Published online Aug 19, 2023. doi: 10.5498/wjp.v13.i8.524
Peer-review started: May 17, 2023
First decision: July 4, 2023
Revised: July 5, 2023
Accepted: July 27, 2023
Article in press: July 27, 2023
Published online: August 19, 2023
Processing time: 92 Days and 4.6 Hours
Antidepressants, particularly selective serotonin reuptake inhibitors, are currently considered the first-line treatment for panic disorder (PD). However, little is known about the relationship between the biomarkers that may predict better treatment.
To compare genome-wide methylation and gene expression patterns between responsive and non-responsive patients with PD after 4 wk of escitalopram treatment.
Thirty patients with PD were enrolled in this study (responders = 13; non-responders = 17). All patients were assessed using the PD Severity Scale-Chinese version before and after treatment. The Illumina Infinium MethylationEPIC (850k) BeadChip for genome-wide methylation screening and mRNA sequencing was used in all patients with PD.
A total of 701 differentially methylated positions (DMPs) were found between responders and non-responders (|Δβ| ≥ 0.06, q < 0.05), and the hyper- and hypomethylated CpG sites were 511 (72.9%) and 190 (27.1%), respectively. Relative to non-responders, there were 59 differential transcripts, of which 20 were downregulated and 39 were upregulated (q < 0.05). However, no differentially expressed genes were identified by mRNA sequencing after correcting for multiple testing (|log2(FC)| > 1, q > 0.05).
This preliminary study showed that DMPs might be associated with the treatment response to escitalopram in PD; however, these DMPs need to be verified in large samples.
Core Tip: No genome-wide methylation studies or mRNA sequencing have been conducted to identify early response biomarkers in patients with panic disorder (PD). This study aimed to compare genome-wide methylation and gene expression patterns between responsive and non-responsive patients with PD after 4 wk of escitalopram treatment. A total of 701 differentially methylated positions (DMPs) were found between responders and non-responders, and the hyper- and hypomethylated CpG sites were 511 (72.9%) and 190 (27.1%), respectively. This preliminary study showed that DMPs might be associated with the treatment response to escitalopram in PD.
- Citation: Zou ZL, Zhang Y, Huang YL, Wang JY, Zhou B, Chen HF. Pilot study of genome-wide DNA methylation and gene expression for treatment response to escitalopram in panic disorder. World J Psychiatry 2023; 13(8): 524-532
- URL: https://www.wjgnet.com/2220-3206/full/v13/i8/524.htm
- DOI: https://dx.doi.org/10.5498/wjp.v13.i8.524
Panic disorder (PD) is a common anxiety disorder characterized by recurrent and unexpected panic attacks. The estimated 12-month and lifetime prevalences of PD are 2.4% and 3.8%, respectively[1,2]. A cross-national epidemiological study reported that the lifetime prevalence of panic attacks was 13.2%[3]. Patients with PD experience symptoms such as tachycardia, chest pain, breathlessness, and dizziness. Consequently, they most frequently seek care in medical settings, such as emergency departments, and their condition is often undiagnosed[4]. PD causes substantial suffering and increases economic costs for both patients and society[5]. Hence, the availability of effective treatment may not only benefit individual patients but also provide economic returns to society.
Antidepressants, particularly selective serotonin reuptake inhibitors (SSRIs), are currently considered first-line treatments for PD. A meta-analysis suggested that SSRIs result in high remission rates with a minimal risk of adverse events in the treatment of PD[6,7]. However, not all patients benefit from the antidepressant therapy. For example, in a 12-week clinical trial of sertraline, citalopram, escitalopram, and paroxetine, 11.1%–18% of patients with PD did not respond to treatment[8,9]. Thus, predictive biomarkers of antidepressant response could greatly benefit clinical practice by decreasing the duration of drug efficacy evaluations.
Previous studies have found that common genetic variants could explain 42% of the individual differences in antidepressant response[10]. In addition, gene polymorphism studies focusing on 5-HTT, 5-HTR1A, 5-HTR2A, COMT, BDNF, and P450 (CYP) have been conducted in several large-scale studies on antidepressant drug responses in PD[11-14]. However, no consistent findings have been obtained from these studies. For example, with regard to treatment response to SSRIs in patients with PD, Zou et al[13] found that 5-HTTLPR polymorphism, rather than 5-HTR1A, may be an early predictor of response to sertraline in 2020, whereas Yevtushenko et al[11] indicated the importance of a 5-HT1A receptor gene polymorphism in 2010. Therefore, genetic variation in a single gene cannot fully explain individual differences in the response to treatment.
Emerging evidence from human and animal studies suggests a key role of epigenetic markers, including DNA methylation and histone modifications, in the prediction of antidepressant response[15]. Methylation of some candidate genes, such as SLC6A4, BDNF, and IL11, has shown promising results as a biomarker for predicting antidepressant responses in major depressive disorder (MDD). However, the research methods and results have been heterogeneous[16]. Genome-wide methylation analysis can accurately determine the location of DNA methylation in the genome and screen for effect-related differentially methylated genes. Currently, 850k methylation BeadChip is a new generation of DNA methylation chips developed based on the original 450k methylation chip[17]. The newly developed Human Methylation 850 BeadChip covers over 850k CpG methylation sites and is the most useful tool for analyzing the DNA methylation profile of the human genome. In addition, DNA methylation is a key epigenetic mechanism involved in the developmental regulation of gene expression[18]. Nevertheless, no genome-wide methylation studies or mRNA sequencing have been conducted to identify early response biomarkers in patients with PD.
Hence, a pilot study of genome-wide DNA methylation and gene expression analysis was conducted in patients with PD using the Illumina 850k BeadChip and mRNA sequencing to identify possible predictors of treatment response to escitalopram.
All patients with PD were recruited from the inpatient and outpatient departments of Sichuan Provincial People’s Hospital between March 2019 and December 2020. All patients satisfied the following inclusion criteria: (1) Primary diagnosis of PD in line with the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) performed by a professional psychiatrist through a standardized structured clinical interview for DSM-IV Axis I disorders[19], with none of the patients having any other psychiatric disorders; (2) All PD patients were medication-naïve and had no history of any antidepressant or other psychotropic medication intake; (3) All subjects were without suicide ideations and attempts; (4) Subjects had no family history of psychiatric disorders in a first-degree relative; (5) All patients were Han Chinese aged 18 to 60 years; (6) All subjects were free of acute or chronic somatic disorders, head trauma, or neurological illnesses, and all subjects were free of alcohol consumption within 2 wk before their examination. Women were non-pregnant and non-nursing; and (7) All patients received escitalopram (10–20 mg qd) for 4 wk. The study was approved by the Sichuan Provincial People’s Hospital ethics committee [reference number: (2021) Ethics Review (313)]. All individuals provided written informed consent prior to the initiation of study procedures.
Patients were assessed at baseline and after 4 wk of treatment using the PD Severity Scale (PDSS). The PDSS comprises seven items, and participants are instructed to rate each item from 0 (none) to 4 (extremely severe) based on the severity of each symptom[20]. The PDSS-Chinese version has good internal consistency (Cronbach’s alpha) with an overall score of 0.83[21]. Treatment response for PD was defined as a reduction in the pretreatment PDSS score of at least 40% at 4 wk[22]. The PD samples were divided into responder and non-responder groups.
A 4-mL ethylenediaminetetraacetic acid-anticoagulated peripheral blood sample was collected from all subjects in a fasting state via venipuncture between 7:00 a.m. and 8:30 a.m. DNA and RNA were extracted from whole blood samples from PD patients at baseline using DNeasy Blood and miRNeasy Mini kits (Qiagen, Hilden, Germany), respectively, according to the manufacturer’s protocol.
DNA was subjected to sodium bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, United States). The Illumina Infinium Human Methylation 850K BeadChip (Illumina, San Diego, CA, United States) was used to assess genome-wide DNA methylation, with a genome-wide coverage of over 850k CpG methylation sites per sample according to the manufacturer’s instructions. DNA quality control, bisulfite conversion, genome-wide methylation analysis, and initial methylation signal detection quality control were performed by Sinotech Genomics Co., Ltd. (Shanghai, China).
The raw intensity data were imported into R (v4.0.0; R Foundation for Statistical Computing, Vienna, Austria) and analyzed using the ChIP (ChAMP) package (v2.18.2) for data preprocessing, normalization, and comparison. β-values (ranging from 0 to 1) were used to determine the DNA methylation levels at each CpG site. Probes were filtered, including probes with a detection p-value of < 0.01, probes with less than three beads in at least 5% of the samples, non-CpG probes, and multi-hit probes. Beta-Mixture Quantile was used to normalize β-value matrices to adjust for type I and II probe biases[23]. In addition, we used singular value decomposition analysis to analyze the batch effect caused by the BeadChip Slide and Array and applied Combat to correct this batch effect[24]. Finally, all CpG sites were annotated using EPICanno software. ilm10b5. hg19.
An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) was used to assess RNA quality. A Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and NanoDrop One spectrophotometer (Thermo Fisher Scientific) were used to quantify total RNA, after which the library was constructed. The library for polymerase chain reaction product purification by paired-end libraries was synthesized using the mRNA-seq Lib Prep Kit for Illumina (ABclonal, Wuhan, China) following the sample preparation guide. Libraries were quantified using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific) and validated using an Agilent 2100 Bioanalyzer (Agilent Technologies) to calculate the molar concentration and insert size. Clusters were generated using the cBot user guide and sequenced using an Illumina NovaSeq 6000 system (Illumina). Library construction and sequencing were performed by Sinotech Genomics Co. Ltd.
Hisat2 (2.1.0) was used to map the paired-end sequence files (fastq) to the reference genome (GRCh38.91). The output sequencing alignment/map files were converted into binary alignment/map files and sorted using SAM tools (v1.7). Gene abundance was expressed as fragments per kilobase of exons per million reads mapped (FPKM). StringTie software was used to count the fragments within each gene, and the TMM algorithm was used for normalization (https://www.ncbi.nlm.nih.gov/COG/). Finally, the FPKM value of each gene was calculated. EdgeR software was used to conduct differential expression analysis of the mRNA[25].
Data were analyzed using SPSS software (version 18.0; SPSS Inc., Chicago, IL, United States). Intergroup comparisons of continuous variables were conducted using Student’s t-test, and categorical variables were analyzed using Pearson’s chi-square test. Differentially methylated CpG positions were identified using ChAMP. Δβ was calculated as the difference of mean β-values between responders and non-responders. We set the criteria for differential methylation positions (DMPs) as the calling significance of an absolute change in Δβ-value between groups (|Δβ| ≥ 0.06 and q < 0.05). Differentially expressed mRNAs were defined as |log2(FC)| > 1 and q < 0.05. To identify genes with the same function and pathway in DNA methylation and gene expression profiling, we performed gene ontology (GO) (http://geneontology.org/) analysis for biological processes, cellular components, and molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.kegg.jp/) for differentially methylated and expressed genes. For all statistical analyses, a two-tailed p-value < 0.05 indicated statistical significance. The q-value cut-off of 0.05 was corrected using the Benjamini–Hochberg method for multiple hypotheses[26].
PD samples were divided into 13 responders (6 men and 7 women; 33.54 ± 11.64 years old) and 17 non-responders (8 men and 9 women; 33.88 ±9.65 years old) at 4 wk. No statistically significant differences were found between the responders and non-responders in terms of sex (χ2 = 0.002, p > 0.05) or age (t = -0.088, p > 0.05). There were 4 smokers (30.8%) and 9 nonsmokers (69.2%) in the responder group, and 5 smokers and 12 nonsmokers in the non-responder group. The number of drinkers and nondrinkers among the responders was 3 (23.1%) and 10 (76.9%) in the responder group, and 5 drinkers and 12 nondrinkers in the non-responder group. No statistically significant group difference was observed in terms of smoking and drinking (χ2 = 0.006, 0.151; p > 0.05). In addition, the average PDSS total score was 13.08 ± 4.25 in responders and 14.94 ± 3.25 in non-responders at the start of treatment, and no statistically significant group difference was observed (t = -1.363, p > 0.05). The average PDSS total score for responders was 4.54 ± 2.11, which was significantly lower than that for non-responders 10.35 ± 2.00 after 4 wk of treatment (p < 0.001).
Compared with non-responders, there were 701 DMPs (|Δβ| ≥ 0.06, q < 0.05), and the hyper- and hypomethylated CpG sites were 511 (72.9%) and 190 (27.1%), respectively (Supplementary Table 1). The distribution of DMPs in the CpG islands was as follows: N-shore (34.6%), S-shore (24.5%), N-shelf (16.5%), island (14.4%), and S-shelf (10.1%). These DMPs in different regions of the gene were in the following order: Body region (64.4%), transcriptional start site (TSS) 1500 (15.3%), 5′-untranslated region (UTR) region (12.5%), 1st Exon (3.4%), TSS 200 (3.1%), and the 3′-UTR region (1.3%). According to the University of California Santa Cruz annotation, 437 unique genes were identified, including 311 hypermethylated and 122 hypomethylated genes; the coexistence of hyper- and hypomethylation sites was found in four genes (Supplementary Table 2).
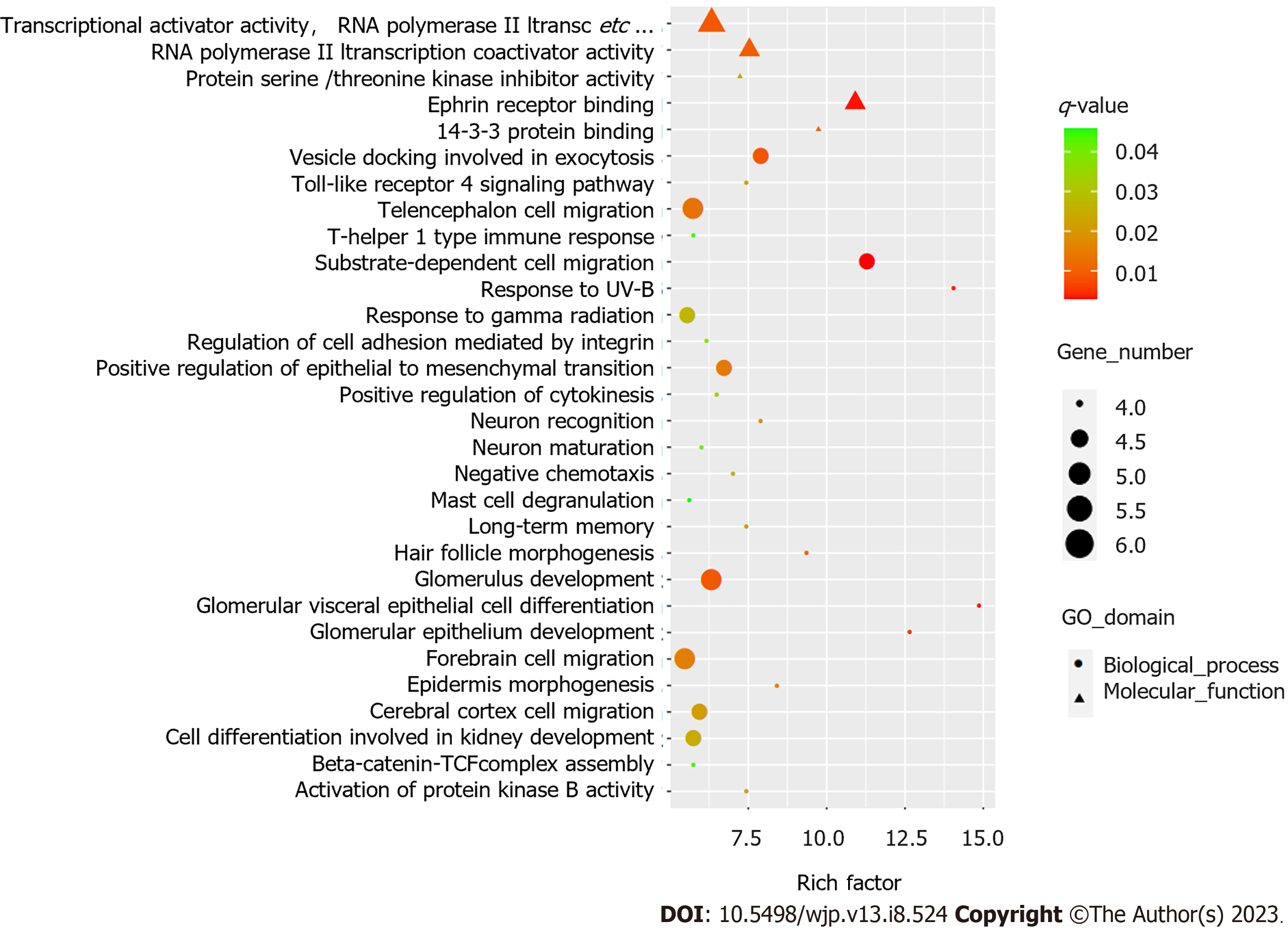
GO analysis revealed distinct functional categories for the associated genes, and approximately 226 biological processes, 14 cellular components, and 22 molecular functions were identified in differentially methylated genes (q < 0.05). Figure 1 shows the top 30 GO annotations for the differentially methylated genes. KEGG analysis showed that these differentially methylated genes were involved in 43 significant pathways (p < 0.05). However, no statistically significant pathways were found after adjusting using the Benjamini–Hochberg method (q > 0.05) (Supplementary Table 3).
Compared with non-responders, there were 59 differential transcripts after adjusting using the Benjamini–Hochberg method, of which 20 were downregulated and 39 were upregulated (q < 0.05) (Figure 2). After the data were analyzed using the screening procedure, the results of mRNA sequencing showed that 132 differentially expressed genes were identified (p < 0.05). However, no statistically significant group differences between responders and non-responders were observed after adjusting using the Benjamini–Hochberg method (q > 0.05) (Supplementary Table 4).
DNA methylation plays an important role not only in the diagnosis of diseases but also in the prediction of efficacy[16,27]. In this study, a pilot investigation of genome-wide DNA methylation in the early response to antidepressants in patients with PD was conducted using the Illumina 850k BeadChip. To our knowledge, this is the first genome-wide DNA methylation study of the treatment response to escitalopram in PD. According to the screening criteria for differential methylation sites, 701 DMPs were screened among responders and non-responders in patients with PD, and these DMPs were located within 437 unique genes. Similarly, one study assessed genome-wide DNA methylation using the Infinium MethylationEPIC BeadChip in patients with MDD for escitalopram treatment response. They identified 2571 significant DMPs, and 303 DMPs with an absolute change in Δβ-value between groups larger than 0.2[28]. These findings indicate that DMPs are potential peripheral predictors of antidepressant treatment response, and present an important opportunity to improve symptoms through prediction of medication response. Not only that, there is also some emerging evidence to suggest that PD patients have aberrant DNA methylations[29]. Psychiatry as a medical discipline, a diagnosis identifying a disorder should lead to an effective therapy[30]. Hence, epigenetic factors contributing to antidepressant response will be a unique and promising opportunity to implement personalized medicine in PD treatment. In addition, in the present study, most DMPs with an absolute change in value between groups were lower than 0.1. Furthermore, Ju et al[28] found that many DMPs showed very small differences in methylation (Δβ < 0.5%), making it difficult to fully explain efficacy prediction from a single CpG site. The treatment efficacy may be largely determined by the combined effects of multiple loci.
Notably, the current study identified 10 DMPs with an absolute change in Δβ-value larger than 0.2 (|Δβ| ≥ 0.2) between groups, and these genes included HLA-DPB1, HLA-DPA1, PDE1A, COL23A1, RUFY4, FRMD5, and SOHLH1. For example, the HLA-DPB1 site (cg12865025) is located in the TSS 1500 region. The major histocompatibility complex class II molecule consists of non-covalently associated alpha and beta chains, and this class II molecule is a heterodimer consisting of an alpha (DPA) and a beta chain (DPB)[31]. HLA-DPA1 and HLA-DPB1 are expressed on the surfaces of antigen-presenting cells; they play a central role in the adaptive immune system[32] and are associated with the pathophysiology of psychiatric disorders[17]. In recent years, increasing evidence has suggested that aberrations in immune-related pathways contribute to the pathophysiology and prediction of treatment responses in patients with PD[33,34]. Moreover, escitalopram led to a decrease in immune system activation[35], augmented tumor necrosis factor- peripheral secretion, induced faster kinetics of interleukin-1β secretion[36], and decreased Interleukin-17 levels[37]. Therefore, immune-related pathways may play an important role in the response to antidepressants in patients with PD. However, whether these DMPs can be used as molecular markers needs to be verified in larger samples.
In this study, functional enrichment analysis of differentially expressed genes was performed. GO analysis identified 262 GO terms in differentially methylated genes. Notably, the top 30 GO annotations, such as neuron recognition, neuron maturation, and cerebral cortex cell migration, may be associated with the treatment response. These enriched genes, including DAB1, NRP1, DIXDC1, ROBO1, PEX5, APP, ANKS1A, EDNRB, BCL2, and NTM, play central roles in brain development, neuronal remodeling, neurite outgrowth, and adhesion. For example, antidepressants such as citalopram increase APP secretion in primary rat neuronal cultures[38]. In addition, previous findings suggest that citalopram reduces mutant APP, Aβ, and mitochondrial toxicities and may have a protective role against mutant APP and Aβ-induced injuries in patients with depression, anxiety, and Alzheimer’s disease[39]. In addition, these genes have a potential role in the development of many neuropsychiatric diseases. For example, a previous study found that Dab1 knockout mice exhibited behavioral abnormalities, including hyperactivity, decreased anxiety-like behavior, and impaired working memory[40]. Hence, these differentially methylated genes in the GO terms may be involved in the pathophysiology and therapeutic response in PD, and the functions of these genes need further exploration. Further studies are needed to explore whether differential methylation in peripheral blood is similar to that in the brain.
Few studies have explored the association between gene expression and the prediction of therapeutic response in patients with PD. In this study, mRNA sequencing was compared between responders and non-responders among patients with PD. Unfortunately, no statistically significant group differences were found in differentially expressed genes. Furthermore, no candidate genes were identified based on integrative analysis of differential DNA methylation and expression. This may be due to the following reasons. First, the sample size was small. The different clinical features of patients with PD are closely related to the heterogeneity of heredity. Second, the use of whole blood, which is a mixed cell type sample, may limit the identification of mRNA expression changes. Finally, gene expression is affected by many factors, such as environmental and other epigenetic factors, especially non-coding RNA, which represents a promising source of peripheral biomarkers of antidepressant responses[41]. Ultimately, it is likely that combinations and not individual biomarkers will have the greatest utility in predicting antidepressant responses. In the future, researchers should integrate multiple types of information, including genetic, epigenetic, and gene expression data, to identify the most meaningful panel of biomarkers.
In conclusion, this preliminary study showed that DMPs may be associated with an early response to antidepressants in patients with PD. However, the results of our study should be considered in light of the following limitations. Since this is the first study to investigate genome-wide DNA methylation and gene expression in the prediction of early response to antidepressants in PD, it would be valuable to replicate our findings in a larger cohort. Furthermore, these DMPs must be verified using larger sample sizes. Lastly, it is necessary to test the association between treatment outcomes and changes in DNA methylation and gene expression from pre- to post-treatment with a long-term follow-up.
Selective serotonin reuptake inhibitors are currently considered the first-line treatment for panic disorder (PD). However, not all patients benefit from the antidepressant therapy.
No genome-wide methylation studies or mRNA sequencing have been conducted to identify early response biomarkers in patients with PD.
To compare genome-wide methylation and gene expression patterns between responsive and non-responsive patients with PD after 4 wk of escitalopram treatment.
Thirty patients with PD were enrolled in this study (responders = 13; non-responders = 17). All patients were assessed using the PD Severity Scale-Chinese version before and after treatment. The Illumina Infinium MethylationEPIC (850k) BeadChip for genome-wide methylation screening and mRNA sequencing was used in all patients with PD.
A total of 701 differentially methylated positions (DMPs) were found between responders and non-responders (|Δβ| ≥ 0.06, q < 0.05), and the hyper- and hypomethylated CpG sites were 511 (72.9%) and 190 (27.1%), respectively. Relative to non-responders, there were 59 differential transcripts, of which 20 were downregulated and 39 were upregulated (q < 0.05).
This preliminary study showed that DMPs might be associated with the treatment response to escitalopram in PD, however, these DMPs need to be verified in large samples.
DNA methylation contributing to antidepressant response will be a unique and promising opportunity to implement personalized medicine in PD treatment.
The authors would like to thank all participants in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sobanski T, Germany; Stoyanov D, Bulgaria S-Editor: Lin C L-Editor: A P-Editor: Chen YX
| 1. | Kessler RC, Chiu WT, Jin R, Ruscio AM, Shear K, Walters EE. The epidemiology of panic attacks, panic disorder, and agoraphobia in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2006;63:415-424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 473] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 2. | Kessler RC, Petukhova M, Sampson NA, Zaslavsky AM, Wittchen H -U. Twelve-month and lifetime prevalence and lifetime morbid risk of anxiety and mood disorders in the United States. Int J Methods Psychiatr Res. 2012;21:169-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2008] [Cited by in RCA: 1811] [Article Influence: 139.3] [Reference Citation Analysis (0)] |
| 3. | de Jonge P, Roest AM, Lim CC, Florescu SE, Bromet EJ, Stein DJ, Harris M, Nakov V, Caldas-de-Almeida JM, Levinson D, Al-Hamzawi AO, Haro JM, Viana MC, Borges G, O'Neill S, de Girolamo G, Demyttenaere K, Gureje O, Iwata N, Lee S, Hu C, Karam A, Moskalewicz J, Kovess-Masfety V, Navarro-Mateu F, Browne MO, Piazza M, Posada-Villa J, Torres Y, Ten Have ML, Kessler RC, Scott KM. Cross-national epidemiology of panic disorder and panic attacks in the world mental health surveys. Depress Anxiety. 2016;33:1155-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 4. | Sung SC, Rush AJ, Earnest A, Lim LEC, Pek MPP, Choi JMF, Ng MPK, Ong MEH. A Brief Interview to Detect Panic Attacks and Panic Disorder in Emergency Department Patients with Cardiopulmonary Complaints. J Psychiatr Pract. 2018;24:32-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Batelaan N, Smit F, de Graaf R, van Balkom A, Vollebergh W, Beekman A. Economic costs of full-blown and subthreshold panic disorder. J Affect Disord. 2007;104:127-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Andrisano C, Chiesa A, Serretti A. Newer antidepressants and panic disorder: a meta-analysis. Int Clin Psychopharmacol. 2013;28:33-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Chawla N, Anothaisintawee T, Charoenrungrueangchai K, Thaipisuttikul P, McKay GJ, Attia J, Thakkinstian A. Drug treatment for panic disorder with or without agoraphobia: systematic review and network meta-analysis of randomised controlled trials. BMJ. 2022;376:e066084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Bandelow B, Behnke K, Lenoir S, Hendriks GJ, Alkin T, Goebel C, Clary CM. Sertraline versus paroxetine in the treatment of panic disorder: an acute, double-blind noninferiority comparison. J Clin Psychiatry. 2004;65:405-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Kamijima K, Kuboki T, Kumano H, Burt T, Cohen G, Arano I, Hamasaki T. A placebo-controlled, randomized withdrawal study of sertraline for panic disorder in Japan. Int Clin Psychopharmacol. 2005;20:265-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A, Wendland JR, Lewis CM, McGuffin P, Uher R. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73:679-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 162] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 11. | Yevtushenko OO, Oros MM, Reynolds GP. Early response to selective serotonin reuptake inhibitors in panic disorder is associated with a functional 5-HT1A receptor gene polymorphism. J Affect Disord. 2010;123:308-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | He Q, Yuan Z, Liu Y, Zhang J, Yan H, Shen L, Luo X, Zhang Y. Correlation between cytochrome P450 2C19 genetic polymorphism and treatment response to escitalopram in panic disorder. Pharmacogenet Genomics. 2017;27:279-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Zou Z, Huang Y, Wang J, Min W, Zhou B. The association between serotonin-related gene polymorphisms and susceptibility and early sertraline response in patients with panic disorder. BMC Psychiatry. 2020;20:388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Yang J, Li S, Lv H, Wang W, Zhang J, Chu L, Zhang Y. CREB1 and BDNF gene polymorphisms are associated with early treatment response to escitalopram in panic disorder. J Affect Disord. 2021;278:536-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Hack LM, Fries GR, Eyre HA, Bousman CA, Singh AB, Quevedo J, John VP, Baune BT, Dunlop BW. Moving pharmacoepigenetics tools for depression toward clinical use. J Affect Disord. 2019;249:336-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Lisoway AJ, Zai CC, Tiwari AK, Kennedy JL. DNA methylation and clinical response to antidepressant medication in major depressive disorder: A review and recommendations. Neurosci Lett. 2018;669:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Morgan LZ, Rollins B, Sequeira A, Byerley W, DeLisi LE, Schatzberg AF, Barchas JD, Myers RM, Watson SJ, Akil H, Bunney WE Jr, Vawter MP. Quantitative Trait Locus and Brain Expression of HLA-DPA1 Offers Evidence of Shared Immune Alterations in Psychiatric Disorders. Microarrays (Basel). 2016;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4367] [Cited by in RCA: 4245] [Article Influence: 193.0] [Reference Citation Analysis (0)] |
| 19. | First MB, Spitzer RL, Gibbon M, William, JBW. Structured Clinical Interview For DSM-IV Axis I Disorders (SCID-I), Clinical Version. Washington DC: American Psychiatric Press Inc, 1997. |
| 20. | Shear MK, Brown TA, Barlow DH, Money R, Sholomskas DE, Woods SW, Gorman JM, Papp LA. Multicenter collaborative panic disorder severity scale. Am J Psychiatry. 1997;154:1571-1575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 605] [Cited by in RCA: 623] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 21. | Xiong HF, Li ZJ, Han HY, Xu ZY, Guo ZJ, Yao SM, Guo M, Jiang CQ. A Reliability Study of the Chinese Version of the Panic Disorder Severity Scale. Zhonghua Jingshenke Zazhi. 2012;45:285-288. [DOI] [Full Text] |
| 22. | Furukawa TA, Katherine Shear M, Barlow DH, Gorman JM, Woods SW, Money R, Etschel E, Engel RR, Leucht S. Evidence-based guidelines for interpretation of the Panic Disorder Severity Scale. Depress Anxiety. 2009;26:922-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 970] [Cited by in RCA: 1207] [Article Influence: 92.8] [Reference Citation Analysis (0)] |
| 24. | Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4620] [Cited by in RCA: 5490] [Article Influence: 288.9] [Reference Citation Analysis (0)] |
| 25. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 29136] [Article Influence: 1821.0] [Reference Citation Analysis (0)] |
| 26. | Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2111] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 27. | Webb LM, Phillips KE, Ho MC, Veldic M, Blacker CJ. The Relationship between DNA Methylation and Antidepressant Medications: A Systematic Review. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 28. | Ju C, Fiori LM, Belzeaux R, Theroux JF, Chen GG, Aouabed Z, Blier P, Farzan F, Frey BN, Giacobbe P, Lam RW, Leri F, MacQueen GM, Milev R, Müller DJ, Parikh SV, Rotzinger S, Soares CN, Uher R, Li Q, Foster JA, Kennedy SH, Turecki G. Integrated genome-wide methylation and expression analyses reveal functional predictors of response to antidepressants. Transl Psychiatry. 2019;9:254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Zou Z, Zhang Y, Huang Y, Wang J, Min W, Xiang M, Zhou B, Li T. Integrated genome-wide methylation and expression analyses provide predictors of diagnosis and early response to antidepressant in panic disorder. J Affect Disord. 2023;322:146-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Stojanov D, Korf J, de Jonge P, Popov G. The possibility of evidence-based psychiatry: depression as a case. Clin Epigenetics. 2011;2:7-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Hughes AL, Nei M. Evolutionary relationships of class II major-histocompatibility-complex genes in mammals. Mol Biol Evol. 1990;7:491-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 30] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Yamamoto F, Suzuki S, Mizutani A, Shigenari A, Ito S, Kametani Y, Kato S, Fernandez-Viña M, Murata M, Morishima S, Morishima Y, Tanaka M, Kulski JK, Bahram S, Shiina T. Capturing Differential Allele-Level Expression and Genotypes of All Classical HLA Loci and Haplotypes by a New Capture RNA-Seq Method. Front Immunol. 2020;11:941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Petersen CL, Chen JQ, Salas LA, Christensen BC. Altered immune phenotype and DNA methylation in panic disorder. Clin Epigenetics. 2020;12:177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 34. | Quagliato LA, Nardi AE. Cytokine profile in drug-naïve panic disorder patients. Transl Psychiatry. 2022;12:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Ho PS, Yeh YW, Huang SY, Liang CS. A shift toward T helper 2 responses and an increase in modulators of innate immunity in depressed patients treated with escitalopram. Psychoneuroendocrinology. 2015;53:246-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Avitsur R, Paley S, Franko M, Wolff N, Eyal N, Doron R. Escitalopram or novel herbal treatments differentially alter cytokine and behavioral responses to immune challenge. J Neuroimmunol. 2017;309:111-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Munzer A, Sack U, Mergl R, Schönherr J, Petersein C, Bartsch S, Kirkby KC, Bauer K, Himmerich H. Impact of antidepressants on cytokine production of depressed patients in vitro. Toxins (Basel). 2013;5:2227-2240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 38. | Pákáski M, Bjelik A, Hugyecz M, Kása P, Janka Z, Kálmán J. Imipramine and citalopram facilitate amyloid precursor protein secretion in vitro. Neurochem Int. 2005;47:190-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Reddy AP, Yin X, Sawant N, Reddy PH. Protective effects of antidepressant citalopram against abnormal APP processing and amyloid beta-induced mitochondrial dynamics, biogenesis, mitophagy and synaptic toxicities in Alzheimer's disease. Hum Mol Genet. 2021;30:847-864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Imai H, Shoji H, Ogata M, Kagawa Y, Owada Y, Miyakawa T, Sakimura K, Terashima T, Katsuyama Y. Dorsal Forebrain-Specific Deficiency of Reelin-Dab1 Signal Causes Behavioral Abnormalities Related to Psychiatric Disorders. Cereb Cortex. 2017;27:3485-3501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Fiori LM, Lin R, Ju C, Belzeaux R, Turecki G. Using Epigenetic Tools to Investigate Antidepressant Response. Prog Mol Biol Transl Sci. 2018;158:255-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |