Published online Jul 19, 2022. doi: 10.5498/wjp.v12.i7.904
Peer-review started: January 28, 2022
First decision: April 18, 2022
Revised: May 2, 2022
Accepted: June 17, 2022
Article in press: June 17, 2022
Published online: July 19, 2022
Processing time: 171 Days and 17.6 Hours
Schizophrenia (SCZ) is a complex disease which can be affected by both genetic and environmental factors. Prenatal famine exposure may cause changes in DNA methylation levels of genes. Meanwhile, maternal nutrition during pregnancy is a pivotal environmental factor in the development of SCZ. DNA methylation may be an intermediate factor mediating exposure to famine during pregnancy and SCZ, and DNA methylation quantitative trait loci might serve as a promising tool for linking SCZ and prenatal famine.
To analyze the association between prenatal famine exposure and SCZ risk in Northeast Han Chinese through analysis of DNA methylation related loci.
A total of 954 Han Chinese from Northeast China were recruited, including 443 patients with SCZ and 511 healthy controls. The participants were further divided into famine (born in 1960-1962) and non-famine (born in 1963-1965) groups to investigate the effect of prenatal famine exposure. Four single-nucleotide polymorphisms (SNPs) selected according to the relevant literature were genotyped, namely, rs11917047 in PTPRG, rs2239681 in IGF2, rs3842756 in INSIGF, and rs61955196 in ABCB9. DNA were extracted from peripheral blood samples, and the genotypes of these SNP loci were detected using the improved Multiple Ligase Detection Reaction multiple SNP typing technique. The associations of the DNA methylation related SNPs with SCZ risk and prenatal famine, and their interactions were analyzed using logistic regression analysis and generalized multifactor dimensionality reduction (GMDR) software.
Based on the sequencing data, genotype distributions and allele frequencies of the four selected SNPs were determined. All genotype frequencies of the four SNPs in the healthy control group were tested for deviation from Hardy-Weinberg equilibrium (P > 0.05). Logistic regression analysis showed that rs61955196 was significantly associated with SCZ risk in the log-additive model [odds ratio (OR): 1.22; 95% confidence interval (CI): 1.01-1.48; P = 0.040]. We also found that the rs61955196 allele was related with an enhanced risk of SCZ (G>C, OR: 1.22; 95%CI: 1.01-1.47; P = 0.042). However, no associations were observed between rs11917047, rs2239681, or rs3842756 and SCZ risk. Under the optimal genetic model, no significant association of famine with the four SNPs was seen. Though the gene–gene interactions between rs2239681 and rs61955196 were found in GMDR analysis, none of the gene-gene interactions and gene-famine interactions were associated with the risk of SCZ.
Our study suggested that rs61955196 in ABCB9 is associated with SCZ susceptibility in Northeast Han Chinese, providing insight into genetic effects on SCZ.
Core Tip: Prenatal famine exposure may cause changes in DNA methylation levels of genes, while maternal nutrition is a pivotal environmental factor for schizophrenia (SCZ). To analyze the association between prenatal famine exposure and SCZ risk, we recruited 443 SCZ patients and 511 healthy controls with four single-nucleotide polymorphisms genotyped, which were previously identified as DNA methylation quantitative trait loci. Our study observed significant differences in rs61955196 genotype distribution and allele frequency between SCZ patients and healthy controls for the first time, suggesting that rs61955196 in ABCB9 was associated with SCZ susceptibility among the Northeast Han Chinese population.
- Citation: Li XW, Zhang MY, Li ZJ, Ai LZ, Jin MD, Jia NN, Xie MT, Yang YQ, Li WZ, Dong L, Yu Q. ABCB9 polymorphism rs61955196 is associated with schizophrenia in a Chinese Han population. World J Psychiatry 2022; 12(7): 904-914
- URL: https://www.wjgnet.com/2220-3206/full/v12/i7/904.htm
- DOI: https://dx.doi.org/10.5498/wjp.v12.i7.904
Schizophrenia (SCZ) is a complex disease affected by both genetic and environmental factors, which is often characterized by symptoms such as hallucinations, social withdrawals, delusions, and cognitive dysfunction[1,2]. The global point prevalence of SCZ was estimated to be 0.28% (0.24%–0.31%) in 2016 which contributes 13.4 (95% uncertainty interval: 9.9–16.7) million years of life lived with disability to burden of disease globally[3]. And China was assessed to show the highest prevalence of 0.42% among global countries, which raises necessity to conduct research enrolling local Chinese participants to reveal the practical status and underlying biological mechanisms of SCZ for its management and treatment.
DNA methylation is a heritable epigenetic modification which alters gene expression[4]. Studies have demonstrated that overall DNA hypomethylation is evident in SCZ patients, while treatment with haloperidol might increase methylation[5,6]. In other words, DNA methylation, which can regulate gene expression, is closely associated with the risk of SCZ [7-10].
Although the peak incidence rate of SCZ appears in adolescence and early adulthood, many believe that its etiological origin exists much earlier in one’s life, which includes exposure to environmental and genetic factors. The exposure occurring in the early stages of life development along with a cumulative effect during the later stages may eventually lead to the appearance of symptoms[11]. Among the environmental factors, maternal nutrition during pregnancy plays an early and vital role in the occurrence and development of SCZ[12,13]. Studies have shown that prenatal famine exposure may cause changes in DNA methylation levels of genes. Malnutrition during pregnancy, especially the lack of maternal protein and folic acid, seriously affects fetal development which will result in changes in DNA methylation[14]. Empirical studies of the Great Famine of China in 1959-1961 and the Dutch famine in 1944-1945 both showed that prenatal famine exposure led to an obviously increased risk of SCZ[15-17]. It was found that those who were born during the famine are twice as likely to have SCZ in their later years as normal people[16]. Therefore, we proposed that prenatal nutritional deficiencies may increase the risk of SCZ by altering DNA methylation status.
In recent years, genome-wide association studies (GWAS) have been effectively used for studying genetic variation associated with SCZ[18,19]. As DNA methylation tends to be sensitive to environmental factors, DNA methylation quantitative trait loci (meQTL) seems more promising. They can be derived by GWAS mapping levels of DNA methylation in genotyped individuals and define loci at which DNA methylation is influenced by genetic variation[20], with a superiority of higher consistency throughout one’s life than DNA methylation itself. There have already been reports revealing the role of meQTLs in SCZ risk, which promote the feasibility of them serving as a useful tool for SCZ-related research[21,22]. However, the results from GWAS studies are often not repeatable due to the enormous number for detection and heterogeneity of genetic information regarding people from different races and regions[23]. Given the high SCZ prevalence in China and current lack of available genetic data covering native patients, we find it necessary to conduct research collecting genetic data among Chinese individuals.
Here we intended to analyze the associations between single-nucleotide polymorphisms (SNPs) identified as meQTLs with the risk of SCZ and prenatal famine exposure among a Han population in Northeast China. We recruited SCZ patients and healthy controls (HCs) with comparable age including individuals born between 1959 and 1961 with prenatal famine exposure, and collected their peripheral blood samples for genotyping. We selected four SNPs which were previously reported as meQTLs, and determined their associations with SCZ and prenatal famine along with their interactions. We hope our work may provide more practical reference in management of SCZ.
A desired sample size of 417 patients was calculated using the software Quanto with a proper power before the recruitment of participants, with a unmatched case-control rate of 1.2, an estimated population risk of 1% for SCZ, a log-additive model gene with allele frequency of 0.1, genetic effect of 1.5, and a type I error rate of 0.05 by two-sided test. According to the desired sample size and the inclusion and exclusion criteria, a total of 954 Han Chinese from Northeast China were finally recruited between 2010 and 2012, including 443 SCZ patients and 511 healthy people. The patients were recruited from the Siping Psychiatric Hospital and Sixth Hospital of Changchun City (Jilin, China). Each patient was diagnosed according to the Tenth Revision of International Classification of Diseases-10 for SCZ and confirmed by at least two experienced psychiatrists. Those with neurological disorders, severe organic lesions, and drug dependence were excluded. Subjects in the HC group matching the patients by gender and age were recruited from the Changchun Municipal Centre for Disease Control and Prevention, in order to get a comparable proportion of famine-exposed individuals between two groups and a similar ratio of gender. The healthy subjects were required to have no history of mental illness and were in good health without any known disease at the time of recruitment. Furthermore, subjects who were in uterus between 1959 and 1961 were regarded to be exposed to famine. And then they were divided into two groups, namely, famine group (born in 1960-1962) and non-famine group (born in 1963-1965), according to whether they were exposed to famine before birth. All methods were performed in accordance with the relevant guidelines and regulations. The study adhered to the tenets of the Declaration of Helsinki, and was approved by the Ethics Committee of the School of Public Health of Jilin University (Approval No: 2014-03-11). All participants provided informed consent.
In the first step, we collected peripheral blood samples from the participants and extracted genomic DNA. Then, DNA content and purity were determined based on the ratio of OD260/OD280. Combining the feasibility of the detection method and the previous publications, we selected four SNPs (rs11917047 in PTPRG, rs2239681 in IGF2, rs3842756 in INSIGF, and rs61955196 in ABCB9) which have been confirmed as meQTLs, and the SNPs themselves or the genes that they belong to were assessed to be associated with SCZ[21,24-26].
Then, the genotypes of these SNP locus were detected using the improved Multiplex Ligase Detection Reaction multiple SNP typing technique (Shanghai Tian Hao Biological Technology Co. Ltd.). Using the Assay Design software 3.1, we successfully designed primers for the four meQTL SNPs. And the primer sequences for each SNP are as follows: rs11917047-F, AGATGAAAGATTGGGGTGTGGGTA and rs11917047-R, GCTGGTACCCAACCAGGAACAC; rs2239681-F, ATGGGCAAATCAGCCTGAAGAG and rs2239681-R, GTGTGCAAGAGGGGTGAAAGGT; rs3842756-F, TCCACAGGGACTCCATCAGAAA and rs3842756-R, CCTGTGGCTCAGGGTCCAGTAT; and rs61955196-F, GCTGCAAGGTCGGAGCTGAG and rs61955196-R TGGGAGGAGTTTGCCACAGG.
Deviation of the genotypes from Hardy-Weinberg equilibrium (HWE) between the SCZ patients and healthy individuals was assessed using a χ2 goodness-of-fit test. Logistic regression analysis was used to examine the relationship between SNPs and the risk of SCZ as well as the association of famine with SNPs with age and sex adjusted as covariates. The online genetic analysis software, SNPStats, was used to select the optimal genetic model according to the Akaike information criterion (AIC) and Bayesian information criterion. Generalized multifactor dimensionality reduction (GMDR) analysis was conducted to analyze the gene-gene interactions, which are rather critical in investigating genetic information for multifactorial diseases, and the gene-environment interactions were analyzed by crossover analysis based on logistic regression analysis. Except for the above specified, all statistical analyses were performed with SPSS 24.0 software. A P-value < 0.05 was considered statistically significant.
Based on the SNP sequencing data, the genotype distributions and allele frequencies of the four selected SNPs in SCZ patients (n = 443) and HCs (n = 511) were determined and the detailed data are shown in Table 1. All genotype frequencies of the four SNPs in the HC group were in accordance with HWE (P > 0.05). Logistic regression analysis showed that, compared with those carrying the wild-type homozygote (CC) of rs61955196, subjects carrying the mutant homozygote (GG) had a higher risk of SCZ [odds ratio (OR): 1.54; 95% confidence interval (CI): 1.03-2.30; P = 0.037]. We also found that the rs61955196 allele was related with an enhanced risk of SCZ (OR: 1.22; 95%CI: 1.01-1.47; P = 0.042). The frequency of the rs61955196 G allele was 40.5% in the case group, which was significantly higher than that of the control group (36.6%; P < 0.05). No associations were observed between SCZ patients and HC subjects regarding different genotypes or alleles of the rest three SNPs.
| SNP | Genotype/allele | SCZ (n = 443) | HC (n = 511) | P value | OR (95%CI) | HWE test for controls |
| rs11917047 (PTPRG) | AA | 219 (49.4) | 259 (50.7) | 1.00 (ref) | 0.151 | |
| AG | 183 (41.3) | 200 (39.1) | 0.431 | 1.12 (0.85-1.47) | ||
| GG | 41 (9.3) | 52 (10.2) | 0.713 | 0.92 (0.58-1.45) | ||
| A | 621 (70.1) | 718 (70.3) | 1.00 (ref) | |||
| G | 265 (29.9) | 304 (29.7) | 0.885 | 1.02 (0.83-1.24) | ||
| rs2239681 (IGF2) | AA | 156 (35.2) | 177 (34.6) | 1.00 (ref) | 0.104 | |
| AG | 211 (47.6) | 232 (45.4) | 0.667 | 1.07 (0.80-1.43) | ||
| GG | 76 (17.2) | 102 (20.0) | 0.447 | 0.87 (0.59-1.26) | ||
| A | 523 (59.0) | 586 (57.3) | 1.00 (ref) | |||
| G | 363 (41.0) | 436 (42.7) | 0.567 | 0.95 (0.79-1.14) | ||
| rs3842756 (INSIGF) | CC | 405 (91.4) | 475 (93.0) | 1.00 (ref) | 0.409 | |
| CT | 38 (8.6) | 36 (7.0) | 0.319 | 1.28 (0.79-2.08) | ||
| TT | - | - | - | - | ||
| C | 848 (95.7) | 986 (96.5) | 1.00 (ref) | |||
| T | 38 (4.3) | 36 (3.5) | 0.329 | 1.27 (0.79-2.04) | ||
| rs61955196 (ABCB9) | CC | 157 (35.4) | 202 (39.5) | 1.00 (ref) | 0.513 | |
| CG | 213 (48.1) | 244 (47.7) | 0.297 | 1.16 (0.88-1.55) | ||
| GG | 73 (16.5) | 65 (12.7) | 0.037a | 1.54 (1.03-2.30) | ||
| C | 527 (59.5) | 648 (63.4) | 1.00 (ref) | |||
| G | 359 (40.5) | 374 (36.6) | 0.042a | 1.22 (1.01-1.47) |
Based on the findings, we dug into the association between genotypes of rs61955196 and SCZ risk using multiple genetic models. As shown in Table 2, a significant association between rs61955196 and SCZ in the log-additive model was revealed (OR: 1.22; 95%CI: 1.01-1.48; P = 0.040). In the codominant model, we also found the association of rs61955196 with SCZ in the GG vs CC genotype comparison. No obvious effect of rs61955196 on the risk of SCZ was found in other models (P > 0.05).
