Copyright
©The Author(s) 2023.
World J Psychiatry. Feb 19, 2023; 13(2): 36-49
Published online Feb 19, 2023. doi: 10.5498/wjp.v13.i2.36
Published online Feb 19, 2023. doi: 10.5498/wjp.v13.i2.36
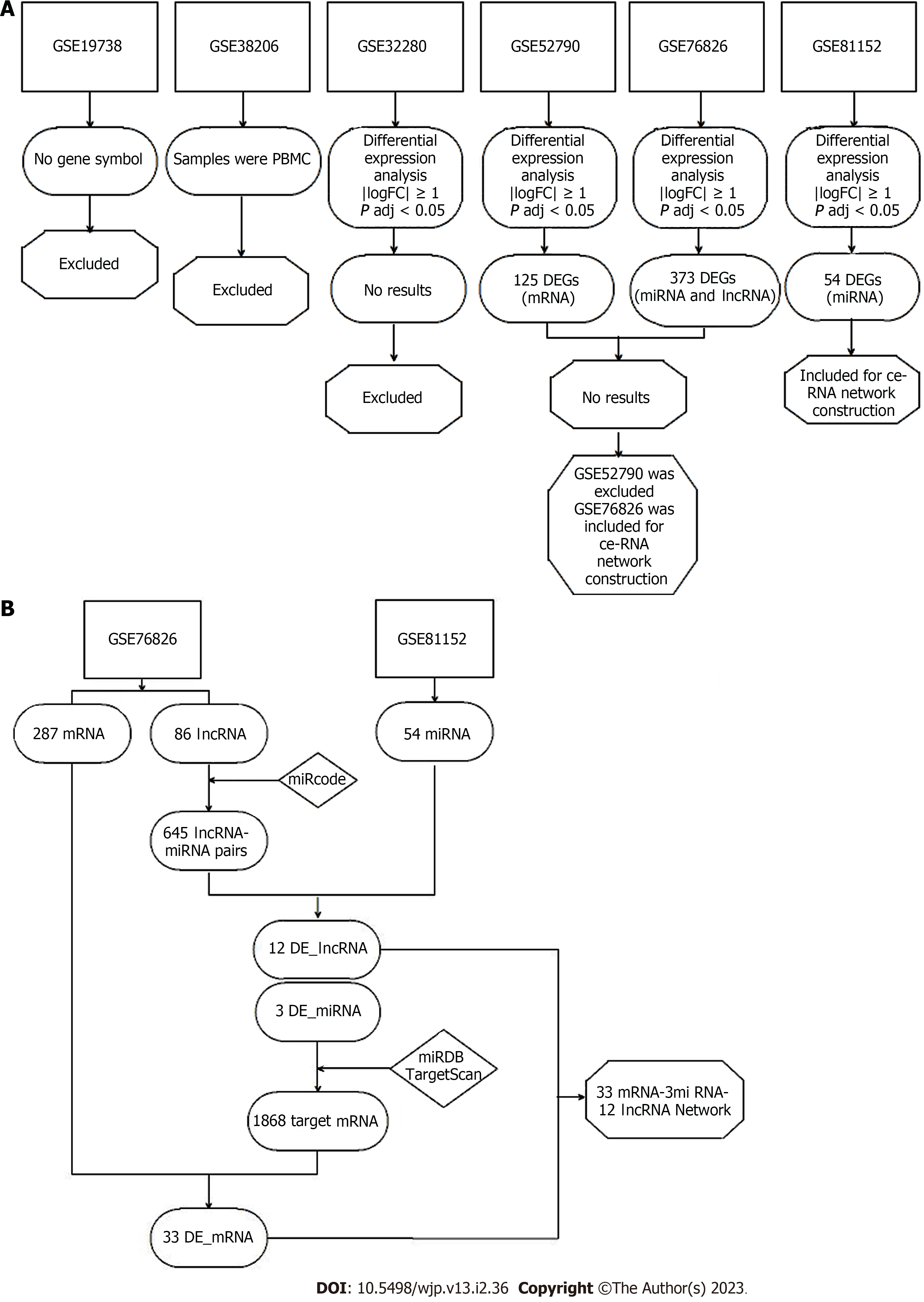
Figure 1 Flowchart of the study.
A: Step 1, dataset screening; B: Step 2, competing endogenous RNA network construction. DEGs: Differentially Expressed Genes; mRNA: Messenger RNA; miRNA: MicroRNA; lncRNA: Long non-coding RNA; ceRNA: Competing endogenous RNAs; miRcode: http://www.mircode.org/; miRDB: http://mirdb.org/; TargetScan: http://www.targetscan.org/vert_72/; DE_mRNA: Differentially expressed mRNA; DE_lncRNA: Differentially expressed lncRNA; DE_miRNA: Differentially expressed miRNA.
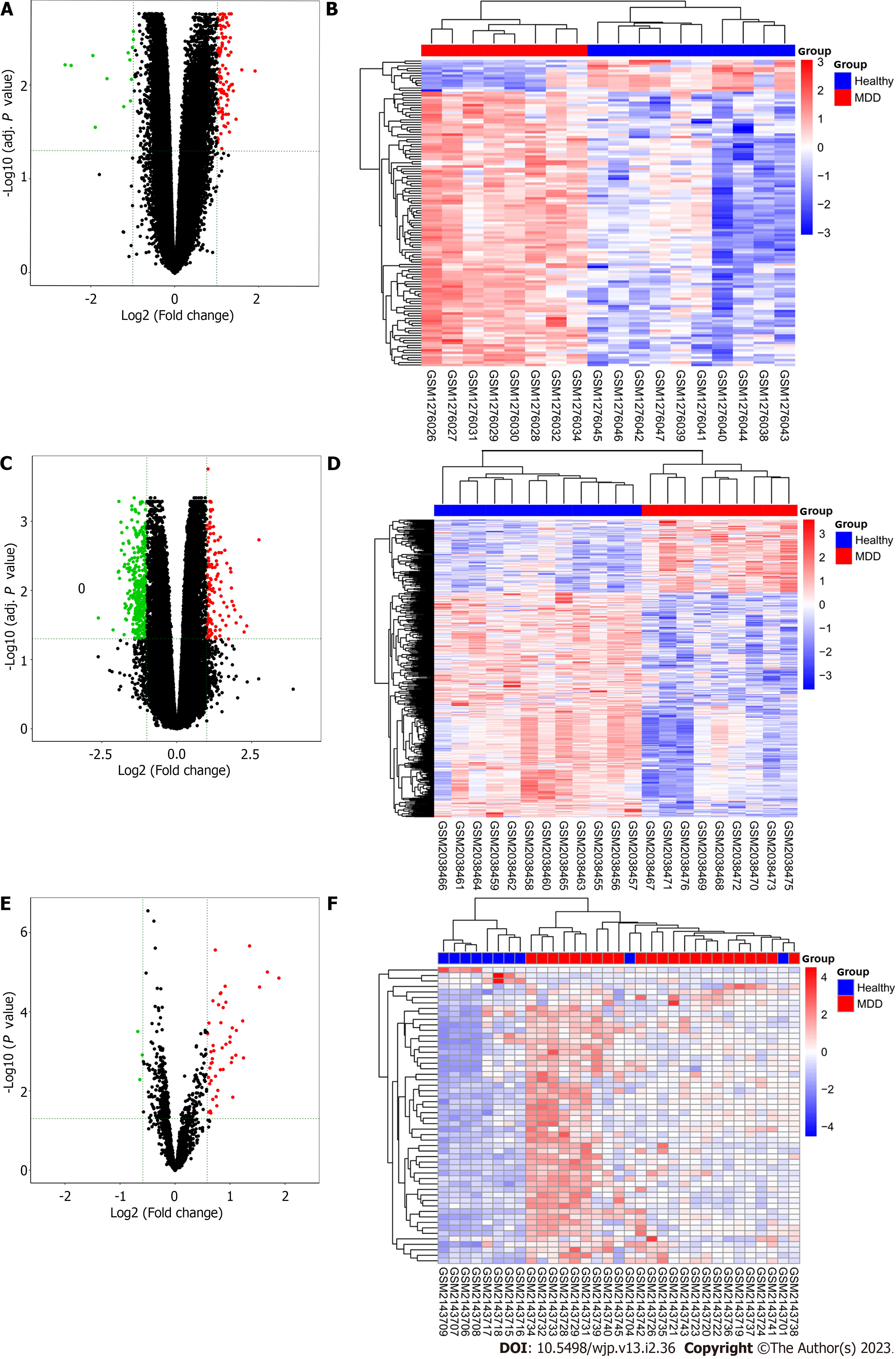
Figure 2 Difference and cluster analyses.
A: Volcanic maps of differentially expressed genes in GSE52790; B: Cluster analysis heatmap of the expression patterns of the differentially expressed genes in GSE52790; C: Volcanic maps of differentially expressed genes in GSE76826; D: Cluster analysis heatmap of the expression patterns of the differentially expressed genes in GSE76826; E: Volcanic maps of differentially expressed genes in GSE81152; F: Cluster analysis heatmap of the expression patterns of the differentially expressed genes in GSE81152. Red indicates upregulated genes; green indicates downregulated genes.
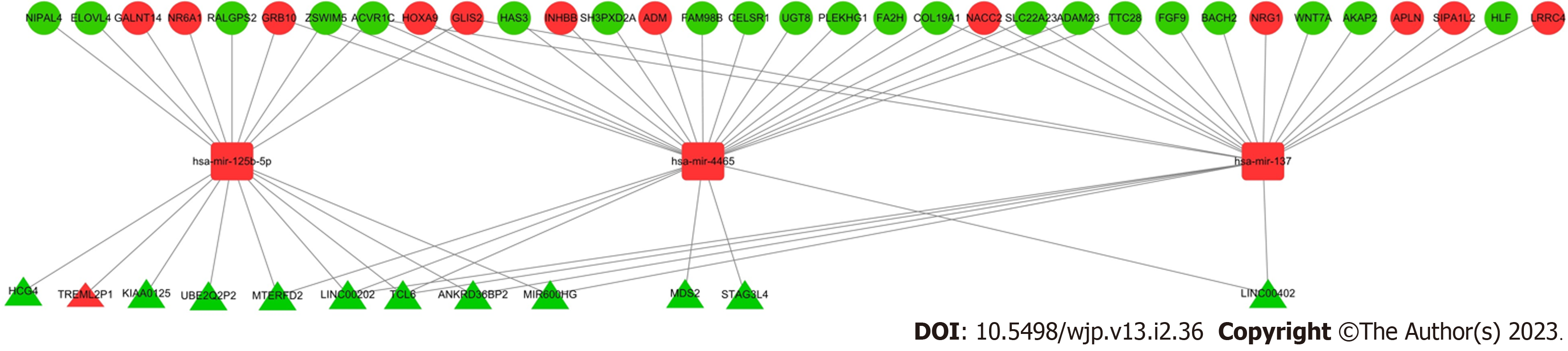
Figure 3 Major depressive disorder-associated enriched competing endogenous RNA network.
Red and green indicate upregulated downregulated genes, respectively. The circles represent differentially expressed RNA (DE_RNA), triangles represent DE long noncoding RNA (DE_lncRNA), and rounded squares represent DE_microRNA.
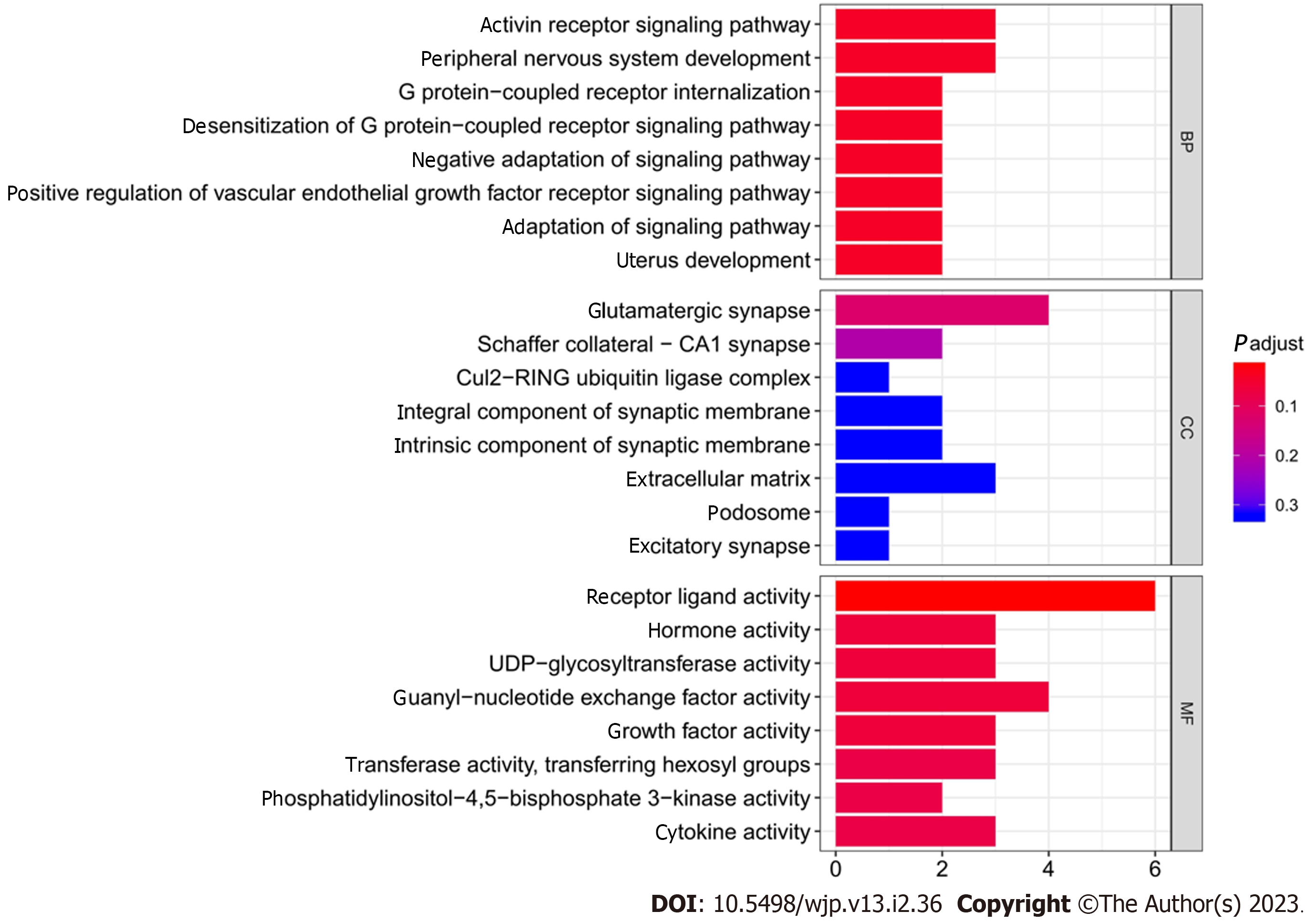
Figure 4
Gene Ontology functional enrichment analysis.
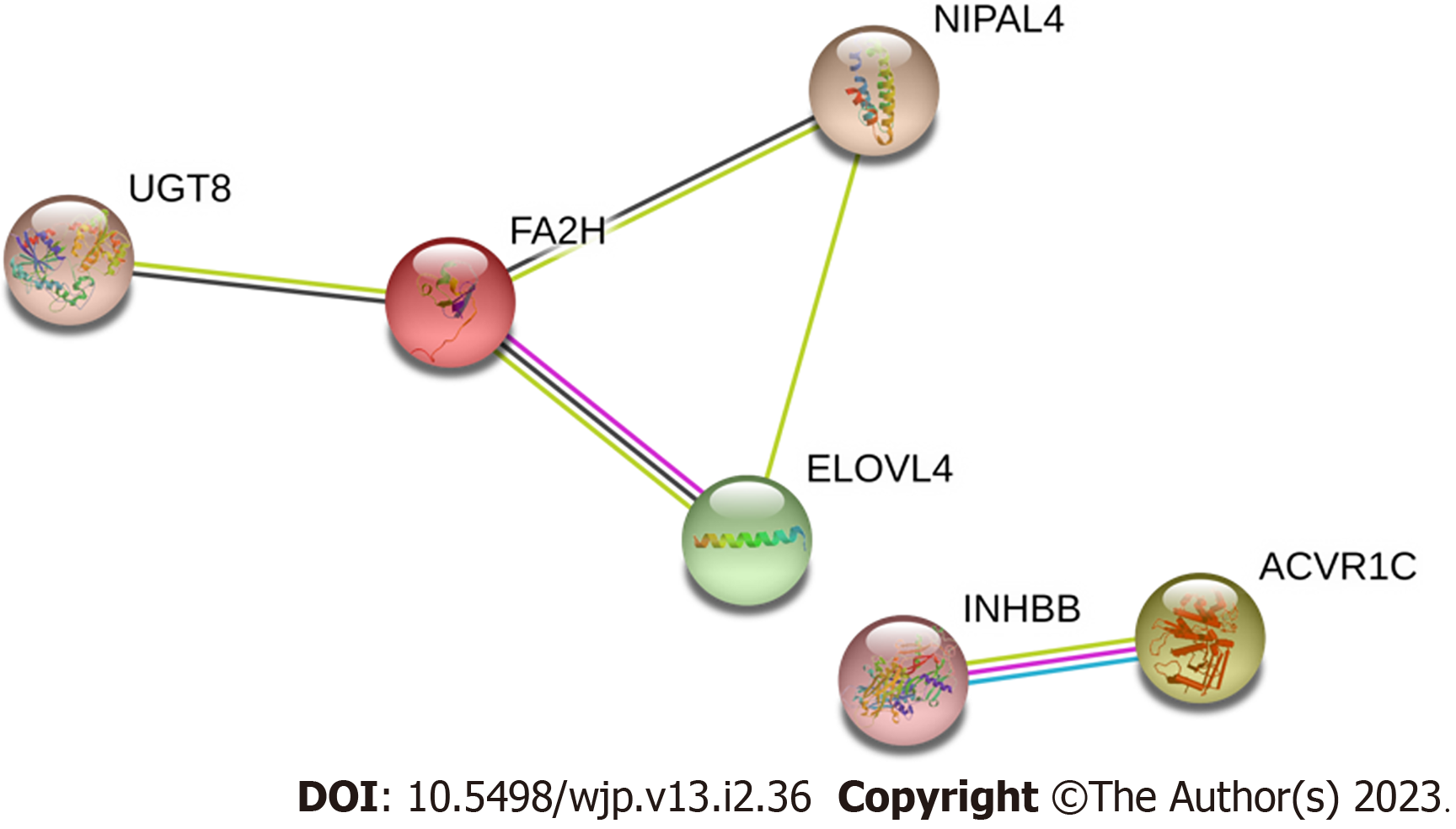
Figure 5 Protein interaction network construction.
FA2H: Fatty acid 2-hydroxylase.
- Citation: Zou ZL, Ye Y, Zhou B, Zhang Y. Identification and characterization of noncoding RNAs-associated competing endogenous RNA networks in major depressive disorder. World J Psychiatry 2023; 13(2): 36-49
- URL: https://www.wjgnet.com/2220-3206/full/v13/i2/36.htm
- DOI: https://dx.doi.org/10.5498/wjp.v13.i2.36