Copyright
©2012 Baishideng.
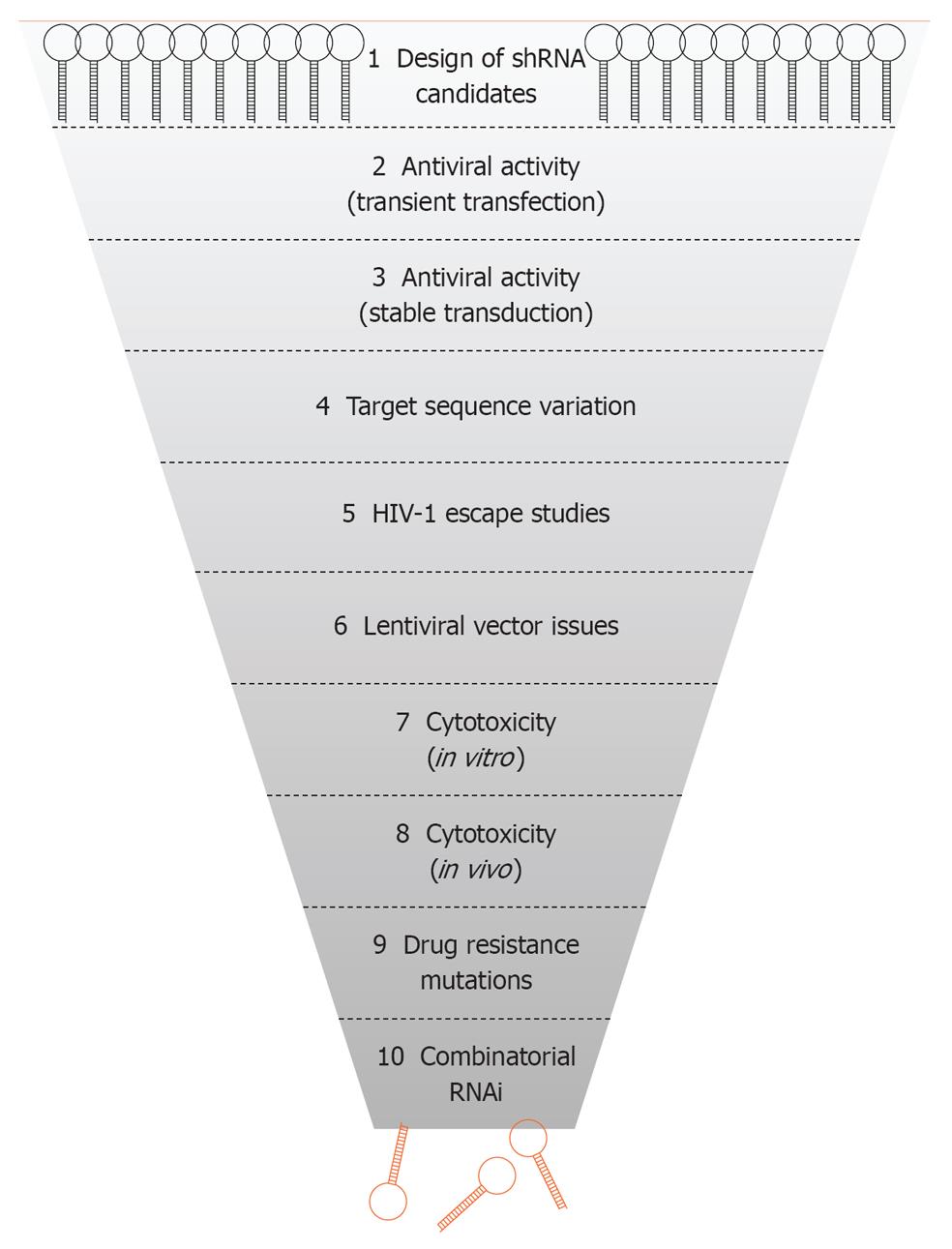
Figure 1 Selection of short hairpin RNAs against human immunodeficiency virus type 1.
Scheme of the 10 steps used for the selection of the best shRNA inhibitors for development of an antiviral RNA interference (RNAi) gene therapy. HIV-1: Human immunodeficiency virus type 1.
