Published online Feb 19, 2023. doi: 10.5498/wjp.v13.i2.36
Peer-review started: October 9, 2022
First decision: November 27, 2022
Revised: December 6, 2022
Accepted: January 23, 2023
Article in press: January 23, 2023
Published online: February 19, 2023
Processing time: 130 Days and 21.4 Hours
Major depressive disorder (MDD) is a common and serious mental illness. Many novel genes in MDD have been characterized by high-throughput methods such as microarrays or sequencing. Recently, noncoding RNAs (ncRNAs) were sug
To investigate the RNA expression datasets downloaded from a public database and construct a network based on differentially expressed long noncoding RNA (lncRNAs), microRNAs (miRNAs), and mRNAs between MDD and controls.
Gene expression data were searched in NCBI Gene Expression Omnibus using the search term “major depressive disorder.” Six array datasets from humans were related to the search term: GSE19738, GSE32280, GSE38206, GSE52790, GSE76826, and GSE81152. These datasets were processed for initial assessment and subjected to quality control and differential expression analysis. Differentially expressed lncRNAs, miRNAs, and mRNAs were determined, Gene Ontology and Kyoto En
After analysis, 3 miRNAs, 12 lncRNAs, and 33 mRNAs were identified in the competing en
The present study presents candidate ncRNAs involved in the neurogenesis and neuroplasticity pathways related to MDD.
Core Tip: Competing endogenous RNAs (ceRNAs) are novel regulatory molecules involved in a wide range of biological processes and diseases. This study explored the potential ceRNA networks (ceRNETs) involved in the pathogenesis of major depressive disorder (MDD) using bioinformatics data mining. A ceRNET comprising 3 miRNAs, 12 lncRNAs, and 33 mRNAs was constructed based on two public datasets obtained from the Gene Expression Omnibus database. Elucidating the correlation of the ceRNET with MDD opens new avenues to discover specific diagnostic biomarkers for MDD and expands our knowledge about this disease.
- Citation: Zou ZL, Ye Y, Zhou B, Zhang Y. Identification and characterization of noncoding RNAs-associated competing endogenous RNA networks in major depressive disorder. World J Psychiatry 2023; 13(2): 36-49
- URL: https://www.wjgnet.com/2220-3206/full/v13/i2/36.htm
- DOI: https://dx.doi.org/10.5498/wjp.v13.i2.36
Major depressive disorder (MDD), characterized by persistent and intense feelings of sadness for extended periods, is one of the most common causes of morbidity and mortality worldwide[1], yearly affecting approximately 5% of adults worldwide[2]. The exact cause of MDD remains unknown; however, the occurrence of MDD is widely believed to involve crosstalk between genetic, environmental, social, and developmental vulnerabilities and resilience factors[2,3]. Numerous risk factors for MDD have been identified from three major perspectives: medical, social, and substance[4,5]. Available treatment options primarily include pharmacotherapy, psychotherapy, and lifestyle changes. However, prevention and treatment of this disease remain difficult because of varying presentations, unpre-dictable course and prognosis, and variable responses to treatment[6]. Therefore, further studies are needed to determine the biological mechanisms underlying MDD to develop better patient therapies.
The increasing popularity of high-throughput microarray technologies has facilitated the identification of genome variations in MDD, improving the understanding of its pathogenesis and deve-lopment, and revealing promising biomarkers. Gao et al[7] identified 241 differentially expressed genes (DEGs) in the hippocampus (hip), 218 DEGs in the prefrontal cortex (pfc), and 327 DEGs in the striatum (str) from patients with MDD. These DEGs were enriched in glycan biosynthesis, RIG-I-like receptor signaling, and pyrimidine metabolism pathways, which significantly contribute to MDD pathogenesis. Additionally, the DEGs AR, PTK2, IRAK1, IL12A, GALNT12, GALNT2, CD19, and PTDSS2 were identified as novel therapeutic targets[7,8]. Moreover, Segman et al[9] found 73 DEGs between patients with postpartum major depression and controls using peripheral blood cells; the immune response, transcriptional effects on cell proliferation, and DNA replication and repair were significantly enriched in these patients. Although novel genes have been discovered and well-studied, genetic predisposition to MDD only partly explains the occurrence of this illness. With the development of epigenetic theory and technology, non-coding RNAs (ncRNAs) have been identified to help explain the interaction between genetics and the environment. Li et al[10] found an inverse relationship between serum brain-derived neurotrophic factor (BDNF) levels and miR-132/miR-182 levels in depression, supporting the notion that miR-182 is a putative regulatory microRNA (miRNA) of BDNF. Zhou et al[11] found that miRNAs act as key regulators of synaptic plasticity in MDD pathogenesis. Liu et al[12] detected four long non-coding ribonucleic acids (lncRNAs) related to the expression of mRNAs associated with MDD. Although ncRNAs have been proposed to play important roles in MDD, the regulatory network among RNA species remains unclear.
The competing endogenous RNA (ceRNA) network proposed by Salmena et al[13] has been verified in an increasing number of studies. This complicated post-transcriptional regulatory network plays a critical role in the progression and pathogenesis of various illnesses, including cancer. However, to date, no ceRNA network has been proposed for MDD. Here, we investigated RNA expression datasets downloaded from the Gene Expression Omnibus (GEO) database and constructed a network based on differentially expressed lncRNAs, miRNAs, and mRNAs between MDD and controls. This network was used to mine for functional ncRNAs that may contribute to the epigenetic mechanism underlying MDD pathogenesis and may be useful as potential therapeutic targets and biomarkers for MDD.
Gene expression data were searched in NCBI GEO using the search term “major depressive disorder”. Six array datasets from humans were related to the search term: GSE19738 (expression profiling by array, n = 42; MDD, 21; control, 21), GSE32280 (expression profiling by array, n = 24; MDD, 8; subsyndromal symptomatic depression, SSD, 8; control, 8), GSE38206 (expression profiling by array, n = 18; MDD, 9; control, 9), GSE52790 (expression profiling by array and ncRNA profiling by array, n = 22; MDD, 10; control, 12), GSE76826 (expression profiling by array, n = 22; MDD, 10; control, 12), and GSE81152 (ncRNA profiling by array, n = 50; MDD, 31; control, 19). These datasets were processed for initial assessment; if they met the requirements, they were subjected to quality control and differential expression analysis.
The differentially expressed mRNAs, lncRNAs, or miRNAs were determined based on the threshold criteria of |log2 (fold-change)| ≥ 1 and an adjusted P value < 0.05 using the R limma package (version 3.40.6). The differentially expressed lncRNAs (DE_lncRNAs) obtained from GSE76826 were entered into the miRcode database (http://www.mircode.org/) that provides an integrated, searchable map of putative target sites of miRNAs, to predict the interactions of DE_lncRNAs and DE_miRNAs. Only DE_lncRNAs for which miRNAs were successfully predicted were selected for ceRNA network construction. The miRNAs obtained were overlapped with differentially expressed miRNAs obtained from GSE81152, and the overlapping miRNAs were identified as a differentially expressed miRNA (DE_miRNA). The name of the DE_miRNA was entered into the miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/vert_72/) databases, both of which are online databases used to predict miRNA targets. The mRNAs obtained above were overlapped with differentially expressed RNAs identified from GSE76826. The DE_lncRNAs, DE_miRNAs, and DE_mRNAs obtained in the manner described above were used for ceRNA network construction.
Cytoscape 3.4.0 was used to visualize the DE_lncRNA/DE_miRNA/DE_mRNA network construction.
To help identify the potential biological functions of these genes in the MDD ceRNA network, Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway enrichment analyses were performed using the clusterProfiler package in R. GO terms have three categories; biological process, cellular compartment, and molecular function. P ≤ 0.05 indicated a significant difference in GO terms and KEGG pathways.
The protein interaction (PPI) network was constructed to evaluate the interactions among genes. The DE_mRNAs were selected to construct a PPI network based on Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) ver.11.0 (https://string-db.org/). In the PPI network, a node represents a gene; the undirected link between two nodes is an edge, representing the interaction between two genes, and the degree of a node corresponds to the number of interactions of a gene with other genes in the network.
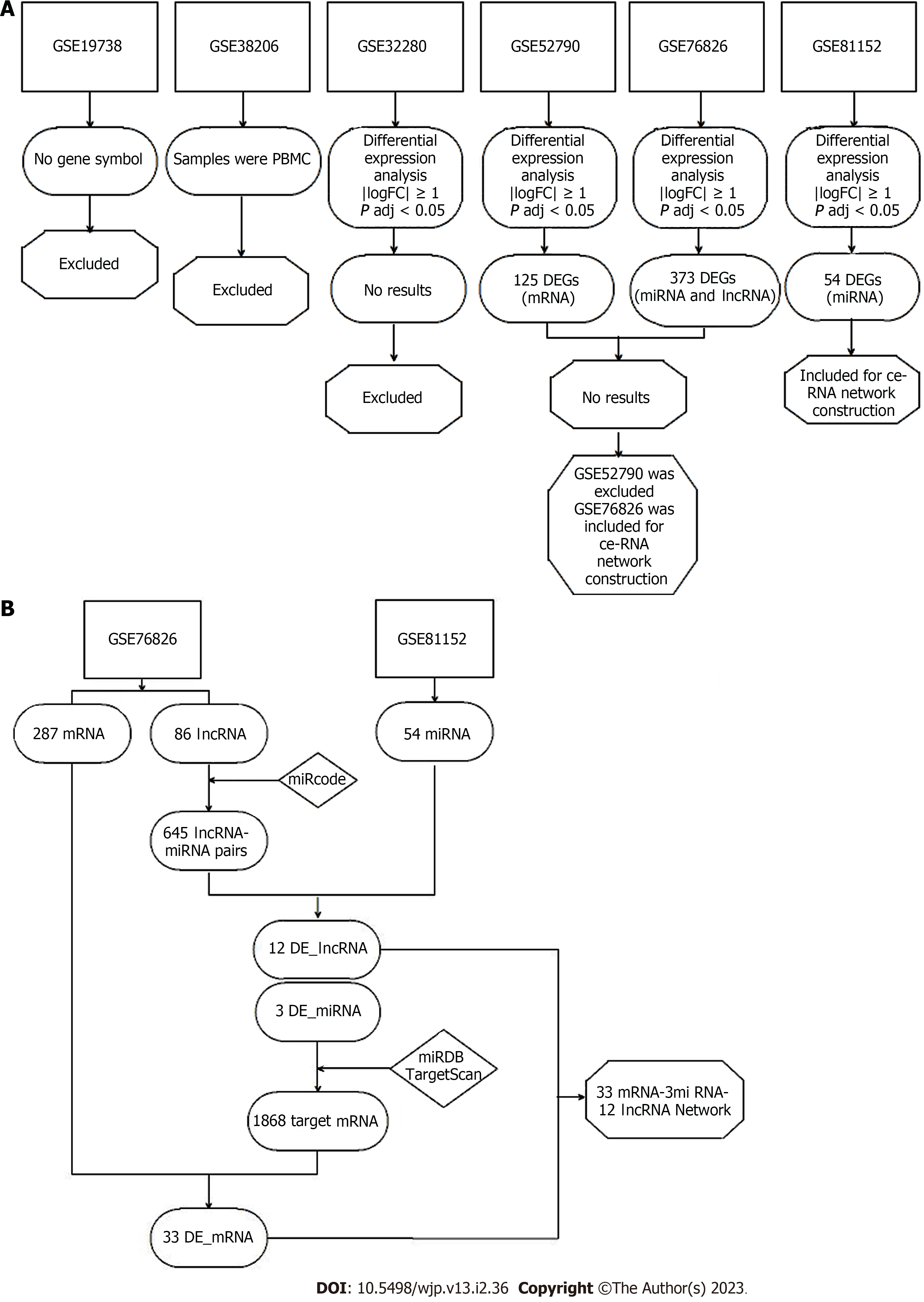
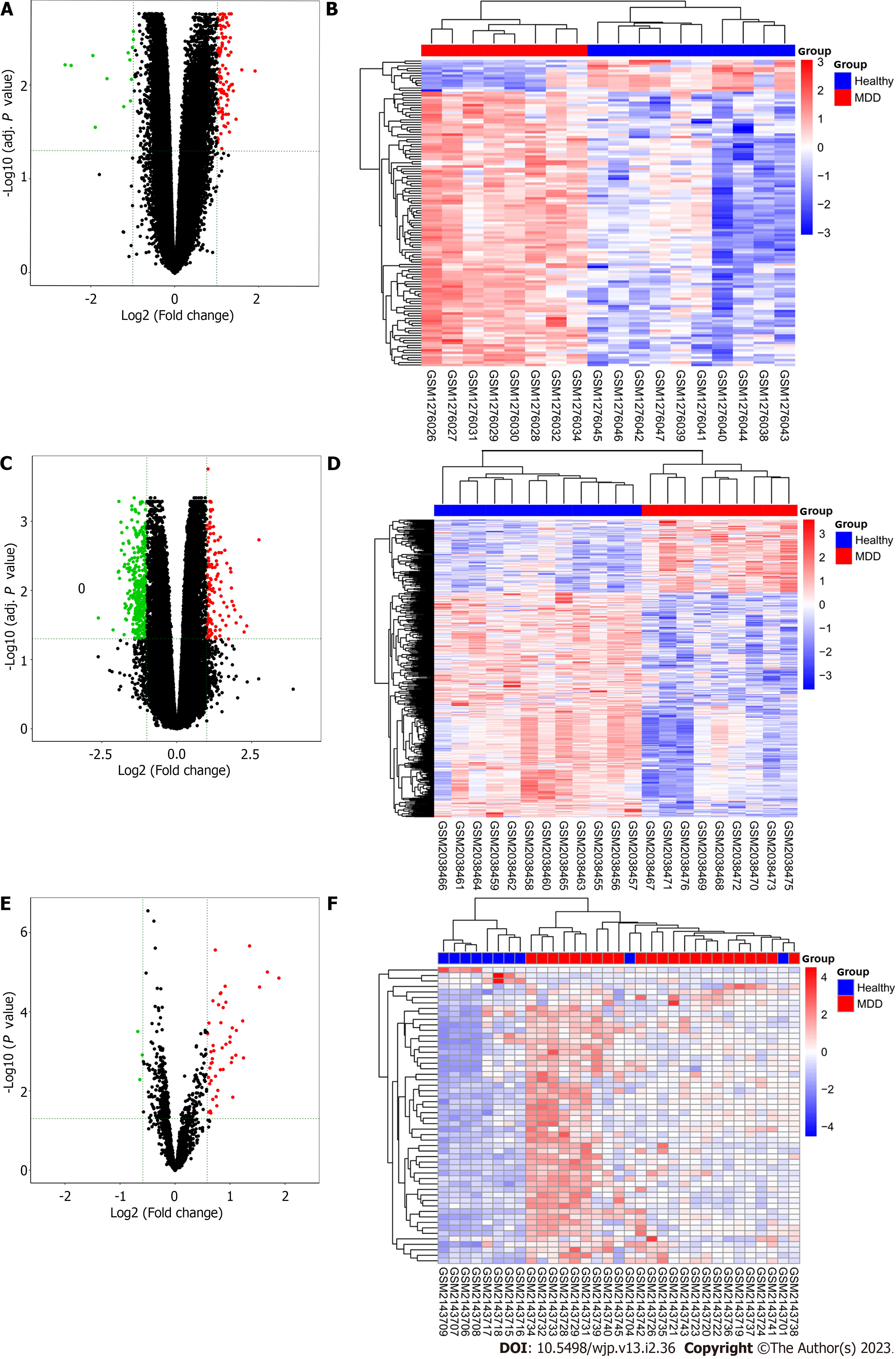
The workflow of the analysis is shown in Figure 1. Because the GSE19738 dataset lacks documents to annotate gene symbols and the GSE32280 dataset uses peripheral blood mononuclear cells rather than whole blood, both these datasets were excluded from analysis at the first stage. Next, sample-level quality control was performed using principal component analysis to ensure that the cases and controls were well-separated and to identify sample outliers. After removing the outliers, there were 10 controls and 8 MDD cases in GSE52790, 12 controls and 9 MDD cases in GSE76826, and 10 controls and 23 MDD cases in GSE81152. The four remaining datasets, GSE32280, GSE52790, GSE76826, and GSE81152, were utilized to identify differentially expressed mRNAs, lncRNAs, and miRNAs using the limma R package with the threshold set to an adjusted P value < 0.05 and |log2 (fold-change)| ≥ 1. A hierarchical cluster heatmap showing the expression patterns of the DEGs between MDD and controls is shown in Figure 2. As no DEGs were found in GSE32280, this dataset was excluded from subsequent analysis. A total of 125, 373, and 54 DEGs were identified in the GSE52790, GSE76826, and GSE81152 datasets, respectively. There were 112 upregulated and 13 downregulated genes among the 125 DEGs in GSE52790, 109 upregulated genes (including 89 mRNAs and 10 lncRNAs) and 264 downregulated genes (including 198 mRNAs and 66 LncRNAs) in the 373 DEGs of GSE76826, and 51 upregulated and 3 downregulated genes in the 54 DEGs of GSE81152.
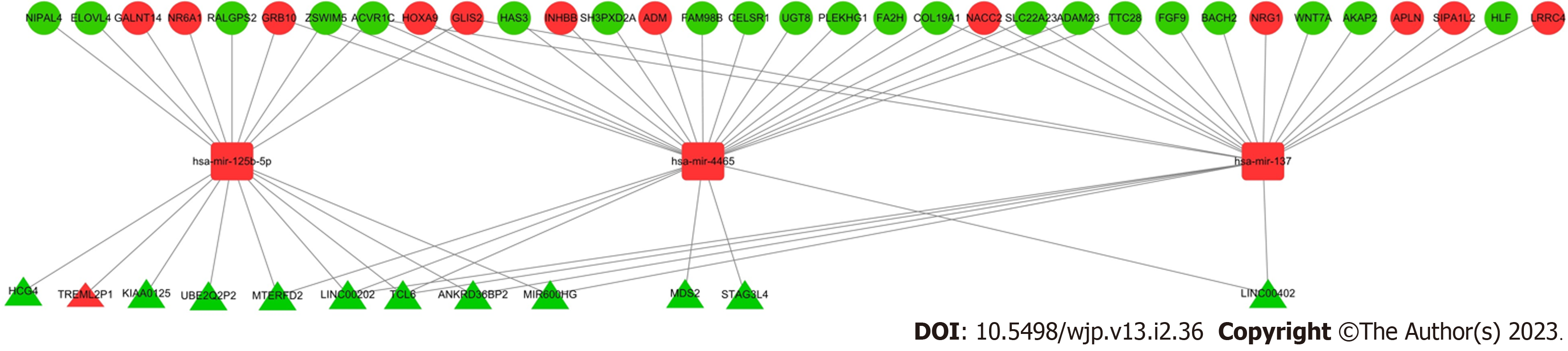
To better understand the effect of lncRNAs on mRNAs mediated by combinations with miRNAs in MDD, we constructed a ceRNA network based on the GSE76826 and GSE81152 data. Eighty-six differentially expressed lncRNAs from GSE76826 were retrieved and entered into the miRcode database to predict the potential interacting miRNAs. Eighty-four miRNAs (645 LncRNA-miRNA pairs) were identified in the miRcode database. Next, we overlapped these 84 miRNAs to the differentially expressed miRNAs identified in GSE81152, generating 3 shared miRNAs (the corresponding lncRNAs were reduced to 12) (Table 1). Thereafter, we entered these 3 miRNAs into the miRDB and TargetScan databases to identify their potential target mRNAs and combined the results from these two databases to achieve high true-positive coverage. A total of 1,868 mRNAs was generated from the two databases; when the 287 differentially expressed mRNAs identified from GSE76826 were overlapped, 33 shared mRNAs were identified (Table 2). Finally, we assembled all identified ceRNA pairs to generate an mRNA-miRNA-lncRNA network in MDD (Figure 3).
| lncRNA | miRNA |
| STAG3L4 | hsa-miR-4465 |
| MTERFD2 | hsa-miR-4465 |
| TCL6 | hsa-miR-4465 |
| MDS2 | hsa-miR-4465 |
| LINC00202 | hsa-miR-4465 |
| LINC00402 | hsa-miR-4465 |
| TCL6 | hsa-miR-137 |
| ANKRD36BP2 | hsa-miR-137 |
| LINC00202 | hsa-miR-137 |
| LINC00402 | hsa-miR-137 |
| MIR600HG | hsa-miR-137 |
| MTERFD2 | hsa-miR-125b-5p |
| HCG4 | hsa-miR-125b-5p |
| TCL6 | hsa-miR-125b-5p |
| UBE2Q2P2 | hsa-miR-125b-5p |
| TREML2P1 | hsa-miR-125b-5p |
| KIAA0125 | hsa-miR-125b-5p |
| ANKRD36BP2 | hsa-miR-125b-5p |
| LINC00202 | hsa-miR-125b-5p |
| MIR600HG | hsa-miR-125b-5p |
| miRNA | mRNA |
| hsa-miR-4465 | FAM98B |
| hsa-miR-4465 | ADM |
| hsa-miR-4465 | INHBB |
| hsa-miR-4465 | FA2H |
| hsa-miR-4465 | HOXA9 |
| hsa-miR-4465 | GRB10 |
| hsa-miR-4465 | UGT8 |
| hsa-miR-4465 | ACVR1C |
| hsa-miR-4465 | COL19A1 |
| hsa-miR-4465 | TTC28 |
| hsa-miR-4465 | HAS3 |
| hsa-miR-4465 | SLC22A23 |
| hsa-miR-4465 | ADAM23 |
| hsa-miR-4465 | NACC2 |
| hsa-miR-4465 | SH3PXD2A |
| hsa-miR-4465 | CELSR1 |
| hsa-miR-4465 | PLEKHG1 |
| hsa-miR-4465 | ZSWIM5 |
| hsa-miR-137 | AKAP2 |
| hsa-miR-137 | HLF |
| hsa-miR-137 | FGF9 |
| hsa-miR-137 | GLIS2 |
| hsa-miR-137 | SIPA1L2 |
| hsa-miR-137 | WNT7A |
| hsa-miR-137 | NRG1 |
| hsa-miR-137 | ADAM23 |
| hsa-miR-137 | LRRC4 |
| hsa-miR-137 | SLC22A23 |
| hsa-miR-137 | COL19A1 |
| hsa-miR-137 | TTC28 |
| hsa-miR-137 | APLN |
| hsa-miR-137 | NACC2 |
| hsa-miR-137 | BACH2 |
| hsa-miR-137 | ACVR1C |
| hsa-miR-125b-5p | NIPAL4 |
| hsa-miR-125b-5p | NR6A1 |
| hsa-miR-125b-5p | ZSWIM5 |
| hsa-miR-125b-5p | GALNT14 |
| hsa-miR-125b-5p | GRB10 |
| hsa-miR-125b-5p | ELOVL4 |
| hsa-miR-125b-5p | ACVR1C |
| hsa-miR-125b-5p | RALGPS2 |
| hsa-miR-125b-5p | GLIS2 |
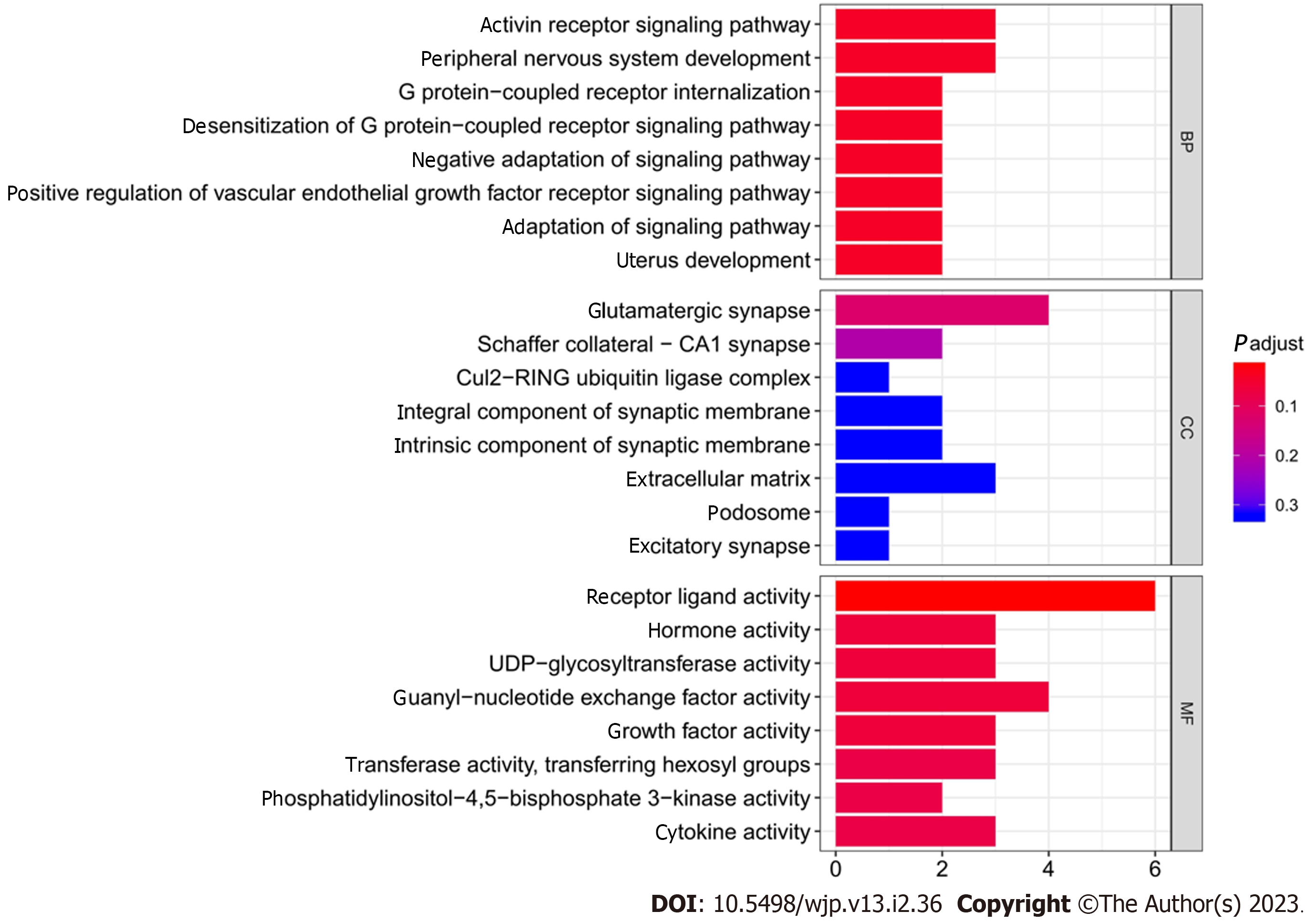
Enrichment analysis indicated that the 33 differentially expressed mRNAs were significantly associated with the GO terms for 887 biological process, 105 molecular function, and 55 cell component terms. For the KEGG pathways, the only significant pathway was the mammalian target of rapamycin (mTOR) signaling pathway. Table 3 and Figure 4 list the top 10 most significant biological processes, molecular functions, and cellular components from the GO terms.
| Category | Term description | Count of genes | P value |
| Biology process | Activin receptor signaling pathway | 3 | 5.54E-05 |
| Biology process | Peripheral nervous system development | 3 | 0.000292 |
| Biology process | G protein-coupled receptor internalization | 2 | 0.0003 |
| Biology process | Desensitization of G protein-coupled receptor signaling pathway | 2 | 0.000487 |
| Biology process | Negative adaptation of signaling pathway | 2 | 0.000487 |
| Biology process | Positive regulation of vascular endothelial growth factor receptor signaling pathway | 2 | 0.000487 |
| Biology process | Adaptation of signaling pathway | 2 | 0.000597 |
| Biology process | Uterus development | 2 | 0.000656 |
| Biology process | Sphingolipid biosynthetic process | 3 | 0.000679 |
| Biology process | Reproductive structure development | 5 | 0.00078 |
| Cellular component | Glutamatergic synapse | 4 | 0.002395 |
| Cellular component | Schaffer collateral-CA1 synapse | 2 | 0.00786 |
| Cellular component | Cul2-RING ubiquitin ligase complex | 1 | 0.024149 |
| Cellular component | Integral component of synaptic membrane | 2 | 0.025933 |
| Cellular component | Intrinsic component of synaptic membrane | 2 | 0.029786 |
| Cellular component | Extracellular matrix | 3 | 0.044750 |
| Cellular component | Podosome | 1 | 0.047732 |
| Cellular component | Excitatory synapse | 1 | 0.075303 |
| Cellular component | Integral component of postsynaptic density membrane | 1 | 0.079823 |
| Cellular component | Intrinsic component of postsynaptic density membrane | 1 | 0.084321 |
| Molecular function | Receptor ligand activity | 6 | 0.000191 |
| Molecular function | Hormone activity | 3 | 0.001352 |
| Molecular function | UDP-glycosyltransferase activity | 3 | 0.002314 |
| Molecular function | Guanyl-nucleotide exchange factor activity | 4 | 0.002772 |
| Molecular function | Growth factor activity | 3 | 0.003326 |
| Molecular function | Transferase activity, transferring hexosyl groups | 3 | 0.006074 |
| Molecular function | Phosphatidylinositol-4,5-bisphosphate 3-kinase activity | 2 | 0.006746 |
| Molecular function | Cytokine activity | 3 | 0.007189 |
| Molecular function | Phosphatidylinositol bisphosphate kinase activity | 2 | 0.007334 |
| Molecular function | Transcription factor activity, RNA polymerase II proximal promoter sequence-specific DNA binding | 4 | 0.008356 |
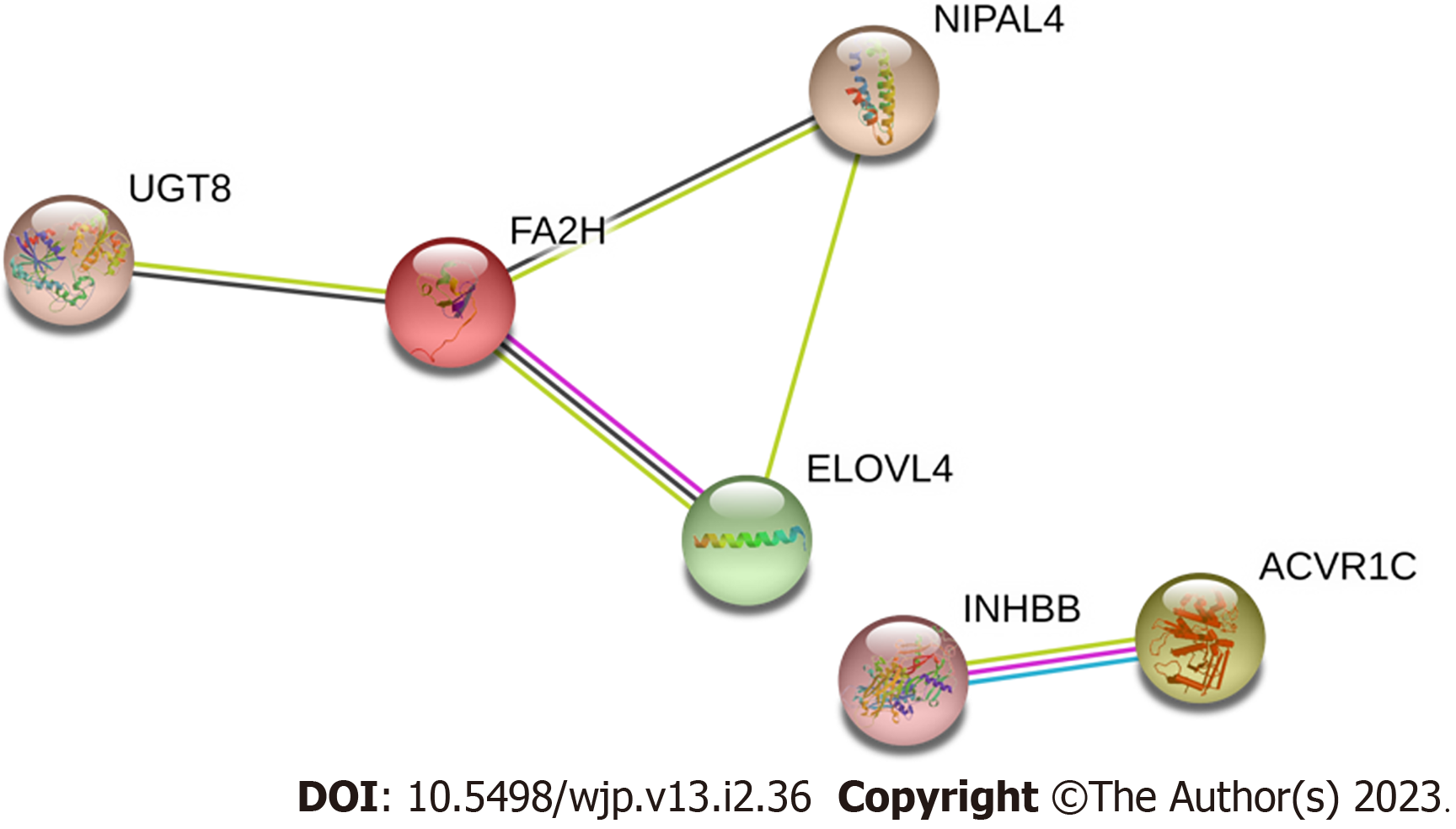
A total of 33 DEG mRNAs were entered into the STRING database to build a PPI network. Gene nodes with the highest W values in the network were ACVR1C, ELOVL4, fatty acid 2-hydroxylase (FA2H), INHBB, NIPAL4, and UGT8 (Figure 5).
Considerable progress has been made in understanding the pathophysiology of MDD; however, no single model or mechanism can fully explain all aspects of the disease[6]. MDD is believed to involve the following: Reduced levels of monoamines, changes in the hypothalamic-pituitary-adrenal axis, inflammation, limited neurogenesis, changes in brain structure and function, heredity factors, environmental milieu, and epigenetics[6]. Epigenetics, a field focusing on gene-environment interactions, has gained attention over the past decades. The recent application of microarray-based genome-wide expression analysis has enabled the identification of MDD-associated genes, including novel ncRNAs. Although numerous studies have demonstrated that ncRNAs play important roles in MDD pathogenesis, the mechanisms of how these genes regulate MDD have not been well-characterized[14,15]. Thus, we performed ceRNA network analysis by data mining to determine the potential regulatory mechanisms of MDD.
Multiple GO terms were identified in enrichment analysis, reflecting the complexity of the disorder. We observed that protein-coding genes in the network were highly correlated with cellular growth and developmental differentiation, including the activin receptor signaling pathway, peripheral nervous system development, G protein-coupled receptor internalization, receptor-ligand activity, hormone activity, growth factor activity, and cellular growth. The term “signaling by activins” in our results is consistent with previous studies showing that activin can modulate depression and anxiety-related behavior[16-21]. Activins are members of the transforming growth factor-β superfamily that are expressed in various tissues, including neuronal cells, and are involved in proliferation, differentiation, metabolism, homeostasis, apoptosis, immune response, and tissue repair. Postnatal neurogenesis, which leads to the production of new neurons in the adult brain, can influence the replacement of damaged neurons, stress responses, memory formation, and depression, among others[22]. Previous studies have reported that the activin activity in the adult forebrain influences locomotor activity, anxiety-related behavior, and hippocampal neurogenesis, which is also associated with age-related cognitive decline[23]. Additionally, consistent with our results, utilizing a genome-wide association study, Smeeth et al[23] suggested a complex relationship among reproductive hormones, hippocampal neurogenesis, and depression. Based on transcriptome array results analysis of Chinese patients with MDD, Liu et al[12] reported the enrichment of differentially expressed coding genes in the signal transduction pathway and basic metabolic process associated with neurodevelopmental disease[12,13]. Our KEGG results also showed that the mTOR signaling pathway is a part of the regulatory network of MDD. This agrees with previous studies which showed that the mTOR signaling pathway, which senses and integrates diverse extracellular stimuli to promote cellular growth or limit catabolic processes, contributes to normal neuronal growth and is associated with neurogenesis[24]. Notably, the mTOR signaling pathway has become an important target for treating depression using drugs such as ketamine[25,26].
The use of ncRNAs as biomarkers of psychological disorders is gaining momentum. In the ceRNA network developed in this study, there were 12 lncRNAs, 3 miRNAs, and 33 mRNAs specific to MDD. Consistent with our results, hsa-mir-137, as a brain-enriched miRNA, was earlier confirmed as a gene related to MDD, bipolar disorder, schizophrenia, and Parkinson’s disease susceptibility[27-30], whose underlying mechanisms regulate synaptic plasticity. Moreover, Zhao et al[31] detected significantly lower hsa-miR-137 Levels in the brain and peripheral blood in post-stroke depression rats, and Kim et al[32] found that it can be used as a diagnostic marker for methamphetamine withdrawal syndrome. Similarly, hsa-miR-125b-5p in the peripheral blood is highly expressed in Alzheimer’s disease compared with that in controls.
In addition to genetic components, different epigenetic mechanisms play an important role in the occurrence and development of MDD. A previous study revealed that the non-protein-coding RNA repressor of NFAT, human accelerated region 1, transcribed antisense of rheelin, and B-secretase-1 are associated with cognitive function, potentially contributing to MDD, and the lncRNA BDNF-AS was reportedly related to synaptic plasticity and potentially associated with MDD[33-36]. In a recent study, Abedpoor and colleagues reported that the lncRNA network could play an indispensable role in regulating depression-like behaviors in mice[37]. Furthermore, two studies reported lower RMRP expression in a mouse model of depression and MDD relative to normal subject samples[14]. In another study, Issler et al[38] observed that LINC00473 was downregulated in the prefrontal cortex of depressed females. Collectively, these results suggest that lncRNAs are potential diagnostic and prognostic biomarkers. Our results revealed 12 different lncRNAs involved in the MDD ceRNA network, suggesting the existence of a regulatory network in MDD.
In the present study, FA2H was highlighted in the PPI network. FA2H encoded by FA2H is highly expressed in the human brain and is involved in the formation of 2-hydroxy galactosylceramides and 2-hydroxy sulfatides in myelin[39,40]. The FA2H gene appears to be related to lipid metabolism and is one of the 10 candidate genes identified in an inherited neurologic disorder known as neurodegeneration with brain iron accumulation and is also involved in hereditary spastic paraplegia SPG35 and leukodystrophy[41,42]. Mutations in FA2H have been identified in autism spectrum disorders[43]. The function of FA2H does not appear to be limited to the brain since elevated FA2H expression has also been found in hepatocellular carcinoma and lung adenocarcinoma involving UGT8[44,45]. However, there have been no reports of FA2H in MDD and thus, this is the first study to suggest that FA2H is related to MDD. Considering its role in myelin formation, this gene may contribute to MDD patho-genesis via impacting neurological development and modulating signal transduction.
Elongation of very long-chain fatty acids-4 (ELOVL4) was another protein of interest identified in the PPI. This protein is essential for the synthesis of very long-chain polyunsaturated (VLC-PUFA) and saturated fatty acids (VLC-SFA) of chain lengths greater than 26 carbons[46]. The VLC-PUFAs play an important role in maintaining neural tissue homeostasis. Studies have suggested that dysregulated ELOVL4 expression may be involved in the lipid alterations observed in neuroblastoma, and mutations in the ELOVL4 gene could cause several distinct neurodegenerative diseases, including stargardt-like macular dystrophy, spinocerebellar ataxia 34, and a neuro-ichthyotic syndrome with severe seizures and spasticity[47,48]. However, like FA2H, none of the studies so far have linked ELOVL4 and MDD, and therefore, future validation benchmark experiments are needed.
There were some limitations to our study. The potentially important genes associated with MDD lack gene expression and functional validation, and hence, further in vivo and in vitro studies are required. Furthermore, the two datasets were not from the same platform, which may have resulted in a few deviations. However, the present study provides novel insight into MDD pathogenesis.
To date, MDD diagnosis mainly depends on the patient’s subjective expression, which may be misinterpreted by clinicians. Therefore, identification of a reliable biomarker that can be used to diagnose MDD and guide its treatment is imperative. In conclusion, the present study revealed that hsa-miR-4465, hsa-miR-137, and hsa-miR-125b-5p may be useful genetic biomarkers for MDD as they are potentially involved in the neurogenesis and neuroplasticity pathways in MDD. These results require further validation in future studies.
Major depressive disorder (MDD) is a common and serious mental illness. Many novel genes in MDD have been characterized by high-throughput methods such as microarrays or sequencing. Recently, noncoding RNAs (ncRNAs) were suggested to be involved in the complicated environmental-genetic regulatory network of MDD occurrence.
The interplay among RNA species, including protein-coding RNAs and ncRNAs, in MDD remains unclear.
To investigate the RNA expression datasets and construct a network based on differentially expressed long noncoding RNA (lncRNAs), miRNAs, and mRNAs between MDD and controls through data mining method.
Gene expression data were searched and downloaded from NCBI Gene Expression Omnibus database. Six array datasets from humans were related to the search term: GSE19738, GSE32280, GSE38206, GSE52790, GSE76826, and GSE81152. These datasets were processed for initial assessment and subjected to quality control and differential expression analysis. Differentially expressed lncRNAs, miRNAs, and mRNAs were determined, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed, and protein-protein interaction network was generated. The results were analyzed for their association with MDD.
After analysis, 3 miRNAs, 12 lncRNAs, and 33 mRNAs were identified in the ceRNA network. Two of these miRNAs were earlier shown to be involved in psychiatric disorders, and differentially expressed mRNAs were found to be highly enriched in pathways related to neurogenesis and neuroplasticity as per Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses. The expression of hub gene fatty acid 2-hydroxylase was enriched, and the encoded protein was found to be involved in myelin formation, indicating that neurological development and signal transduction are involved in MDD pathogenesis.
The present study presents candidate ncRNAs involved in the neurogenesis and neuroplasticity pathways related to MDD.
Bioinformatic data mining method can be an cost-effective way to explore potential biomarkers for complicated diseases. However, benchmark experiment is needed in the future to validate the results.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Khosravi M, Iran; Teixeira KN, Brazil S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | Murray CJ, Atkinson C, Bhalla K, Birbeck G, Burstein R, Chou D, Dellavalle R, Danaei G, Ezzati M, Fahimi A, Flaxman D, Foreman, Gabriel S, Gakidou E, Kassebaum N, Khatibzadeh S, Lim S, Lipshultz SE, London S, Lopez, MacIntyre MF, Mokdad AH, Moran A, Moran AE, Mozaffarian D, Murphy T, Naghavi M, Pope C, Roberts T, Salomon J, Schwebel DC, Shahraz S, Sleet DA, Murray, Abraham J, Ali MK, Bartels DH, Chen H, Criqui MH, Dahodwala, Jarlais, Ding EL, Dorsey ER, Ebel BE, Fahami, Flaxman S, Flaxman AD, Gonzalez-Medina D, Grant B, Hagan H, Hoffman H, Leasher JL, Lin J, Lozano R, Lu Y, Mallinger L, McDermott MM, Micha R, Miller TR, Mokdad AA, Narayan KM, Omer SB, Pelizzari PM, Phillips D, Ranganathan D, Rivara FP, Sampson U, Sanman E, Sapkota A, Sharaz S, Shivakoti R, Singh GM, Singh D, Tavakkoli M, Towbin JA, Wilkinson JD, Zabetian A, Alvardo M, Baddour LM, Benjamin EJ, Bolliger I, Carnahan E, Chugh SS, Cohen A, Colson KE, Cooper LT, Couser W, Dabhadkar KC, Dellavalle RP, Dicker D, Duber H, Engell RE, Felson DT, Finucane MM, Fleming T, Forouzanfar MH, Freedman G, Freeman MK, Gillum RF, Gosselin R, Gutierrez HR, Havmoeller R, Jacobsen KH, James SL, Jasrasaria R, Jayarman S, Johns N, Lan Q, Meltzer M, Mensah GA, Michaud C, Mock C, Moffitt TE, Nelson RG, Olives C, Ortblad K, Ostro B, Raju M, Razavi H, Ritz B, Sacco RL, Shibuya K, Silberberg D, Singh JA, Steenland K, Taylor JA, Thurston GD, Vavilala MS, Vos T, Wagner GR, Weinstock MA, Weisskopf MG, Wulf S; U. S. Burden of Disease Collaborators. The state of US health, 1990-2010: burden of diseases, injuries, and risk factors. JAMA. 2013;310:591-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1691] [Cited by in RCA: 1840] [Article Influence: 153.3] [Reference Citation Analysis (1)] |
| 2. | Herrman H, Patel V, Kieling C, Berk M, Buchweitz C, Cuijpers P, Furukawa TA, Kessler RC, Kohrt BA, Maj M, McGorry P, Reynolds CF 3rd, Weissman MM, Chibanda D, Dowrick C, Howard LM, Hoven CW, Knapp M, Mayberg HS, Penninx BWJH, Xiao S, Trivedi M, Uher R, Vijayakumar L, Wolpert M. Time for united action on depression: a Lancet-World Psychiatric Association Commission. Lancet. 2022;399:957-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 504] [Article Influence: 168.0] [Reference Citation Analysis (0)] |
| 3. | Huang X, Luo YL, Mao YS, Ji JL. The link between long noncoding RNAs and depression. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:73-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Pompili M, Innamorati M, Raja M, Falcone I, Ducci G, Angeletti G, Lester D, Girardi P, Tatarelli R, De Pisa E. Suicide risk in depression and bipolar disorder: Do impulsiveness-aggressiveness and pharmacotherapy predict suicidal intent? Neuropsychiatr Dis Treat. 2008;4:247-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | McCarron RM, Shapiro B, Rawles J, Luo J. Depression. Ann Intern Med. 2021;174:ITC65-ITC80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 269] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 6. | Malhi GS, Mann JJ. Depression. Lancet. 2018;392:2299-2312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1255] [Cited by in RCA: 2479] [Article Influence: 354.1] [Reference Citation Analysis (0)] |
| 7. | Gao L, Gao Y, Xu E, Xie J. Microarray Analysis of the Major Depressive Disorder mRNA Profile Data. Psychiatry Investig. 2015;12:388-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Geng R, Huang X. Identification of major depressive disorder disease-related genes and functional pathways based on system dynamic changes of network connectivity. BMC Med Genomics. 2021;14:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Segman RH, Goltser-Dubner T, Weiner I, Canetti L, Galili-Weisstub E, Milwidsky A, Pablov V, Friedman N, Hochner-Celnikier D. Blood mononuclear cell gene expression signature of postpartum depression. Mol Psychiatry. 2010;15:93-100, 2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Li YJ, Xu M, Gao ZH, Wang YQ, Yue Z, Zhang YX, Li XX, Zhang C, Xie SY, Wang PY. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One. 2013;8:e63648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 11. | Zhou L, Zhu Y, Chen W, Tang Y. Emerging role of microRNAs in major depressive disorder and its implication on diagnosis and therapeutic response. J Affect Disord. 2021;286:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, Wang Y, Zhang K. Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One. 2014;9:e93388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 13. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5522] [Article Influence: 394.4] [Reference Citation Analysis (0)] |
| 14. | Seki T, Yamagata H, Uchida S, Chen C, Kobayashi A, Kobayashi M, Harada K, Matsuo K, Watanabe Y, Nakagawa S. Altered expression of long noncoding RNAs in patients with major depressive disorder. J Psychiatr Res. 2019;117:92-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Hosseini E, Bagheri-Hosseinabadi Z, De Toma I, Jafarisani M, Sadeghi I. The importance of long non-coding RNAs in neuropsychiatric disorders. Mol Aspects Med. 2019;70:127-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Link AS, Zheng F, Alzheimer C. Activin Signaling in the Pathogenesis and Therapy of Neuropsychiatric Diseases. Front Mol Neurosci. 2016;9:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Ganea K, Menke A, Schmidt MV, Lucae S, Rammes G, Liebl C, Harbich D, Sterlemann V, Storch C, Uhr M, Holsboer F, Binder EB, Sillaber I, Müller MB. Convergent animal and human evidence suggests the activin/inhibin pathway to be involved in antidepressant response. Transl Psychiatry. 2012;2:e177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Krieglstein K, Zheng F, Unsicker K, Alzheimer C. More than being protective: functional roles for TGF-β/activin signaling pathways at central synapses. Trends Neurosci. 2011;34:421-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 19. | Zheng F, Adelsberger H, Müller MR, Fritschy JM, Werner S, Alzheimer C. Activin tunes GABAergic neurotransmission and modulates anxiety-like behavior. Mol Psychiatry. 2009;14:332-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Dow AL, Russell DS, Duman RS. Regulation of activin mRNA and Smad2 phosphorylation by antidepressant treatment in the rat brain: effects in behavioral models. J Neurosci. 2005;25:4908-4916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Gergues MM, Yohn CN, Bharadia A, Levinstein MR, Samuels BA. Dentate gyrus activin signaling mediates the antidepressant response. Transl Psychiatry. 2021;11:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Ageta H, Tsuchida K. Multifunctional roles of activins in the brain. Vitam Horm. 2011;85:185-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Smeeth DM, Dima D, Jones L, Jones I, Craddock N, Owen MJ, Rietschel M, Maier W, Korszun A, Rice JP, Mors O, Preisig M, Uher R, Lewis CM, Thuret S, Powell TR. Polygenic risk for circulating reproductive hormone levels and their influence on hippocampal volume and depression susceptibility. Psychoneuroendocrinology. 2019;106:284-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Takei N, Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front Mol Neurosci. 2014;7:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 238] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 25. | Abelaira HM, Réus GZ, Neotti MV, Quevedo J. The role of mTOR in depression and antidepressant responses. Life Sci. 2014;101:10-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 147] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 26. | Pham TH, Gardier AM. Fast-acting antidepressant activity of ketamine: highlights on brain serotonin, glutamate, and GABA neurotransmission in preclinical studies. Pharmacol Ther. 2019;199:58-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 130] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 27. | Liu S, Zhang F, Wang X, Shugart YY, Zhao Y, Li X, Liu Z, Sun N, Yang C, Zhang K, Yue W, Yu X, Xu Y. Diagnostic value of blood-derived microRNAs for schizophrenia: results of a meta-analysis and validation. Sci Rep. 2017;7:15328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 28. | Fries GR, Carvalho AF, Quevedo J. The miRNome of bipolar disorder. J Affect Disord. 2018;233:110-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Li N, Pan X, Zhang J, Ma A, Yang S, Ma J, Xie A. Plasma levels of miR-137 and miR-124 are associated with Parkinson's disease but not with Parkinson's disease with depression. Neurol Sci. 2017;38:761-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 30. | Rahmani S, Kadkhoda S, Ghafouri-Fard S. Synaptic plasticity and depression: the role of miRNAs dysregulation. Mol Biol Rep. 2022;49:9759-9765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 31. | Zhao L, Li H, Guo R, Ma T, Hou R, Ma X, Du Y. miR-137, a new target for post-stroke depression? Neural Regen Res. 2013;8:2441-2448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 32. | Kim B, Tag SH, Kim YS, Cho SN, Im HI. Circulating microRNA miR-137 as a stable biomarker for methamphetamine abstinence. Psychopharmacology (Berl). 2022;239:831-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Guennewig B, Cooper AA. The central role of noncoding RNA in the brain. Int Rev Neurobiol. 2014;116:153-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 34. | Liu T, Huang Y, Chen J, Chi H, Yu Z, Wang J, Chen C. Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1AS expression. Mol Med Rep. 2014;10:1275-1281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 35. | Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570-1573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 579] [Cited by in RCA: 602] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 36. | Li X, Isono K, Yamada D, Endo TA, Endoh M, Shinga J, Mizutani-Koseki Y, Otte AP, Casanova M, Kitamura H, Kamijo T, Sharif J, Ohara O, Toyada T, Bernstein BE, Brockdorff N, Koseki H. Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes. Mol Cell Biol. 2011;31:351-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Abedpoor N, Taghian F, Hajibabaie F. Cross Brain-Gut Analysis Highlighted Hub Genes and LncRNA Networks Differentially Modified During Leucine Consumption and Endurance Exercise in Mice with Depression-Like Behaviors. Mol Neurobiol. 2022;59:4106-4123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 38. | Issler O, van der Zee YY, Ramakrishnan A, Wang J, Tan C, Loh YE, Purushothaman I, Walker DM, Lorsch ZS, Hamilton PJ, Peña CJ, Flaherty E, Hartley BJ, Torres-Berrío A, Parise EM, Kronman H, Duffy JE, Estill MS, Calipari ES, Labonté B, Neve RL, Tamminga CA, Brennand KJ, Dong Y, Shen L, Nestler EJ. Sex-Specific Role for the Long Non-coding RNA LINC00473 in Depression. Neuron. 2020;106:912-926.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 39. | Alderson NL, Rembiesa BM, Walla MD, Bielawska A, Bielawski J, Hama H. The human FA2H gene encodes a fatty acid 2-hydroxylase. J Biol Chem. 2004;279:48562-48568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 40. | Levi S, Finazzi D. Neurodegeneration with brain iron accumulation: update on pathogenic mechanisms. Front Pharmacol. 2014;5:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 41. | Tello C, Darling A, Lupo V, Pérez-Dueñas B, Espinós C. On the complexity of clinical and molecular bases of neurodegeneration with brain iron accumulation. Clin Genet. 2018;93:731-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Kruer MC, Paisán-Ruiz C, Boddaert N, Yoon MY, Hama H, Gregory A, Malandrini A, Woltjer RL, Munnich A, Gobin S, Polster BJ, Palmeri S, Edvardson S, Hardy J, Houlden H, Hayflick SJ. Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann Neurol. 2010;68:611-618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 43. | Scheid I, Maruani A, Huguet G, Leblond CS, Nygren G, Anckarsäter H, Beggiato A, Rastam M, Amsellem F, Gillberg IC, Elmaleh M, Leboyer M, Gillberg C, Betancur C, Coleman M, Hama H, Cook EH, Bourgeron T, Delorme R. Heterozygous FA2H mutations in autism spectrum disorders. BMC Med Genet. 2013;14:124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 44. | Ranjpour M, Wajid S, Jain SK. Elevated Expression of A-Raf and FA2H in Hepatocellular Carcinoma is Associated with Lipid Metabolism Dysregulation and Cancer Progression. Anticancer Agents Med Chem. 2019;19:236-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 45. | Lemay AM, Courtemanche O, Couttas TA, Jamsari G, Gagné A, Bossé Y, Joubert P, Don AS, Marsolais D. High FA2H and UGT8 transcript levels predict hydroxylated hexosylceramide accumulation in lung adenocarcinoma. J Lipid Res. 2019;60:1776-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 46. | Kihara A. Very long-chain fatty acids: elongation, physiology and related disorders. J Biochem. 2012;152:387-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 327] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 47. | Agostini M, Melino G, Habeb B, Calandria JM, Bazan NG. Targeting lipid metabolism in cancer: neuroblastoma. Cancer Metastasis Rev. 2022;41:255-260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 48. | Agbaga MP, Stiles MA, Brush RS, Sullivan MT, Machalinski A, Jones KL, Anderson RE, Sherry DM. The Elovl4 Spinocerebellar Ataxia-34 Mutation 736T>G (p.W246G) Impairs Retinal Function in the Absence of Photoreceptor Degeneration. Mol Neurobiol. 2020;57:4735-4753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |