Copyright
©The Author(s) 2022.
World J Clin Cases. Nov 26, 2022; 10(33): 12077-12088
Published online Nov 26, 2022. doi: 10.12998/wjcc.v10.i33.12077
Published online Nov 26, 2022. doi: 10.12998/wjcc.v10.i33.12077
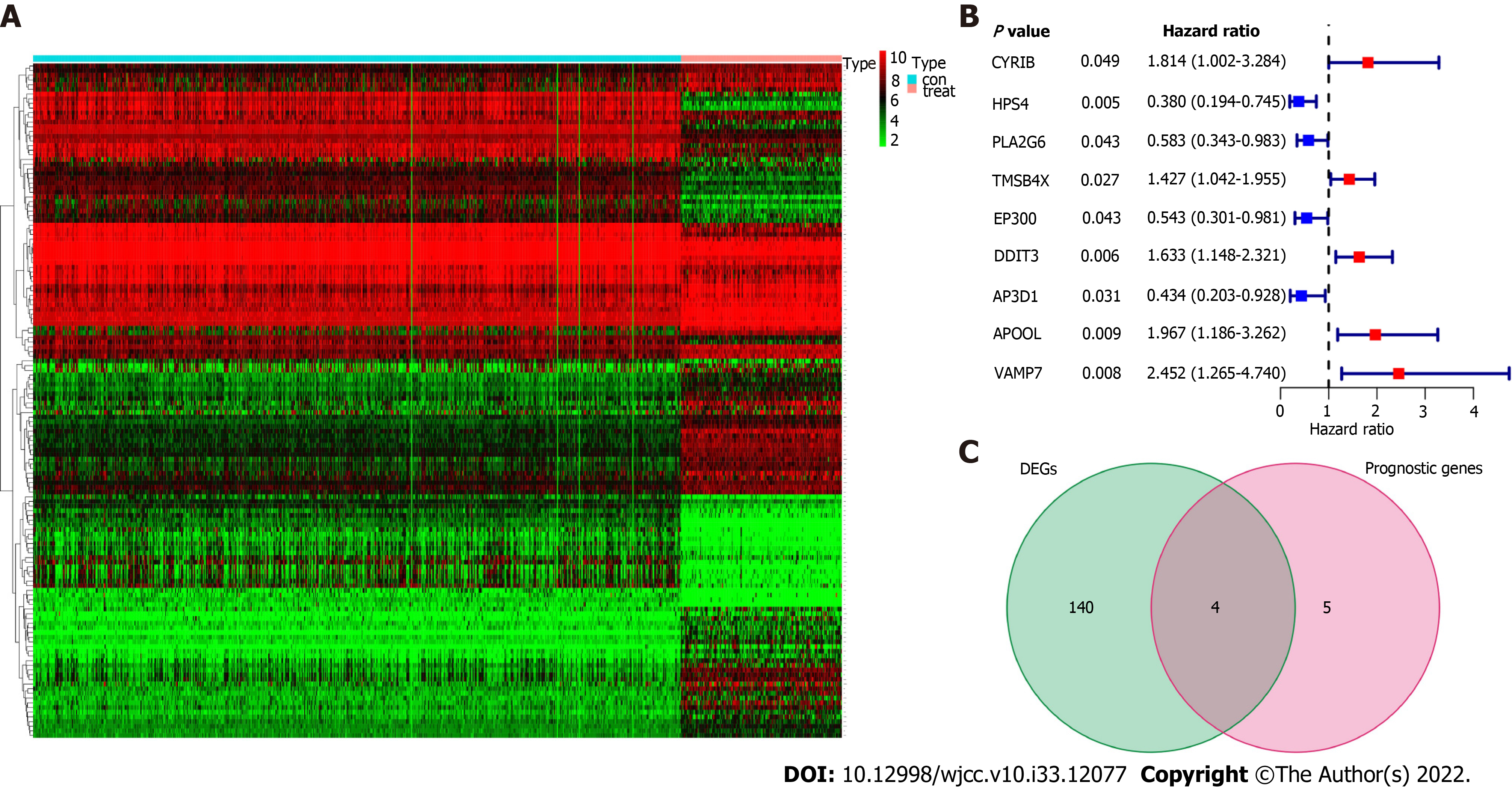
Figure 1 Identification of the candidate platelet-related genes in the esophageal cancer cohort.
A: The heat map showed the expression of 369 platelet-related genes in healthy normal tissues and tumor tissues; B: Forest plots showed the results of the univariate Cox regression analysis between gene expression and overall survival; C: Venn diagram was used to identify differentially expressed genes between tumor and healthy normal tissue that were correlated with overall survival. DEGs: Differentially expressed genes.
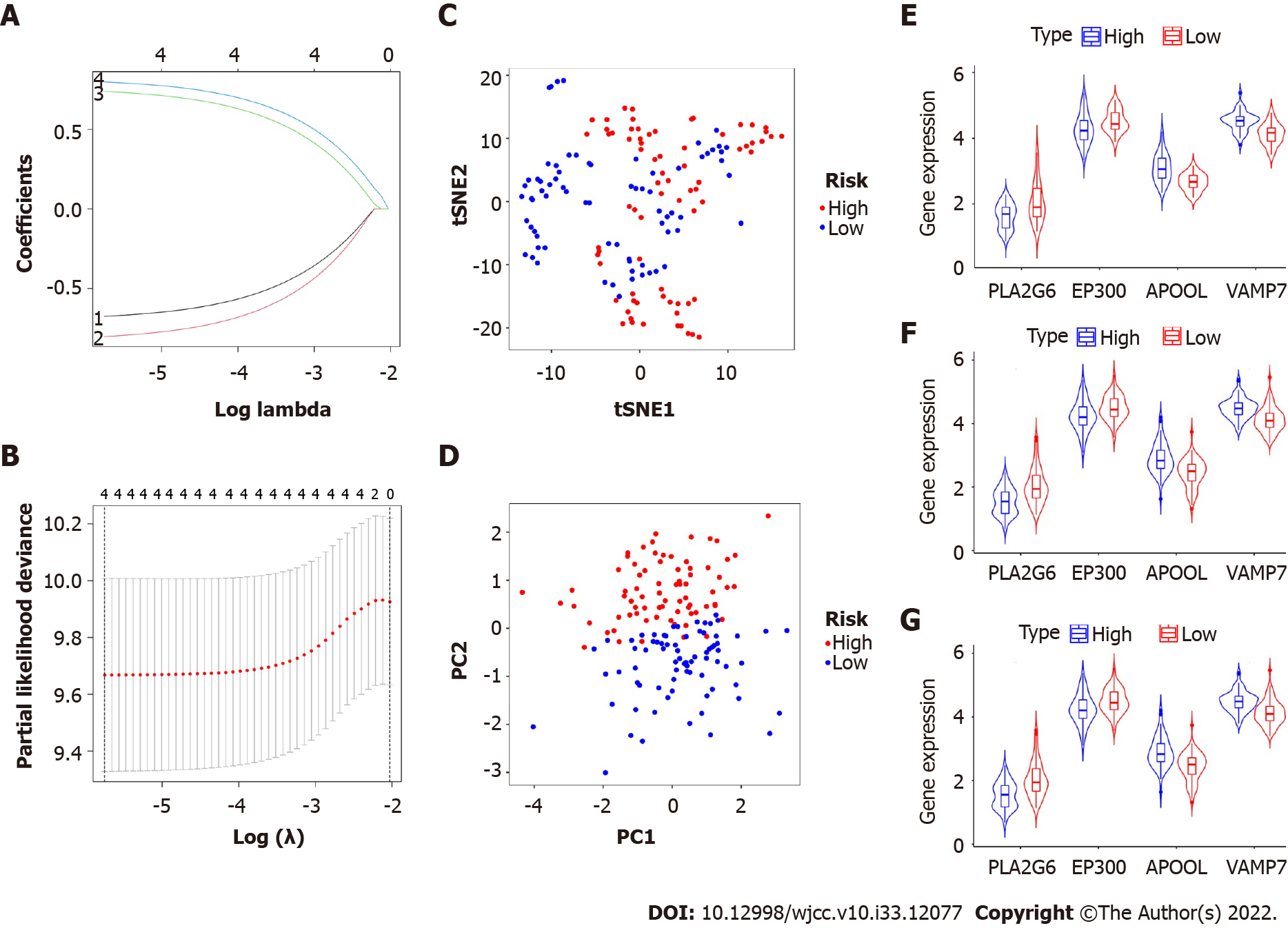
Figure 2 Construction of a prognostic classifier based on platelet-associated genes.
A and B: The results of the least absolute shrinkage and selection operator Cox regression suggest that four platelet-related genes were essential for the classifier; C: t-distributed stochastic neighbor embedding analysis; D: Principal component analysis plot. The expression levels of four biomarkers of the classifier in high- and low-risk group; E: Adenocarcinoma; F: Squamous cell carcinoma; G: All cases. t-SNE: t-distributed stochastic neighbor embedding.
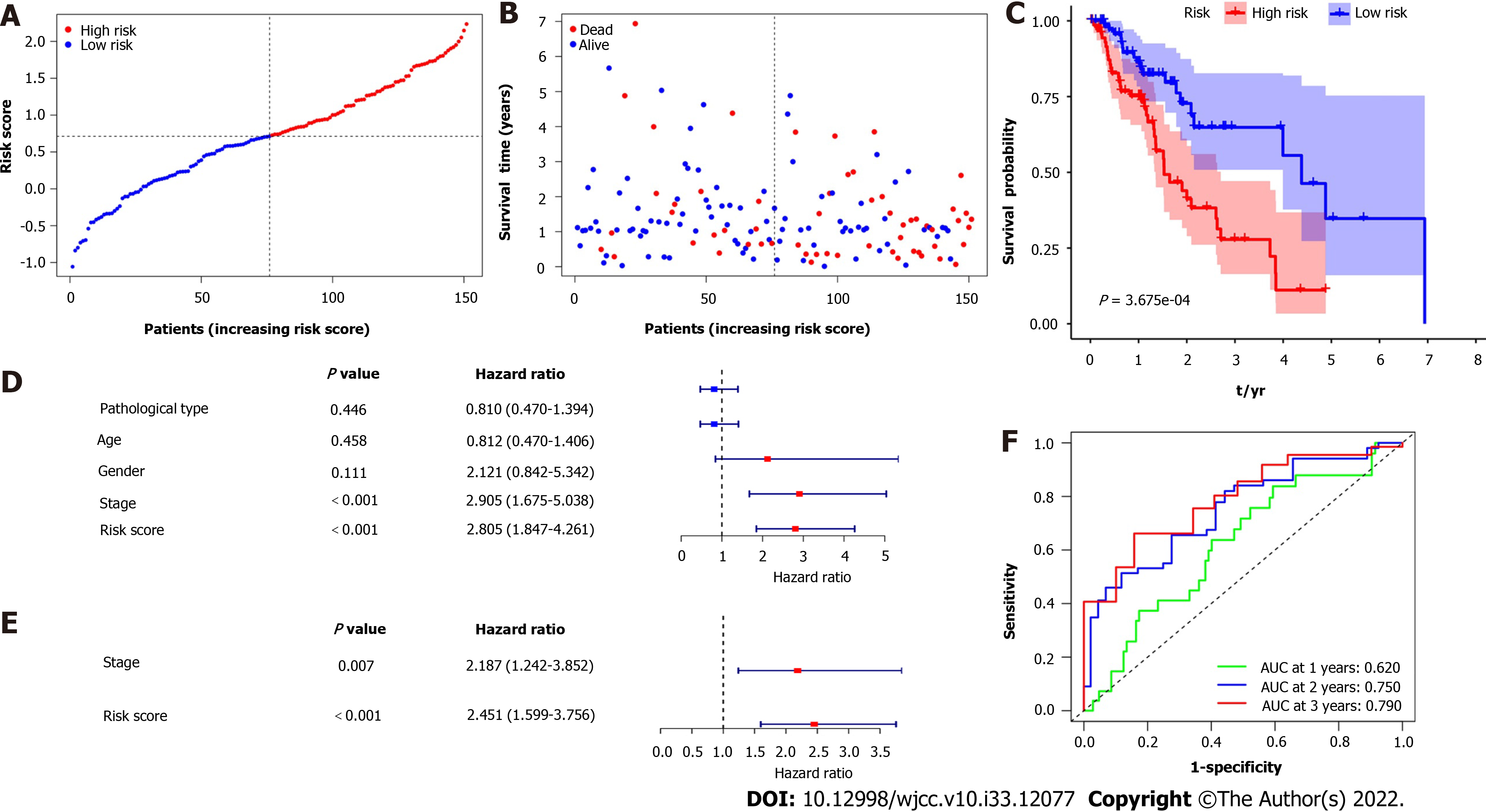
Figure 3 Prognostic analysis of the 4-gene signature model in the esophagus cancer cohort.
A: Distribution and median value of the risk scores; B: Distributions of overall survival status, overall survival, and risk score; C: Kaplan-Meier curves for the overall survival of patients in the high-risk group and low-risk group; D and E: Univariate and multivariate Cox regression analyses regarding overall survival; F: Area under the curve of time-dependent receiver operating characteristic curves.
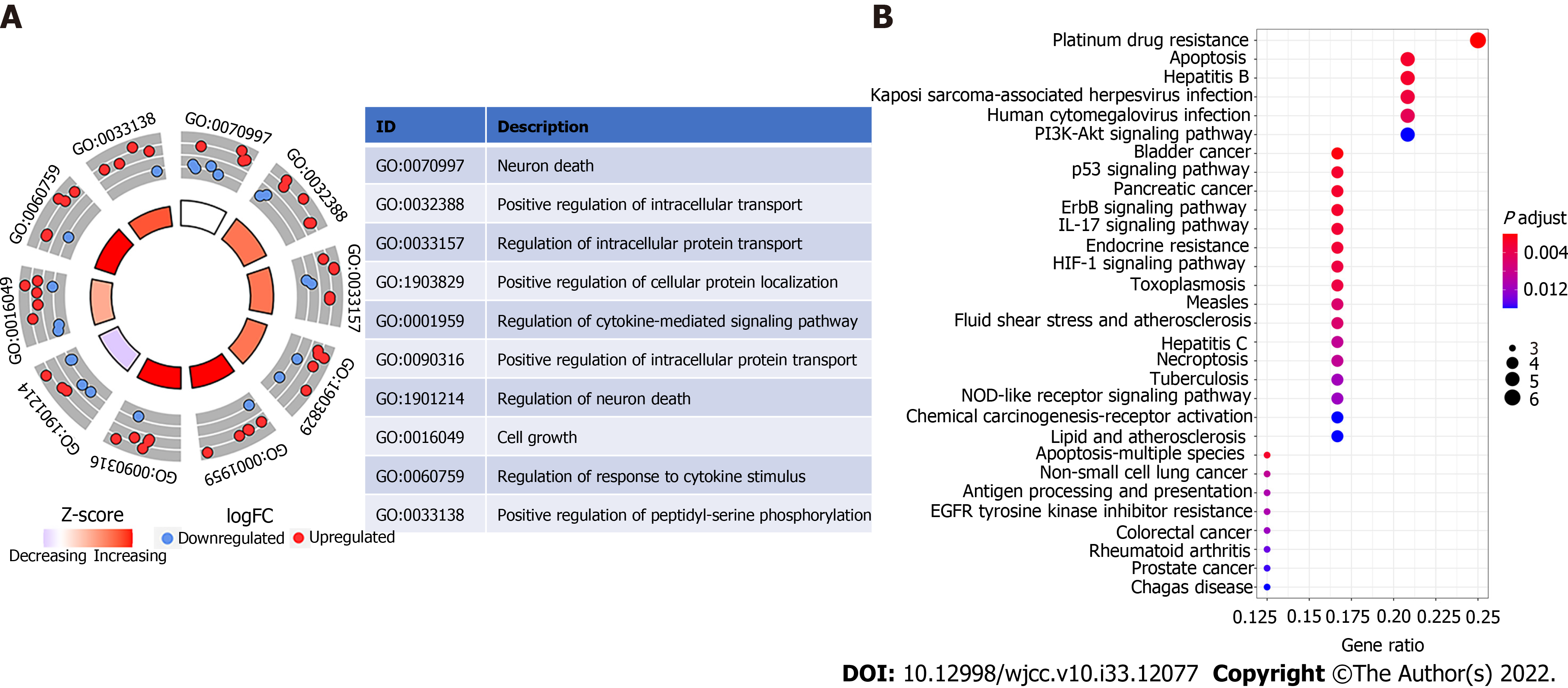
Figure 4 Representative results of Gene Ontology circle plot and Kyoto Encyclopedia of Genes and Genomes analyses.
A: The inner ring is a bar plot where the height of the bar indicates the significance of the term (log10 adjusted P value), and color corresponded to the Z-score. The outer ring displays scatterplots of the expression levels (log fold change) for the genes in each term. The table represents the distribution of platelet-related genes in significant gene ontology terms; B: Most significant or shared Kyoto Encyclopedia of Genes and Genomes pathways. FC: Fold change; GO: Gene ontology.
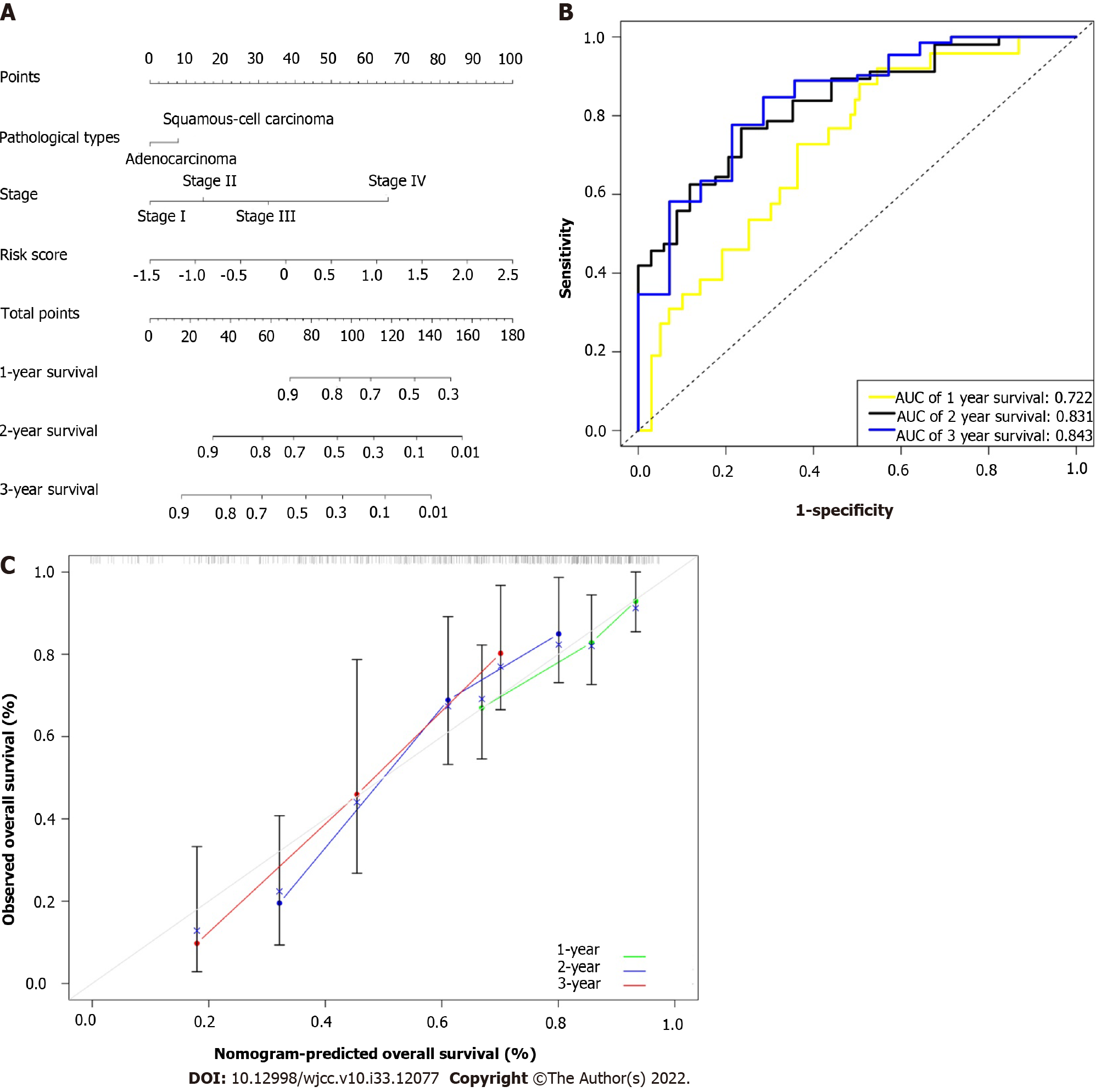
Figure 5 Construction of prognostic nomogram.
A: Nomogram containing the risk score predicted the overall survival in patients with esophageal cancer; B: Receiver operating characteristic curves and area under the curve for 1-, 2-, and 3-year survival of the nomogram; C: Calibration curve of 1-, 2-, and 3-year survival in the nomogram and ideal model.
- Citation: Du QC, Wang XY, Hu CK, Zhou L, Fu Z, Liu S, Wang J, Ma YY, Liu MY, Yu H. Integrative analysis of platelet-related genes for the prognosis of esophageal cancer. World J Clin Cases 2022; 10(33): 12077-12088
- URL: https://www.wjgnet.com/2307-8960/full/v10/i33/12077.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i33.12077