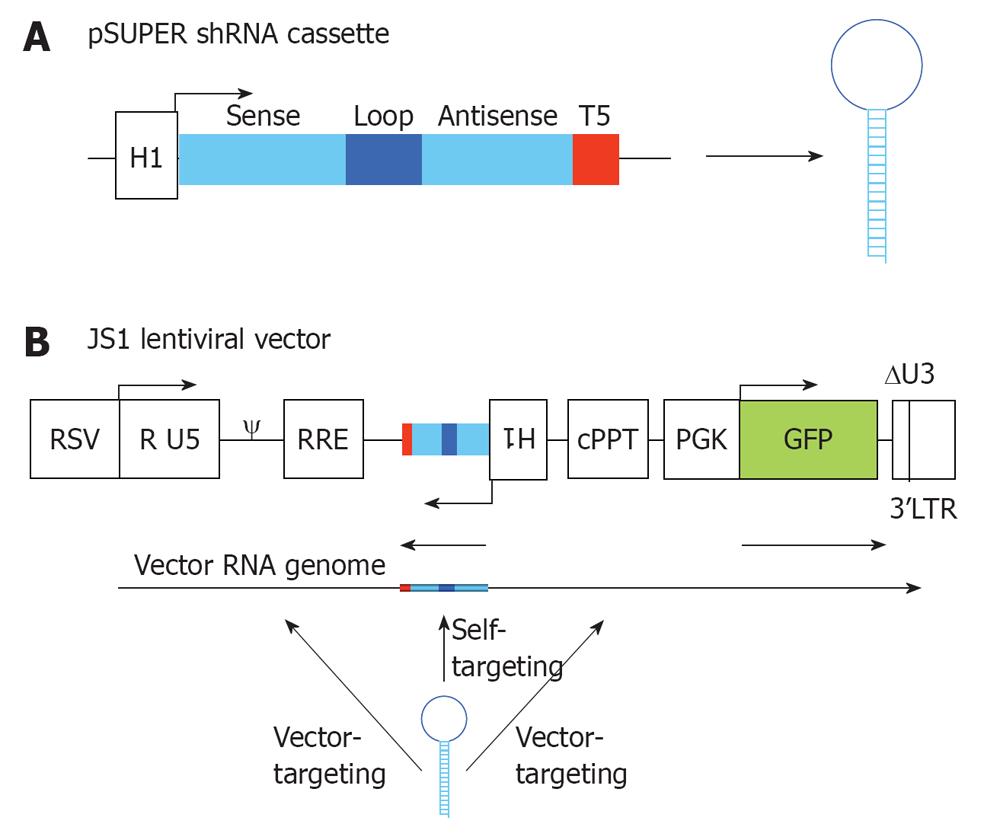
Figure 2 RNA interference vector constructs.
A: Two complementary DNA oligonucleotides are annealed and cloned into pSUPER downstream of the H1 promoter that triggers short hairpin RNA (shRNA) expression. The shRNA cassette encodes a 19 nt sense strand, 9 nt loop, 19 nt antisense strand and a stretch of 5 T’s (T5), which is the termination signal; B: The shRNA cassette was cloned in the lentiviral vector JS1 for stable transduction of human T cells. The shRNA cassette is cloned in antisense direction to avoid promoter interference. During vector production three transcripts are produced from the lentiviral vector: the shRNA, the vector RNA genome and the GFP transcript. The shRNA will have a 100% target match with the shRNA-encoding sequence in the vector RNA genome (self-targeting), and potential targets in the human immunodeficiency virus type 1 (HIV-1) derived sequences of the lentiviral vector (vector-targeting). RSV: Respiratorial syncytial virus promoter; R U5: R and U5 element of HIV-1 promoter; ψ: Packaging signal; RRE: Rev responsive element; cPPT: Central polypurine tract; PGK GFP: Green fluorescent protein driven from a PGK promoter; 3’LTR: 3’ long terminal repeat of HIV-1, with deletion in the U3 region.
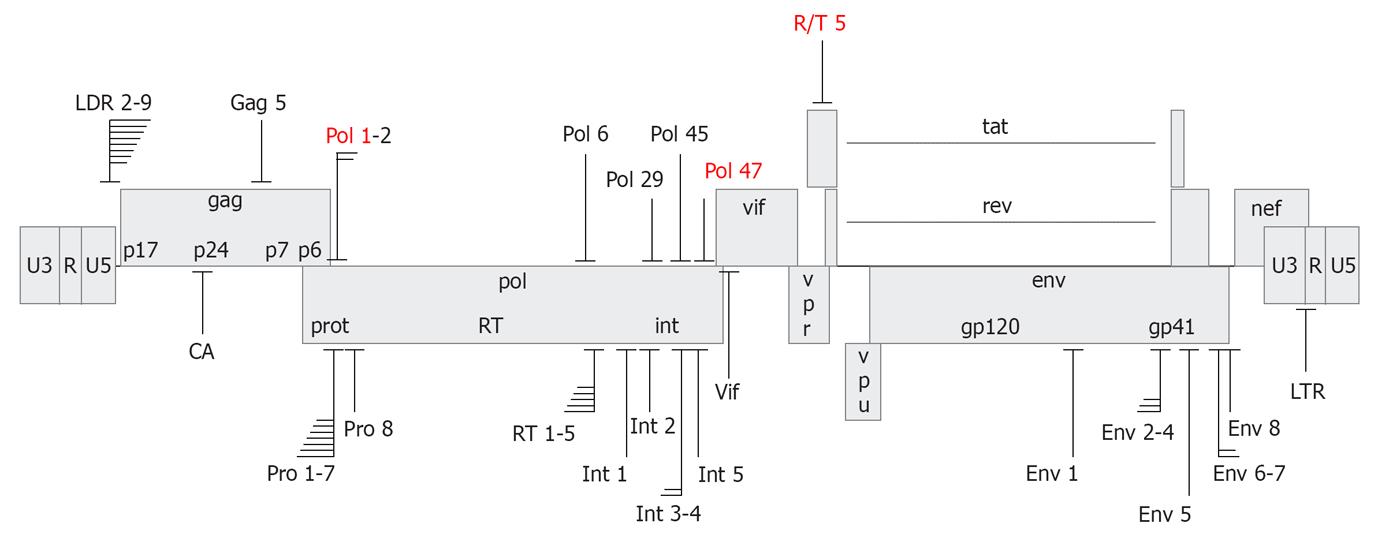
Figure 3 Target sites of anti-human immunodeficiency virus short hairpin RNAs.
Depicted is the human immunodeficiency virus type 1 (HIV-1) LAI proviral genome with short hairpin RNA (shRNA) target sites that yielded > 80% suppression of HIV-1 production. Several clusters of overlapping shRNA targets are indicated. The shRNAs have been designed based on conservedness of the target sequence (upper panel) or accessibility in the structured HIV-1 RNA genome (lower panel). shRNAs selected for the R3 combinatorial RNA interference vector are marked in red.
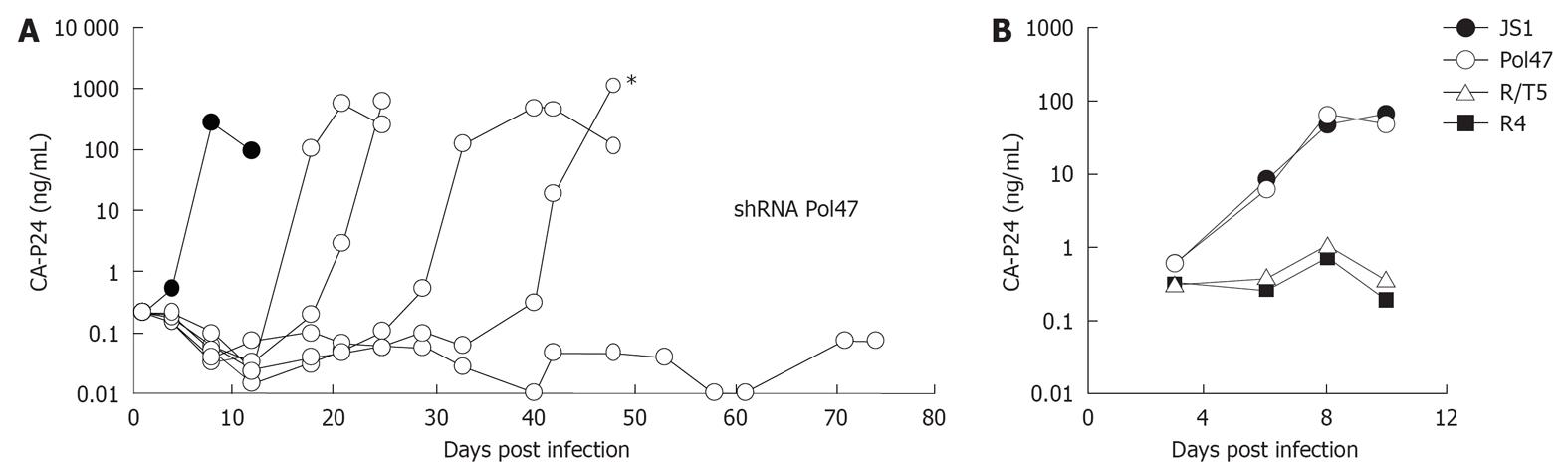
Figure 4 Long-term short hairpin RNA activity in human immunodeficiency virus type 1 replication studies.
A: SupT1 T cells expressing short hairpin RNA (shRNA) Pol47 were infected with HIV-1 and virus replication was monitored in five cultures by measuring CA-p24 production for up to 75 d. Cells transduced with the empty lentiviral vector JS1 served as control (dark circles); B: Supernatant from the indicated culture (asterisk in panel A) was passaged on new cells to test the escape phenotype. The virus replicated on control and shRNA Pol47 cells, but not on cells that express another antiviral shRNA (R/T5) or the R4 construct. Adapted from[16].
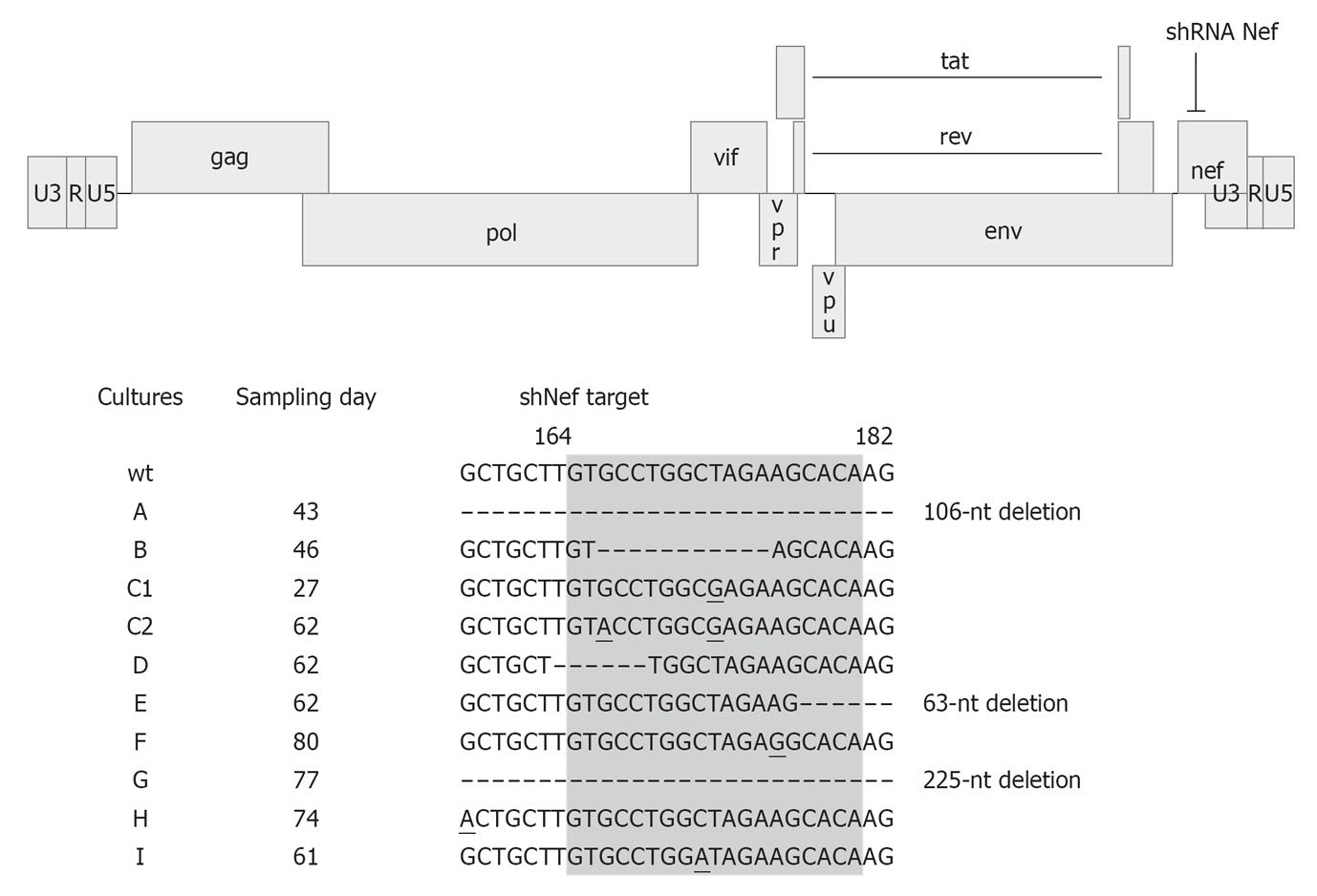
Figure 5 Human immunodeficiency virus type 1 escapes from short hairpin RNA Nef.
The human immunodeficiency virus type 1 (HIV-1) LAI proviral genome and the short hairpin RNA (shRNA) Nef target site are indicated. SupT1 cells expressing shRNA Nef were infected with HIV-1 and passaged twice a week until viral escape occurred. Nine different cultures were examined in parallel and the day of sampling is indicated. Part of the nef gene is shown with the shRNA Nef target site highlighted in gray. Numbers refer to the nucleotide position in the nef gene. Escape was apparent by (1) one or more escape mutations in the target sequence; (2) mutations outside the target region; and (3) complete or partial deletions of the target region. Mutations are underlined. Adapted from[13].
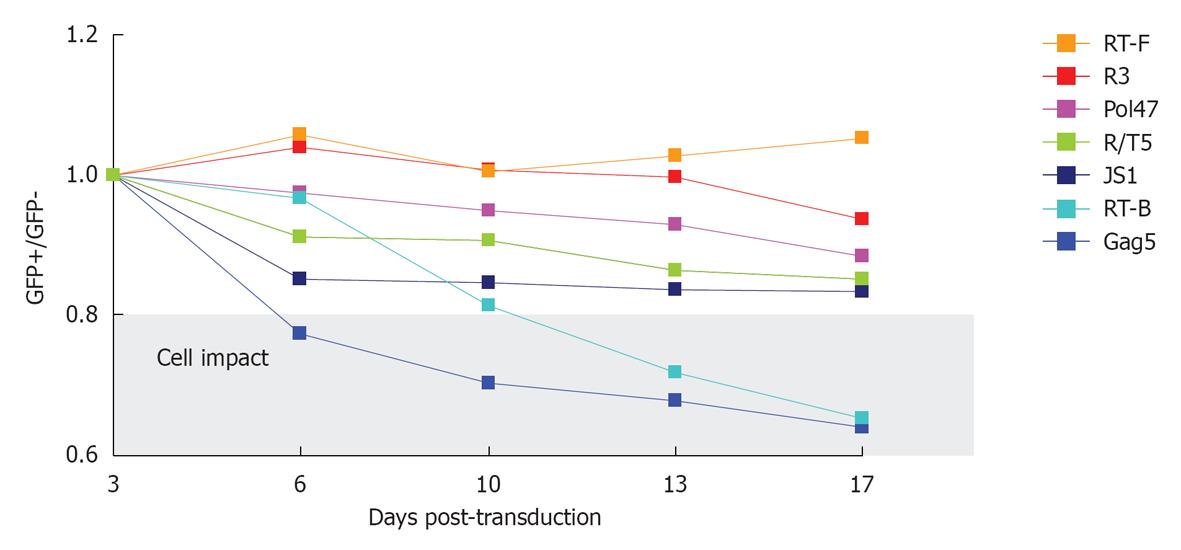
Figure 6 Competitive cell growth assay.
SupT1 T cells were transduced with a short hairpin RNA (shRNA)-expressing lentiviral vector, yielding a cell population with approximately 40% GFP-positive cells. The ratio of GFP-positive cells at day 3 after transduction was set at 1 and measured longitudinally. The cells were passaged and analyzed via FACS measurement twice a week. JS1 represents the empty lentiviral vector without shRNA expression. The gray window highlights shRNAs that have a significant adverse effect on cell growth.
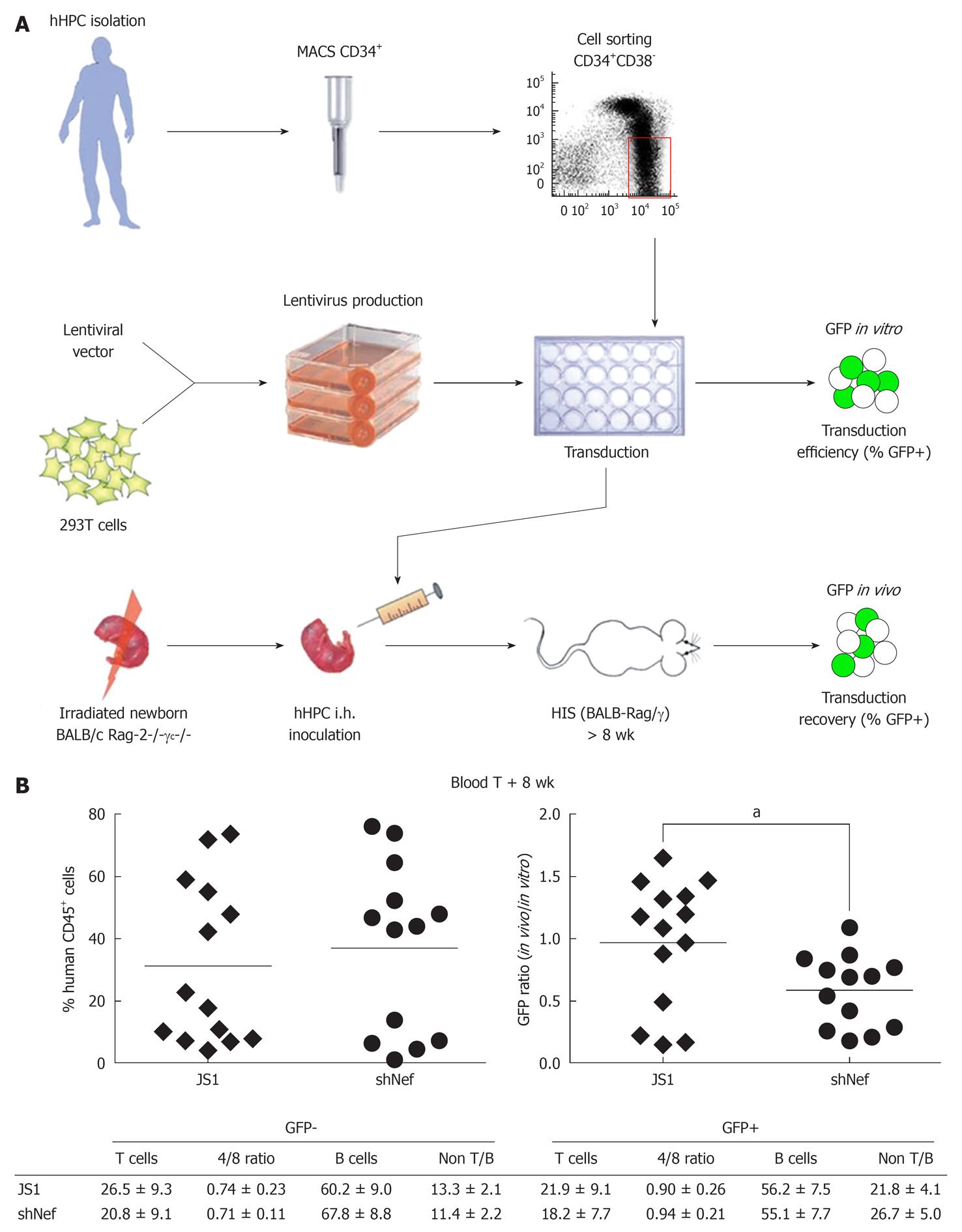
Figure 7 In vivo safety studies in the HIS mouse model.
A: Cell suspensions enriched for human hematopoietic progenitor cells (hHPC) are prepared from fetal liver tissue. Live nucleated CD34+ cells are magnetically sorted and further enriched for the hHPC (CD34+CD38- fraction) using fluorescence activated cell sorting. Lentiviral supernatants are produced on 293T cells. hHPC are transduced ex vivo with the shRNA-expressing lentiviral vector and injected intrahepatically into sublethally irradiated newborn BALB/c Rag-2-/-γc-/- mice. The transduction efficiency is evaluated based on GFP expression after 3.5 d in culture (GFP in vitro); B: The HIS (BALB-Rag/γ) mice are analyzed in the blood and the organs after at least 8 wk post-transplantation for the presence of human cells (%CD45+ cells) (left graph), which were analyzed for GFP recovery (right graph). The GFP recovery is the ratio between the frequency of human GFP+ cells measured in the animals (GFP in vivo) and the frequency of GFP+ hHPC injected in the newborn mice (GFP in vitro, transduction efficiency). The major subsets of the human immune system in the blood are also analyzed for their frequency and absolute number in the human GFP+ and GFP- population. Adapted from[24,70]. aP < 0.05.
- Citation: Knoepfel SA, Centlivre M, Liu YP, Boutimah F, Berkhout B. Selection of RNAi-based inhibitors for anti-HIV gene therapy. World J Virol 2012; 1(3): 79-90
- URL: https://www.wjgnet.com/2220-3249/full/v1/i3/79.htm
- DOI: https://dx.doi.org/10.5501/wjv.v1.i3.79