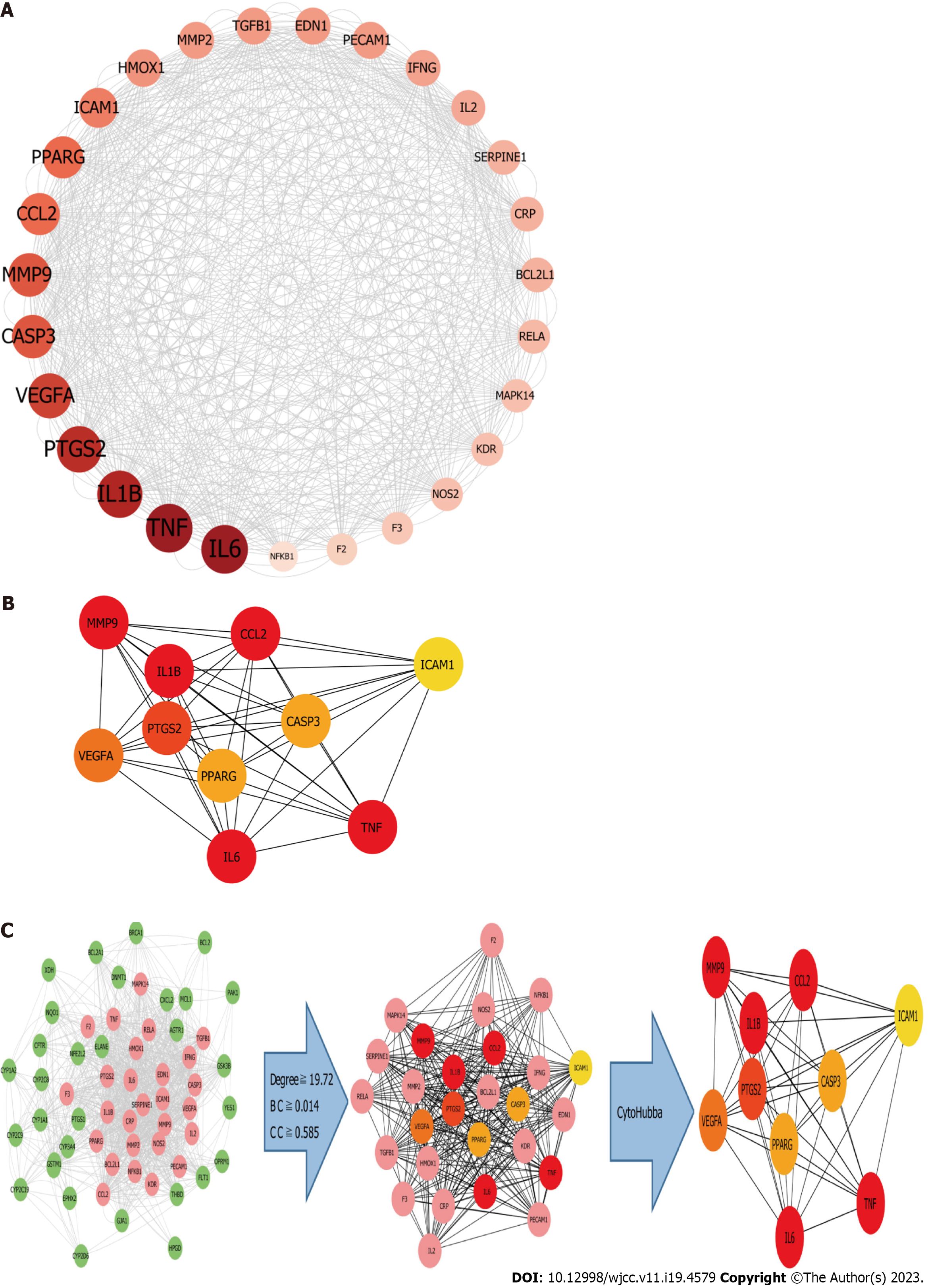
Figure 11 Cluster analysis.
A: The 27 active ingredient-disease- targets of after topology analysis; B: The top 10 active ingredient-disease- targets after cytoHubba and MCC tool; C: Identifcation of candidate Polygoni Cuspidati Rhizoma et Radix -related targets for acute lung injury treatment through protein–protein interaction network. 27 candidate targets are finally predicted. BC: Betweenness centrality; CC: Closeness centrality.
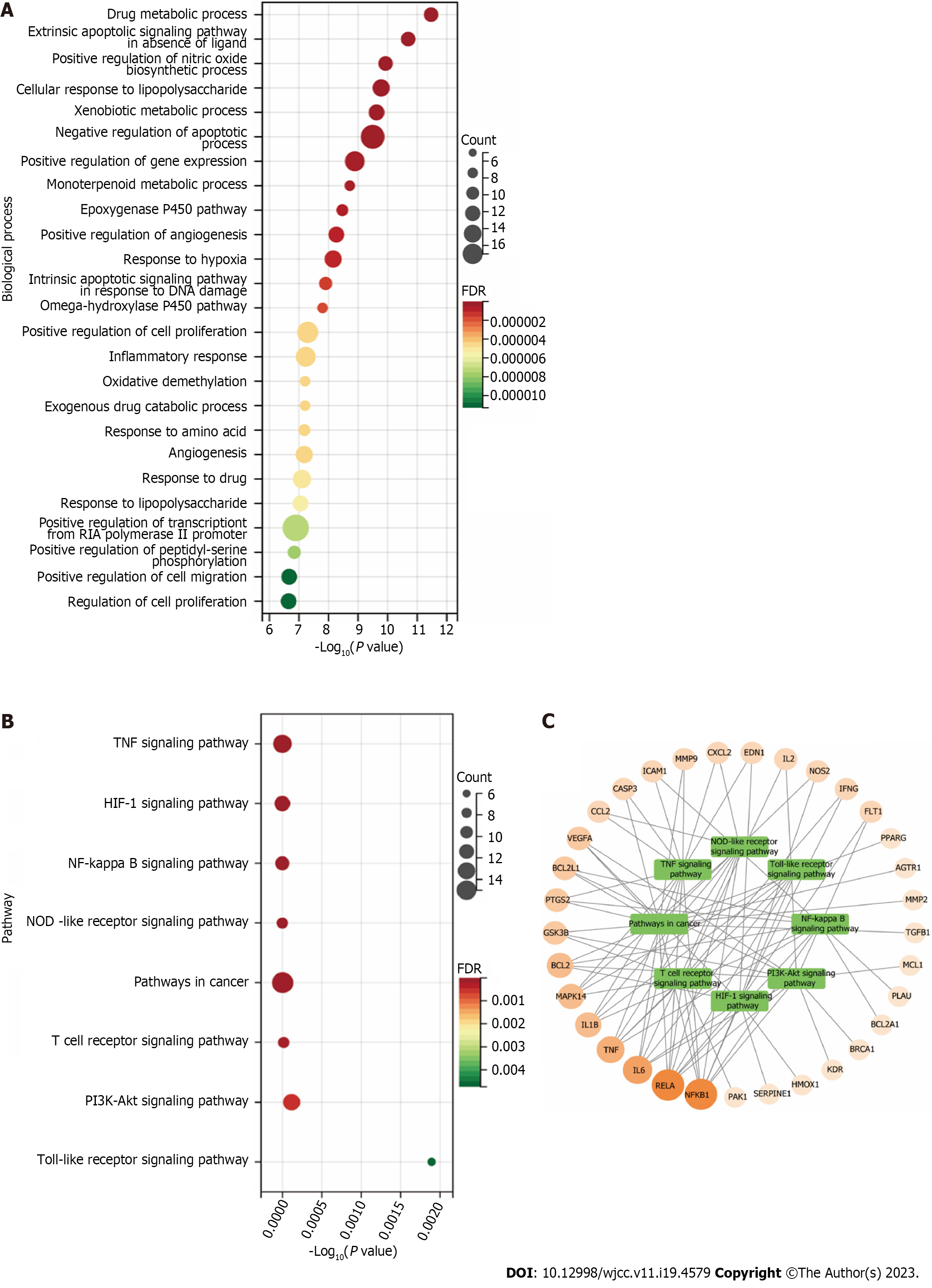
Figure 12 Gene ontology biological process and Kyoto encyclopedia of genes and genomes pathway enrichment analysis.
A: The gene ontology Biological process enrichment analysis of 58 targets. The top 25 significantly enriched terms in Biological process; B: The Kyoto encyclopedia of genes and genomes pathway enrichment analysis of 60 targets. The top 8 significant pathways for 60 targets; C: Target-Pathway Network. The orange rounds represent Polygoni Cuspidati Rhizoma Et Radix-related targets. The node size is proportional to the target degree in the network. The green round rectangles represent the related pathways. TNF: Tumor necrosis factor; NOD: NOD-like receptor; NF-kappa B: Nuclear factor kappa-B; HIF: Hypoxia-inducible factor-1.
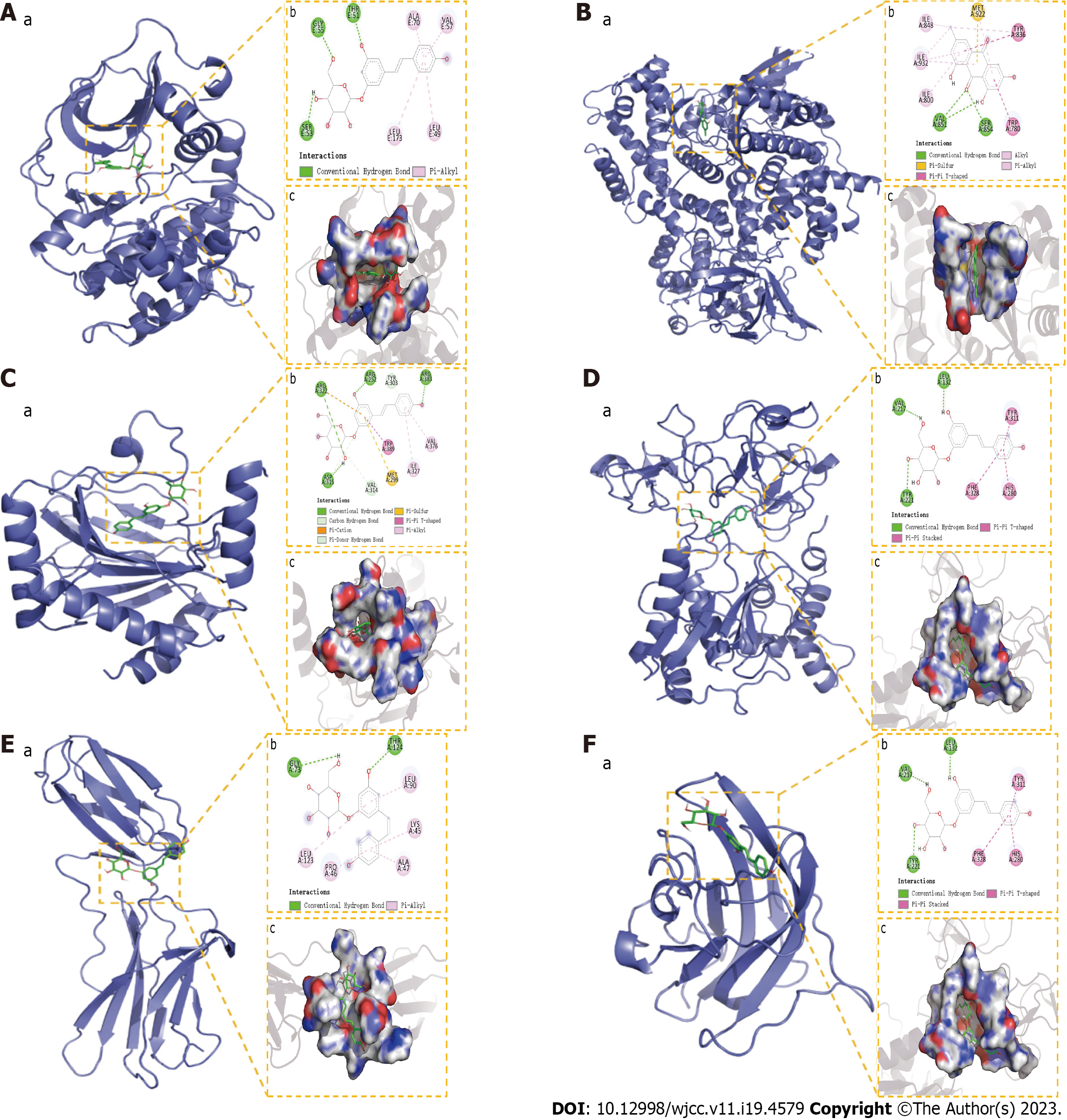
Figure 13 Molecular docking results.
A: The binding mode of RAC-alpha serine/threonine-protein kinase with Polydatin; B: The binding mode of Phosphoinositide 3-Kinase with Emodin; C: The binding mode of hypoxia-inducible factor-1a with Polydatin; D: The binding mode of matrix metalloproteinase-9 with Polydatin; E: The binding mode of IL6 with Polydatin; F: The binding mode of VEGFA with Polydatin. “a” is the 3D structure of the complex, “b” shows the hydrogen bond donor receptor network of the complex, “c” is the 2D binding mode of the complex. AKT: RAC-alpha serine/threonine-protein kinase; PI3K: Phosphoinositide 3-Kinase; HIF1a: hypoxia-inducible factor-1a; MMP9: Matrix metalloproteinase-9; IL6: Interleukin-6; VEGFA: Vascular endothelial growth factor A.
- Citation: Zheng JL, Wang X, Song Z, Zhou P, Zhang GJ, Diao JJ, Han CE, Jia GY, Zhou X, Zhang BQ. Network pharmacology and molecular docking to explore Polygoni Cuspidati Rhizoma et Radix treatment for acute lung injury. World J Clin Cases 2023; 11(19): 4579-4600
- URL: https://www.wjgnet.com/2307-8960/full/v11/i19/4579.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i19.4579