| Model | Genotype | SCZ (%) | HC (%) | OR (95%CI) | P value | AIC | BIC |
| Codominant | CC | 157 (35.4) | 202 (39.5) | 1.00 | 1279.8 | 1299.2 | |
| GC | 213 (48.1) | 244 (47.8) | 1.16 (0.88-1.55) | 0.297 | |||
| GG | 73 (16.5) | 65 (12.7) | 1.54 (1.03-2.30) | 0.037a | |||
| Dominant | CC | 157 (35.4) | 202 (39.5) | 1.00 | 0.120 | 1279.7 | 1294.3 |
| GC + GG | 286 (64.6) | 309 (60.5) | 1.24 (0.95-1.62) | ||||
| Recessive | CC + GC | 370 (83.5) | 446 (87.3) | 1.00 | 0.068 | 1278.9 | 1293.5 |
| GG | 73 (16.5) | 65 (12.7) | 1.41 (0.97-2.04) | ||||
| Overdominant | CC + GG | 230 (51.9) | 267 (52.2) | 1.00 | 0.810 | 1282.1 | 1296.7 |
| GC | 213 (48.1) | 244 (47.8) | 1.03 (0.80-1.34) | ||||
| Log-additive | - | - | - | 1.22 (1.01-1.48) | 0.040a | 1278 | 1292.6 |
To investigate the relationship of meQTLs and prenatal famine exposure, we analyzed the associations of the four SNPs with famine. Totally, 492 subjects were exposed to prenatal famine, including 220 SCZ patients and 272 HC subjects. As shown in Table 3, based on the AIC, the inheritance model was recessive for rs11917047 and rs2239681, codominant for rs3842756, and overdominant for rs61955196. SCZ patients and HCs were further divided into a famine group and a non-famine group. Logistic regression analysis indicated that under the optimal genetic model, there was no significant association of famine with the four SNPs in either the SCZ group or HC group (P > 0.05).
| Group | SNP | Genotype | Famine | Non-famine | P value | OR (95%CI) |
| SCZ | rs11917047 | AA + GA | 204 (92.7) | 198 (88.8) | 0.150 | 1.00 |
| (Recessive) | GG | 16 (7.3) | 25 (11.2) | 1.62 (0.84-3.13) | ||
| rs2239681 | AA + GA | 185 (84.1) | 182 (81.6) | 0.550 | 1.00 | |
| (Recessive) | GG | 35 (15.9) | 41 (18.4) | 1.16 (0.71-1.91) | ||
| rs3842756 | CC | 198 (90) | 207 (92.8) | 0.270 | 1.00 | |
| (Codominant) | CT | 22 (10) | 16 (7.2) | 0.68 (0.35-1.34) | ||
| rs61955196 | CC + GG | 117 (53.2) | 113 (50.7) | 0.610 | 1.00 | |
| (Overdominant) | GC | 103 (46.8) | 110 (49.3) | 1.10 (0.76-1.60) | ||
| HC | rs11917047 | AA + GA | 249 (91.5) | 210 (87.9) | 0.160 | 1.00 |
| (Recessive) | GG | 23 (8.5) | 29 (12.1) | 1.50 (0.84-2.68) | ||
| rs2239681 | AA + GA | 222 (81.6) | 187 (78.2) | 0.330 | 1.00 | |
| (Recessive) | GG | 50 (18.4) | 52 (21.8) | 1.24 (0.80-1.92) | ||
| rs3842756 | CC | 257 (94.5) | 218 (91.2) | 0.150 | 1.00 | |
| (Codominant) | CT | 15 (5.5) | 21 (8.8) | 1.65 (0.83-3.27) | ||
| rs61955196 | CC + GG | 144 (52.9) | 123 (51.5) | 0.750 | 1.00 | |
| (Overdominant) | GC | 128 (47.1) | 116 (48.5) | 1.06 (0.75-1.50) |
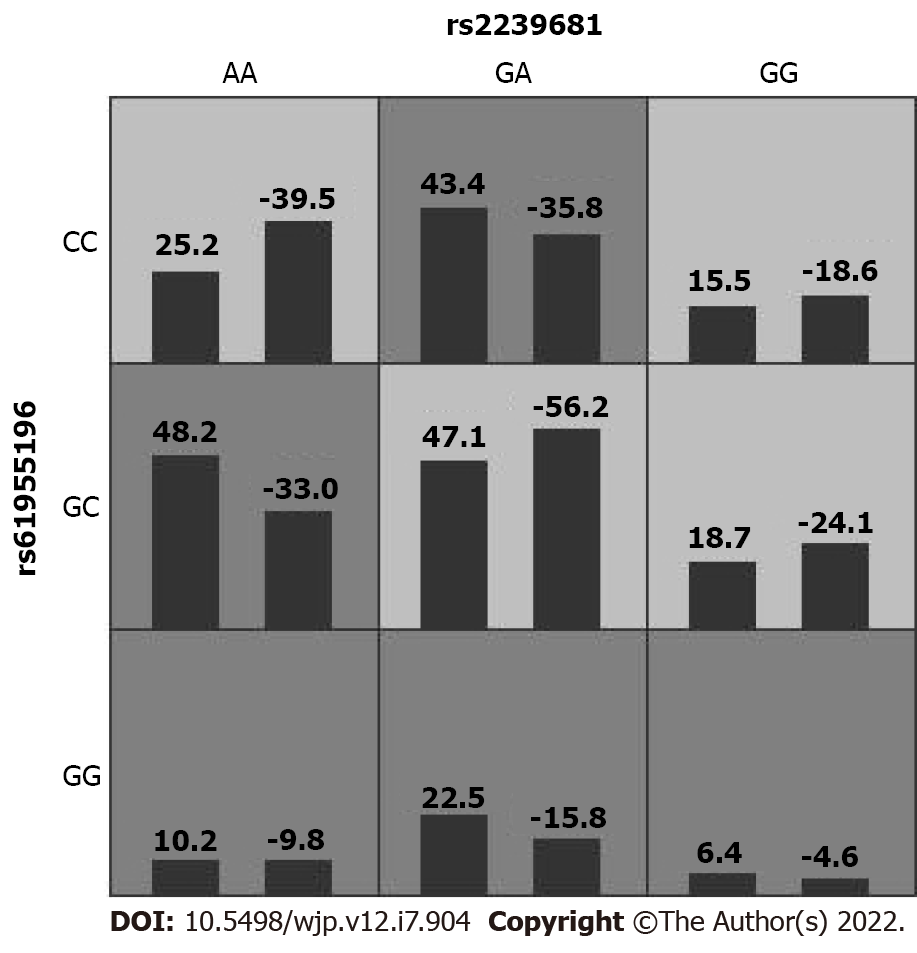
In this study, GMDR was used to import and analyze the interactions between rs11917047, rs2239681, rs3842756, and rs61955196. The impact of gene–gene interaction on the risk of SCZ is summarized in Table 4. The multifactor model 2 (rs2239681 × rs61955196) presented the best cross-validation consistency, which had a testing-balanced accuracy of 55.8%. Figure 1 shows the interaction model of this gene–gene interaction between rs2239681 and rs61955196. However, no significant association of gene-gene interaction with the risk of SCZ was found in this model.
| No. | Best combination | CVC | Te-BA | P value |
| 1 | rs61955196 | 9/10 | 0.5097 | 0.8281 |
| 2 | rs2239681 × rs61955196 | 10/10 | 0.5582 | 0.0547 |
| 3 | rs11917047 × rs2239681 × rs61955196 | 10/10 | 0.5341 | 0.1719 |
| 4 | rs11917047 × rs2239681 × rs3842756 × rs61955196 | 10/10 | 0.5449 | 0.1719 |
Crossover analysis based on a multiplicative model of logistic regression was conducted to determine the interactions between the SNPs and famine in SCZ patients (Table 5). None of the interactions between the genotypes of the four loci of rs11917047/rs2239681/rs3842756/rs61955196 with the risk of famine were statistically significant (P > 0.05).
| SNP | Genotype | Famine | SCZ | HC | OR (95%CI) | P value |
| rs11917047 | AG + GG | + | 121 | 119 | 1.21 (0.85-1.72) | 0.294 |
| AG + GG | - | 103 | 133 | 0.92 (0.65-1.31) | 0.646 | |
| AA | + | 102 | 120 | 1.01 (0.70-1.45) | 0.958 | |
| AA | - | 117 | 139 | 1.00 (ref) | ||
| rs2239681 | AG + GG | + | 144 | 157 | 1.13 (0.78-1.65) | 0.519 |
| AG + GG | - | 143 | 177 | 1.00 (0.69-1.45) | 0.986 | |
| AA | + | 79 | 82 | 1.19 (0.77-1.83) | 0.432 | |
| AA | - | 77 | 95 | 1.00 (ref) | ||
| rs3842756 | CT | + | 16 | 21 | 0.99 (0.50-1.95) | 0.974 |
| CT | - | 22 | 15 | 1.90 (0.96-3.77) | 0.064 | |
| CC | + | 207 | 218 | 1.23 (0.95-1.61) | 0.123 | |
| CC | - | 198 | 257 | 1.00 (ref) | ||
| rs61955196 | CG + GG | + | 147 | 145 | 1.35 (0.94-1.95) | 0.109 |
| CG + GG | - | 139 | 164 | 1.13 (0.78-1.63) | 0.513 | |
| CC | + | 76 | 94 | 1.08 (0.71-1.64) | 0.724 | |
| CC | - | 81 | 108 | 1.00 (ref) |
Based on existing reports, we selected four susceptibility loci of SNPs related to SCZ as the starting point for analysis, which are rs11917047 in PTPRG, rs2239681 in IGF2, rs3842756 in INSIGF, and rs61955196 in ABCB9, respectively. This study analyzed genetic data from representative samples of Northeastern Chinese using meQTL SNPs, and found the difference of rs61955196 genotype distribution with allele frequency between SCZ patients and HC subjects for the first time. rs61955196 is located in the 5' untranslated region of the ABCB9 gene, encoding the ABCB9 protein which belongs to the ATP-binding cassette (ABC) transporter family. The ABC gene can be divided into seven different subfamilies (MRP, ABC1, OABP, ALD, GCN20, MDR/TAP, and White)[27], and the ABCB9 protein is a member of the MDR/TAP subfamily. ABC family and ABCB9 are reported to be involved in progression of multiple malignant tumors and chemoresistance[28-31], but little research has been done on the relationship between ABCB9 gene and SCZ. Recent evidence suggests that ABCB9 is positively associated with the risk of SCZ[32], which is in accordance to our findings to some extent.
Increasing studies have shown that epigenetic modifications are associated with the pathogenesis of SCZ, and DNA methylation is a crucial one regulating gene expression, which may be a key factor in the process[33,34]. Our results showed that the methylation locus rs61955196 increased the risk of SCZ in the log-additive model. However, we did not observe the association between the methylation loci located in the other three genes and SCZ, which is inconsistent with existing studies. For example, Cressant et al[35] discovered that the PTPRG gene containing the rs11917047 locus was associated with SCZ. The receptor protein tyrosine phosphatase PTPRG is a ligand for members of the contact protein family, which are linked to autism spectrum disorders. The interpretation for these disagreements may be due to the disparity in the target population as what we studied is the Han population in Northeast China, which is different from other studies in race, sample size, and geographic location.
It is a pity that we did not find the association of prenatal exposure to famine with DNA methylation loci. A recent study also reported that maternal risk alleles for neurodevelopmental disorders, primarily attention-deficit/hyperactivity disorder, were associated with prenatal exposures, but nor for SCZ or autism spectrum disorder[36]. Nevertheless, there have been much supportive evidence regarding the positive relationship between SCZ and prenatal famine exposure. Waterland[37] discovered that maternal nutritional deficiency may result in permanent abnormal DNA methylation with the potential to affect gene expression. In addition, since human is unable to synthesize folic acid which is necessary for normal DNA methylation, the lack of folic acid which hinders the production of methyl donors might affect gene expression related to neurodevelopmental processes. Prenatal famine leads to undernutrition during fetal development, which is believed to further promote the risk of SCZ in offspring[38]. Wang and Zhang[39] also used data from a nationally representative sample to analyze the association of prenatal famine exposure with the risk of SCZ. The results showed that famine population had a higher risk of SCZ compared to the non-famine cohorts. This pattern was found throughout different subsamples, such as the urban/rural population[40]. Therefore, we still believe that it is vital to continue exploring the association of prenatal famine exposure with DNA methylation and SCZ in the future. Meanwhile, this study had several limitations. First, we only adjusted for gender as we mainly focused on the genetic variants, and we were not able to explore some underlying confounders such as medication as we have directly excluded those who had any medical treatment in the past 3 mo before enrollment. Second, as we did not collect sufficient information from the patients regarding illness-related parameters such as the severity or duration of disease, we could not rule out the possibility that the SNPs could be associated with SCZ under some specific conditions although we got negative results. Third, this is a case-control study and the patients were recruited from hospitals, resulting in inevitable selection bias. Finally, limited by the feasibility and applicability of the detection method, we only selected four SNPs in this study, and the constrained selection may leave out other crucial SNPs related to DNA methylation.
Our study suggested that rs61955196 in ABCB9 could be associated with SCZ susceptibility among the Han population in Northeast China. No association was found between the four meQTL SNPs and prenatal famine. These findings provide insight into genetic effects on SCZ. Future research should be devoted to validating the results, and gathering comprehensive information for additional subgroup analyses may help to reveal the association between prenatal famine and SCZ risk.
Schizophrenia (SCZ) is a severe mental disorder bringing heavy burden, which is closely related with genetic and environmental factors. The effect of prenatal exposure of famine on SCZ risk has been reported with intense interest. DNA methylation may be an intermediate factor mediating prenatal famine and SCZ, and DNA methylation quantitative trait locus (meQTLs) can serve as a promising tool.
The lifetime prevalence of SCZ is approximately 1% around the world, and study has reported the highest age-standardized prevalence of SCZ in China. Meanwhile, the Chinese famine of 1959-1961 is a proper source of study subjects to investigate the effect of prenatal famine on SCZ with little available genetic data. As a result, we intended to conduct analyses for SCZ and prenatal famine using native subjects with collected genetic information, which may provide insights specifically for Chinese researchers and patients.
To investigate the associations of four single-nucleotide polymorphisms (SNPs) identified as meQTLs with the risk of SCZ and prenatal famine exposure along with their interactions among Northeast Han Chinese.
We recruited 954 Han Chinese from Northeast China including 443 patients with SCZ and 511 healthy controls, and their peripheral blood samples were collected. Among them, 492 born in 1960-1962 were further allocated to a famine group. Four SNPs were selected and genotyped, namely, rs11917047 in PTPRG, rs2239681 in IGF2, rs3842756 in INSIGF, and rs61955196 in ABCB9. The associations of the meQTLs with SCZ risk and prenatal famine, and their interactions were analyzed using logistic regression analysis and generalized multifactor dimensionality reduction software.
The genotype distributions along with allele frequencies of the four SNPs were determined among the Chinese participants. We found that rs61955196 was significantly associated with SCZ risk in the log-additive model [odds ratio (OR): 1.22; 95% confidence interval (CI): 1.01-1.48; P = 0.040], and rs61955196 allele was related with an enhanced risk of SCZ (G>C, OR: 1.22; 95%CI: 1.01-1.47; P = 0.042). However, the other three SNPs were not associated with SCZ risk. No association was observed between the SNPs and prenatal famine. Gene-gene interactions were seen between rs2239681 and rs61955196, while no gene-gene or gene-famine interactions were associated with the risk of SCZ.
Our results suggested that rs61955196 in ABCB9 might be involved in SCZ susceptibility among Northeast Han Chinese.
Our study provides a potential functional variant rs61955196 for SCZ susceptibility, and we recommend further research to extend the findings to different populations and verify its function. Although no evidence between SCZ and prenatal famine was found, we believe that gathering comprehensive information for analyses regarding subgroups may help to reveal the association in the future.
We greatly appreciate all subjects who agreed to participate in this study and all staff who contributed to this work for their cooperation and patience.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Aedma K, United States; Goh KK, Taiwan S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Fan JR
| 1. | Potash JB, Bienvenu OJ. Neuropsychiatric disorders: Shared genetics of bipolar disorder and schizophrenia. Nat Rev Neurol. 2009;5:299-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 2. | McCutcheon RA, Reis Marques T, Howes OD. Schizophrenia-An Overview. JAMA Psychiatry. 2020;77:201-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 727] [Article Influence: 145.4] [Reference Citation Analysis (0)] |
| 3. | Charlson FJ, Ferrari AJ, Santomauro DF, Diminic S, Stockings E, Scott JG, McGrath JJ, Whiteford HA. Global Epidemiology and Burden of Schizophrenia: Findings From the Global Burden of Disease Study 2016. Schizophr Bull. 2018;44:1195-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 859] [Cited by in RCA: 944] [Article Influence: 134.9] [Reference Citation Analysis (0)] |
| 4. | Rukova B, Staneva R, Hadjidekova S, Stamenov G, Milanova V, Toncheva D. Whole genome methylation analyses of schizophrenia patients before and after treatment. Biotechnol Biotechnol Equip. 2014;28:518-524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Melas PA, Rogdaki M, Ösby U, Schalling M, Lavebratt C, Ekström TJ. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 2012;26:2712-2718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 6. | Magwai T, Shangase KB, Oginga FO, Chiliza B, Mpofana T, Xulu KR. DNA Methylation and Schizophrenia: Current Literature and Future Perspective. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 7. | Montano C, Taub MA, Jaffe A, Briem E, Feinberg JI, Trygvadottir R, Idrizi A, Runarsson A, Berndsen B, Gur RC, Moore TM, Perry RT, Fugman D, Sabunciyan S, Yolken RH, Hyde TM, Kleinman JE, Sobell JL, Pato CN, Pato MT, Go RC, Nimgaonkar V, Weinberger DR, Braff D, Gur RE, Fallin MD, Feinberg AP. Association of DNA Methylation Differences With Schizophrenia in an Epigenome-Wide Association Study. JAMA Psychiatry. 2016;73:506-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 8. | Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, St Clair D, Mustard C, Breen G, Therman S, Kaprio J, Toulopoulou T, Hulshoff Pol HE, Bohlken MM, Kahn RS, Nenadic I, Hultman CM, Murray RM, Collier DA, Bass N, Gurling H, McQuillin A, Schalkwyk L, Mill J. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 248] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 9. | Ryan J, Saffery R. Crucial timing in schizophrenia: role of DNA methylation in early neurodevelopment. Genome Biol. 2014;15:495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 10. | Hannon E, Dempster EL, Mansell G, Burrage J, Bass N, Bohlken MM, Corvin A, Curtis CJ, Dempster D, Di Forti M, Dinan TG, Donohoe G, Gaughran F, Gill M, Gillespie A, Gunasinghe C, Hulshoff HE, Hultman CM, Johansson V, Kahn RS, Kaprio J, Kenis G, Kowalec K, MacCabe J, McDonald C, McQuillin A, Morris DW, Murphy KC, Mustard CJ, Nenadic I, O'Donovan MC, Quattrone D, Richards AL, Rutten BP, St Clair D, Therman S, Toulopoulou T, Van Os J, Waddington JL; Wellcome Trust Case Control Consortium (WTCCC); CRESTAR consortium, Sullivan P, Vassos E, Breen G, Collier DA, Murray RM, Schalkwyk LS, Mill J. DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia. Elife. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 11. | Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Børglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Böttcher Y, Olesen J, Breuer R, Möller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Réthelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA; Genetic Risk and Outcome in Psychosis (GROUP), Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jönsson EG, Terenius L, Agartz I, Petursson H, Nöthen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1394] [Cited by in RCA: 1282] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 12. | Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc Nutr Soc. 2012;71:154-165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 13. | Kirkbride JB, Susser E, Kundakovic M, Kresovich JK, Davey Smith G, Relton CL. Prenatal nutrition, epigenetics and schizophrenia risk: can we test causal effects? Epigenomics. 2012;4:303-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Xu J, He G, Zhu J, Zhou X, St Clair D, Wang T, Xiang Y, Zhao Q, Xing Q, Liu Y, Wang L, Li Q, He L, Zhao X. Prenatal nutritional deficiency reprogrammed postnatal gene expression in mammal brains: implications for schizophrenia. Int J Neuropsychopharmacol. 2014;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Song S, Wang W, Hu P. Famine, death, and madness: schizophrenia in early adulthood after prenatal exposure to the Chinese Great Leap Forward Famine. Soc Sci Med. 2009;68:1315-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | St Clair D, Xu M, Wang P, Yu Y, Fang Y, Zhang F, Zheng X, Gu N, Feng G, Sham P, He L. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959-1961. JAMA. 2005;294:557-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 454] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 17. | Susser ES, Lin SP. Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944-1945. Arch Gen Psychiatry. 1992;49:983-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 339] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 18. | Seng KC, Seng CK. The success of the genome-wide association approach: a brief story of a long struggle. Eur J Hum Genet. 2008;16:554-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | Mowry BJ, Gratten J. The emerging spectrum of allelic variation in schizophrenia: current evidence and strategies for the identification and functional characterization of common and rare variants. Mol Psychiatry. 2013;18:38-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Hoffmann A, Ziller M, Spengler D. The Future is The Past: Methylation QTLs in Schizophrenia. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, Kleinman JE. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19:40-47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 351] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 22. | Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, Troakes C, Turecki G, O'Donovan MC, Schalkwyk LC, Bray NJ, Mill J. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19:48-54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 294] [Cited by in RCA: 263] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 23. | Huang T, Shu Y, Cai YD. Genetic differences among ethnic groups. BMC Genomics. 2015;16:1093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 24. | Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 315] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 25. | Tobi EW, Slagboom PE, van Dongen J, Kremer D, Stein AD, Putter H, Heijmans BT, Lumey LH. Prenatal famine and genetic variation are independently and additively associated with DNA methylation at regulatory loci within IGF2/H19. PLoS One. 2012;7:e37933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 26. | Pardo M, Cheng Y, Sitbon YH, Lowell JA, Grieco SF, Worthen RJ, Desse S, Barreda-Diaz A. Insulin growth factor 2 (IGF2) as an emergent target in psychiatric and neurological disorders. Review. Neurosci Res. 2019;149:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 27. | Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 672] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 28. | Pasello M, Giudice AM, Scotlandi K. The ABC subfamily A transporters: Multifaceted players with incipient potentialities in cancer. Semin Cancer Biol. 2020;60:57-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 29. | Gong JP, Yang L, Tang JW, Sun P, Hu Q, Qin JW, Xu XM, Sun BC, Tang JH. Overexpression of microRNA-24 increases the sensitivity to paclitaxel in drug-resistant breast carcinoma cell lines via targeting ABCB9. Oncol Lett. 2016;12:3905-3911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Adamska A, Falasca M. ATP-binding cassette transporters in progression and clinical outcome of pancreatic cancer: What is the way forward? World J Gastroenterol. 2018;24:3222-3238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 31. | Hou L, Zhang X, Jiao Y, Li Y, Zhao Y, Guan Y, Liu Z. ATP binding cassette subfamily B member 9 (ABCB9) is a prognostic indicator of overall survival in ovarian cancer. Medicine (Baltimore). 2019;98:e15698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Hauberg ME, Zhang W, Giambartolomei C, Franzén O, Morris DL, Vyse TJ, Ruusalepp A; CommonMind Consortium, Sklar P, Schadt EE, Björkegren JLM, Roussos P. Large-Scale Identification of Common Trait and Disease Variants Affecting Gene Expression. Am J Hum Genet. 2017;100:885-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 33. | Nishioka M, Bundo M, Kasai K, Iwamoto K. DNA methylation in schizophrenia: progress and challenges of epigenetic studies. Genome Med. 2012;4:96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 34. | Liu J, Siyahhan Julnes P, Chen J, Ehrlich S, Walton E, Calhoun VD. The association of DNA methylation and brain volume in healthy individuals and schizophrenia patients. Schizophr Res. 2015;169:447-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Cressant A, Dubreuil V, Kong J, Kranz TM, Lazarini F, Launay JM, Callebert J, Sap J, Malaspina D, Granon S, Harroch S. Loss-of-function of PTPR γ and ζ, observed in sporadic schizophrenia, causes brain region-specific deregulation of monoamine levels and altered behavior in mice. Psychopharmacology (Berl). 2017;234:575-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Leppert B, Havdahl A, Riglin L, Jones HJ, Zheng J, Davey Smith G, Tilling K, Thapar A, Reichborn-Kjennerud T, Stergiakouli E. Association of Maternal Neurodevelopmental Risk Alleles With Early-Life Exposures. JAMA Psychiatry. 2019;76:834-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 37. | Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:1706S-1710S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 186] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 38. | Brown AS, Susser ES. Prenatal nutritional deficiency and risk of adult schizophrenia. Schizophr Bull. 2008;34:1054-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 188] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 39. | Wang C, Zhang Y. Schizophrenia in mid-adulthood after prenatal exposure to the Chinese Famine of 1959-1961. Schizophr Res. 2017;184:21-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 40. | He P, Chen G, Guo C, Wen X, Song X, Zheng X. Long-term effect of prenatal exposure to malnutrition on risk of schizophrenia in adulthood: Evidence from the Chinese famine of 1959-1961. Eur Psychiatry. 2018;51:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |