Published online Aug 28, 2019. doi: 10.5497/wjp.v8.i3.26
Peer-review started: January 23, 2019
First decision: January 30, 2019
Revised: July 5, 2019
Accepted: July 16, 2019
Article in press: July 16, 2019
Published online: August 28, 2019
Processing time: 219 Days and 15.7 Hours
Inflammatory bowel disease (IBD) has been a worldwide health problem. It is characterized by severe intestinal inflammation due to immune responses against the gut microbes in genetically susceptible individuals. The understanding of gut microbiota for its composition and complex interaction in normal and diseased conditions has been assisted by the use of molecular, metagenomics and meta transcriptomics studies. The alteration of intestinal microbiota is the key determinant in the degree of inflammation caused and the prolonged course of disease. The relationship between luminal gut bacteria and innate immunity is also of prime significance. Such developments have further led to the search of specific (including bacteria and fungi) as a causative agent of IBD. Although detailed research has been done for the role of gut microbiota in IBD, molecular mechanisms and related gene expression are still not well understood in this disease, which hampers the generation of effective therapeutic agents for IBD. This paper assessed various factors contributing to IBD, genetic dysbiosis and pathogenic influence in the gut microbiota, interactions such as microbiome-host immune system interaction and microbe-microbe interactions involved in IBD, currently available IBD therapies, followed by a detailed review on bacterial infections that might be involved in IBD, globally and specifically in India.
Core tip: Ulcerative colitis and Crohn’s disease are two clinical forms of inflammatory bowel disease (IBD) causing recurring diarrhea, abdominal bleeding, pain and inflammation. Scientific evidence clearly indicated the role of heredity in IBD, but an accurate cause of IBD is still unclear. However, complex interactions between genes, environmental factors and the immune system could be one of the leading causes in IBD. Gut microbiome is a key link between these factors and progression of IBD. This article reviews all contributing factors and pathogenic association in IBD. Identifying such microbial causes of the onset of IBD can help researchers to develop effective treatment strategies.
- Citation: Chandra N, Srivastava A, Kumar S. Bacterial biofilms in human gastrointestinal tract: An intricate balance between health and inflammatory bowel diseases. World J Pharmacol 2019; 8(3): 26-40
- URL: https://www.wjgnet.com/2220-3192/full/v8/i3/26.htm
- DOI: https://dx.doi.org/10.5497/wjp.v8.i3.26
Inflammatory bowel disease (IBD) is a common gastrointestinal disorder whose pathophysiology is still not completely understood due to its complex and multifactorial nature. It is generally characterized by abdominal pain and recurring diarrhea. It occurs in genetically vulnerable populations or individuals due to inappropriate and intensified immune response to commensal bacteria, which leads to intestinal inflammation[1,2]. The two main clinical forms of IBD according to their location and nature of histological modifications/damage caused in the gastrointestinal wall are ulcerative colitis (UC) and Crohn’s disease (CD). UC is limited to mucosal surfaces of the colon, cecum and rectum. Several microbes such as Salmonella spp., Shigella spp., Fusobacterium spp., and adhesive E. coli have been found in the inflamed colon, but no causative microbial species has yet been made responsible for UC[1-4]. CD exhibits transmural inflammation and the mucosal bacteria concentration in CD patients has been found to be twice as high as that of a healthy individual[5,6]. The most predominant bacterial species found in CD was Bacteroides spp., which composed 80% of the total mucosal bacteria as compared to 15% in UC. Also, the role of Listeria monocytogenes and adherent-invasive E. coli (AIEC) has been determined in CD[5,6]. Both UC and CD cause diarrhea with or without bleeding and patients display a weakened tolerance to antigens present in the intestines[1]. Its occurrence is similar in both sexes (men and women), and most cases are recorded in young adults[6,7].
Depending upon the type of microbes, several unfriendly and mutuality interactions occur between the residing gut microbes. The three main mechanisms by which the bacteria communicate with fellow bacteria around them are: combatting, competing and cooperating. Also, formation of biofilms protects the bacterial population from host immune responses and antimicrobials/antibiotics by secretion of extracellular matrix and binding bacteria together in layers. Thick, dense and resistant biofilm formation is very common in IBD patients and is the prime cause of dysbiosis and resistance to treatments/therapies including antibiotics[5,8]. The residing microbes also regulate their gene expression in response to changing microbial cell population and its concentration through a cell-cell communication known as quorum sensing[9].
UC is the chronic and recurring IBD in which severe inflammation and immune responses (T helper cell and production of cytokines) are generated in the intestinal mucosa. The initiation site for ulcers is distal large intestine, and eventually the inflammation moves towards the proximal bowel. UC can severely disturb the quality of life, and if oral medicines are not effective, surgical removal of parts of ulcer affected intestines is obligatory. Several microbes such as Shigella spp., Fusobacterium spp. and adhesive E. coli have been found in the inflamed colon, but no causative microbial species has yet been made responsible for UC[2-4].
CD exhibits transmural inflammation and epithelioid granulomas in the gut tissues with T-helper cell (Th1) responses and elevated levels of interferon gamma. The mucosal bacteria concentration in CD patients has been found to be twice as high as that of a healthy individual[5,6]. Higher levels of antibody IgG is a characteristic of CD patients. The most predominant bacterial species found was Bacteroides spp., which composed 80% of the total mucosal bacteria as compared to 15% in UC. Also, the role of AIEC has been determined in CD[5,6].
Rural populations are less susceptible to IBD. Environmental factors such as poor sanitation, decreased use of antibiotics, rural environment, consumption of whole unrefined food, etc decrease the risk of IBD. That is increased exposure to microbes or infection will lead to low susceptibility for IBD infection. Also, the mucosal immunity in intestines is changed during IBD. UC and CD patients tend to secrete more antibodies (IgG and IgA) against the commensal intestinal microflora and damage their intestinal mucosa[10,11]. Thus, the pathogenesis of IBD is partly understood, and it has also been discovered that multiple factors are associated with IBD such as genetic vulnerability, environmental factors, host-commensal/pathogenic microbe interaction and disturbed mucosal immune responses[12-14].
The maximum occurrence of IBD has been reported in Northern Europe and North America, whereas it is rare amongst Asians and Africans. Incidences of IBD vary depending on race, and its prevalence increases in regions with industrialization. According to the hygiene hypothesis, the autoimmune and inflammatory responses in the body occur due to the absence or low exposure to pathogens in childhood/infancy due to rigorous sanitation practices.
The effect of smoking was guarding against UC and aggravating for CD. That is smoking may improve one’s condition during UC, but CD patients may suffer with a declined quality of life. Other factors, such as domestic hygiene, prenatal events, oral contraceptives, microbial agents and refined sugar consumption require further evaluation to confirm their involvement in IBD and to describe their strength[15-17]. Another accepted factor to increase IBD inflammation and cause relapse of the disease is psychological stress. Any sort of depression, adverse life situations or chronic stress can deteriorate the course of disease[18,19]. Several clinical studies suggest that IBD is not a psychosomatic disease, but stressful life events and depression are associated with high risk of relapse and increased pathogenesis of IBD[20,21].
Heterogeneous geographical distribution and occurrence of ancestral forms of IBD as well as monozygotic twin studies strongly support the genetic component of IBD. High risk in patients having family members with IBD, relatively high risk of siblings acquiring the disease and high concordance rate of monozygotic twins than dizygotic twins of being affected by IBD further strengthen the hypothesis. Therefore, genetic makeup plays an important role in both forms of IBD. Also, several susceptibility genes have been reported. Autophagy genes such as ATG16L1, IRGM and Card15/NOD-2 have been reported to be associated with CD for innate immunity responses. Other candidate genes involved in UC are HERC2, STAT3 and PTPN2[22,23]. Several other susceptible genes such as IL23R, IBD5, NKX2-3, BSN, IL12B and CCNY have been found to be associated with both UC and CD. Table 1 lists some of the genes and their respective roles in IBD. Mutations in these particular genes can lead to abnormal immune response generation in the gut mucosa and adversely affect healthy microbial population in terms of composition and concentration.
| No. | Related disease | Gene(s) | Role(s) ininflammation | Ref. |
| 1 | UC + CD | NOD2/CARD15, CD19, CD11, IL4R | Detection of cytosolic bacterial components | [105] |
| 2 | UC + CD | IL23R, PTGER4 | Generation and maintenance of Th17 cells; prostaglandins signaling | [23,106] |
| 3 | CD | IRGM, IL12B | Autophagy | [22,23] |
| 4 | CD | STAT3, ORMDL3 | Development of T cell response | [22,23] |
| 5 | CD | IL3, IL4, IL5 and IL13,OCTN1, OCTN2, CSF2, SLC22A5 | Mucosal barrier function, cytokines production, Regulation of inflammation | [106] |
| 6 | UC + CD | MST1, BSN, GNAI2 | Regulation of expression of proinflammatory mediators | [105] |
The complex anaerobic environment of the gut is home to several microbial communities including bacteria, archaea and fungi. This microbial diversity in the gut has been studied by the culture independent 16S rRNA studies. These studies indicate that the gut is mainly inhabited by Gram-positive Firmicutes and Gram-negative Bacteroidetes, while Actinomycetes, methanogens and fungi are present in lower quantities[24]. Most Firmicutes were identified as clostridia and are butyrate-producing bacteria. Several Proteobacteria and Actinomycetes were also identified out of which Bifidobacteria (subgroup of Actinomycetes), which has health promoting utilities was found to be 5% of the microbiota. Archaeal diversity consists of Methanobrevibacter smithii and Methanosphaera stadtmanae. Eukaryotic microbes in the human gut consist of Blastocystis sp., (uni- and multicellular protists) and several fungi belonging to Ascomycetes (53.5%) or Basidiomycetes (46.5%) with the majority belonging to the genera Candida albicans, C. glabrata (6%), Penicillium italicum, P. glabrum, P. sacculum, P. verruculosum (61.5%), Saccharomyces cerevisiae, S. cariocanus and S. bayanus (24.1%)[24-26].
Several studies aiming to distinguish residing microbes in a healthy gut and an IBD gut revealed an overall decline in bacterial diversity in IBD patients. Further, a decrease in methanogen diversity and an increase in fungal diversity were noted in the guts of IBD patients[25,26]. Clustering of microbial communities was found over inflamed gut surfaces, but the microbial communities did not vary over the healthy gut tissues.
Different studies have also reported the presence of pathogenic microbes in the gastrointestinal tract of IBD patients. Saccharomyces cerevisiae, Candida albicans, Listeria monocytogenes, Mycobacterium avium subsp. paratuberculosis, Chlamydia pneumonia and AIEC have been reported as the potentially infectious microbes in the spread of CD. In CD, Bacteroides, Peptostreptococcus and Eubacteria are increased, whereas Bifidobacteria numbers are considerably reduced. Moreover, in UC the presence of facultative anaerobic bacteria is amplified. Table 2 lists the infectious agents (viral, bacterial, fungal and parasitic agents) that have been suspected in IBD etiology. E. coli has been reported to induce the release of cytokines in the inflamed IBD gut. AIEC, which is a facultative pathogen, has been reported to cause CD in genetically susceptible hosts[27-29]. Even though these studies have shown relation between pathogens and IBD, specific pathogenic microbes responsible for CD or UC is still contentious.
| Bacteria | Virus | Fungi | |
| Campylobacter spp. | Adenovirus | Saccharomyces cerevisiae | |
| Escherichia coli | Cytomegalovirus | Candida albicans | |
| Helicobacter spp. | Coronavirus | ||
| Legionella spp. | Rotavirus | ||
| Mycobacterium spp. | Measles virus | ||
| Pseudomonas spp. | Paramyxovirus | ||
| Shigella spp. | Epstein-Barr virus | ||
| Yersinia spp. | Parasite | ||
| Bacteroides vulgatus | Borrelia spp. | ||
| Listeria monocytogenes | Treponema spp. | ||
| Staphylococcus spp. | |||
| Streptococcus spp. | |||
| Enterococcus | |||
| Adherent-Invasive E. coli | |||
| Chlamydia spp. (Chlamydia trachomatis) | |||
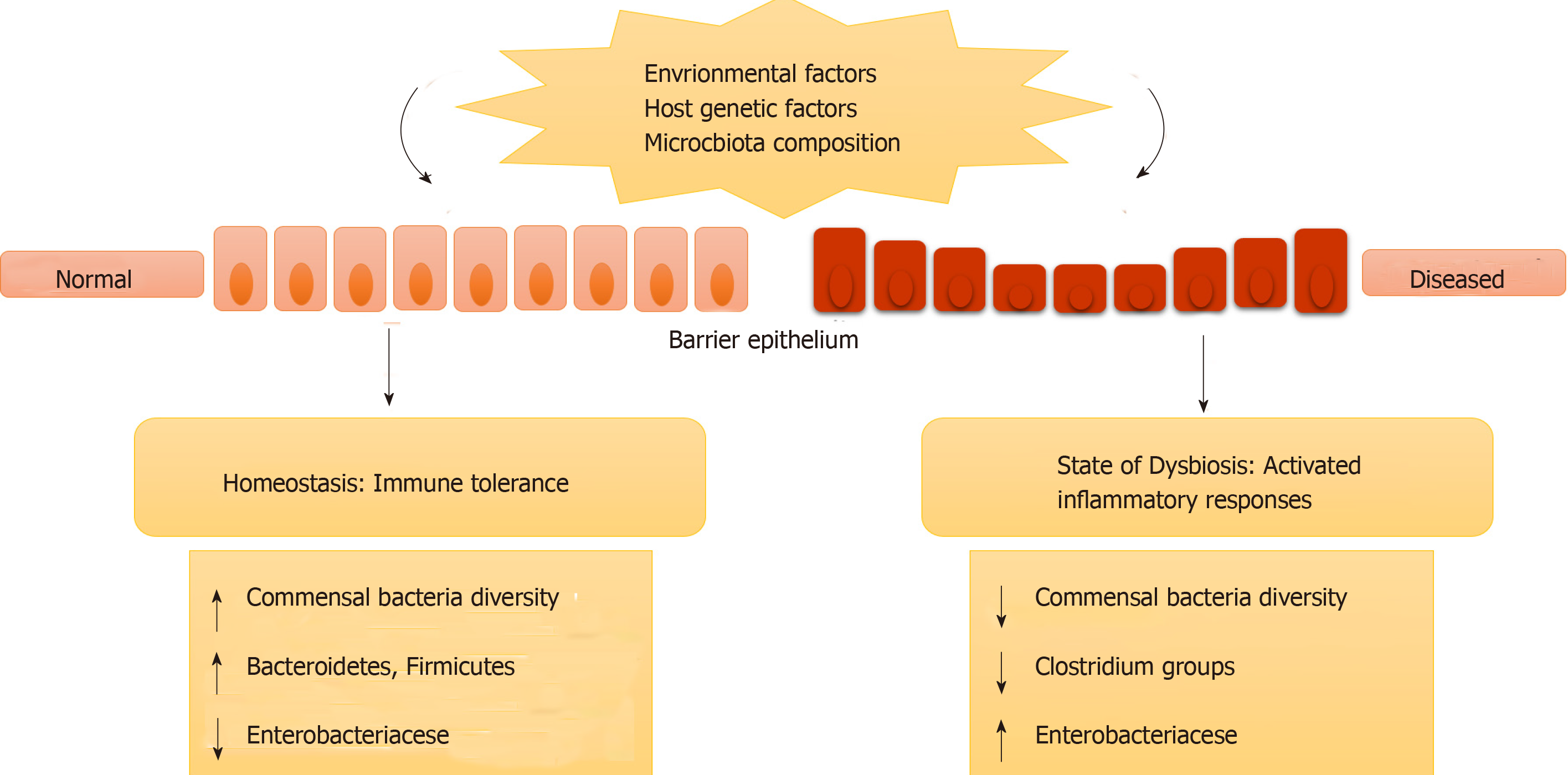
IBD may not be caused by any specific microbial infection; instead it occurs due to a change in the overall residing microbes in the gut (intestinal microbe biofilms). It may also be caused by misrecognition of normal, commensal microbes as foreign leading to immune responses and inflammation. This change in intestinal microbiota (in terms of species and their concentration) or misrecognition by the body is termed genetic dysbiosis[30,31]. This ever-changing gut microbiota can modify the expression of certain genes that are involved in various activities in the intestines. Further, it can cause inflammation and disease in genetically susceptible individuals who have mutations or polymorphism in genes involved in immune responses (as described in previous sections)[30,31]. Figure 1 represents how genetic dysbiosis can lead to diseases.
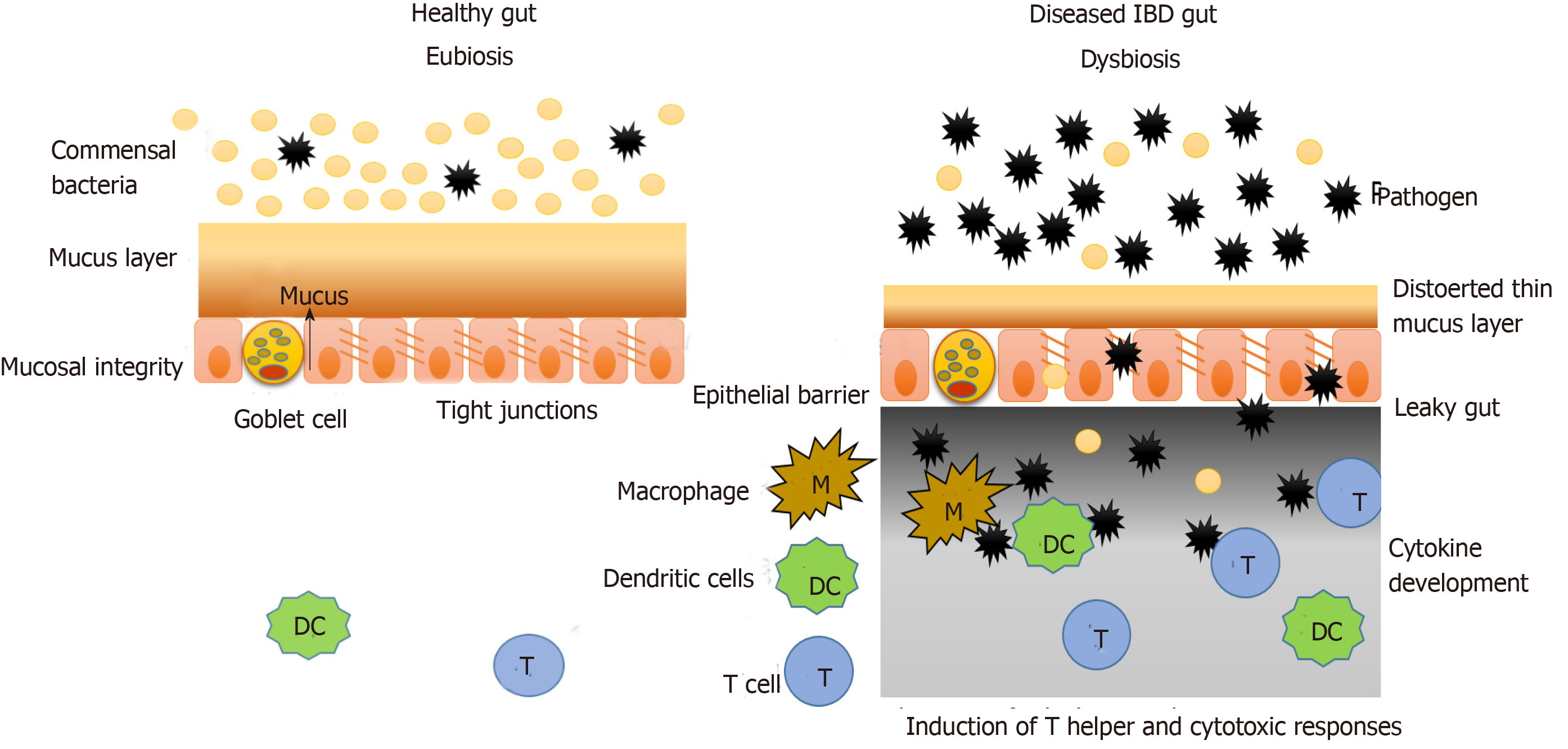
The intestinal mucosal barrier consists of two layers of mucus (inner and outer), which are associated with antimicrobial factors. Under this mucus layer are the gut epithelial cells that are joined together by a network of connecting proteins called tight junctions. Hence, this intestinal mucosal barrier splits the luminal components (food particles, microbes, etc.) from the immune system components (innate and adaptive immunity) (Figure 2)[32].
Under normal conditions the microbiota does not interact with the epithelial cells, other than the controlled interaction in the Peyer’s patches. Under the condition of disturbed microbiota composition or genetic dysbiosis, as is the case with IBD, this mucosal barrier is interrupted, and the interaction between the microbiota and immune system components occurs, which leads to generation of a heightened immune response and inflammation as shown in Figure 2.
In IBD, the mucus layer is permeable and defective due to improper secretion of mucus components by goblet cells (decreased mucin, antimicrobial factors, glycosylation products, etc.). In the case of UC, the goblet cells are depleted in the epithelium, and the mucus layer formed is very thin. Also, the tight junction protein network becomes increasingly penetrable in IBD, which further increases the interaction between immune and luminal components. Such changes are a result of environmental factors and genetic dysbiosis as discussed earlier[33,34].
Various studies have shown that microorganisms that pass the mucus layer and invade the epithelial cells, activate various components of the immune system comprising innate immune responses, adaptive immune responses and autophagy[35,36]. According to Parkes, CD is linked to mutations in autophagy genes such as nucleotide oligomerization domain 2 and ATG16L1. Intracellular pathogens have also been observed to form autophagic vacuoles to prevent interaction with immune components and remain protected[37].
Microorganisms that pass through the damaged tight junctions mesh and reach the lower surface of epithelial cells are recognized by the pattern recognition molecules similar to toll-like receptors present there. This interaction then activates the innate immune responses (including phagocytic cells, natural killer cells, dendritic cells, inflammation-related proteins, cytokines and antimicrobial peptides such as defensins and cathelicidins) and adaptive immune response including several cytokine profiles[38,39]. Fernandes et al[40] reported that in IBD, the expression of regulatory molecules of immune system and toll-like receptors is different from that in a healthy tissue.
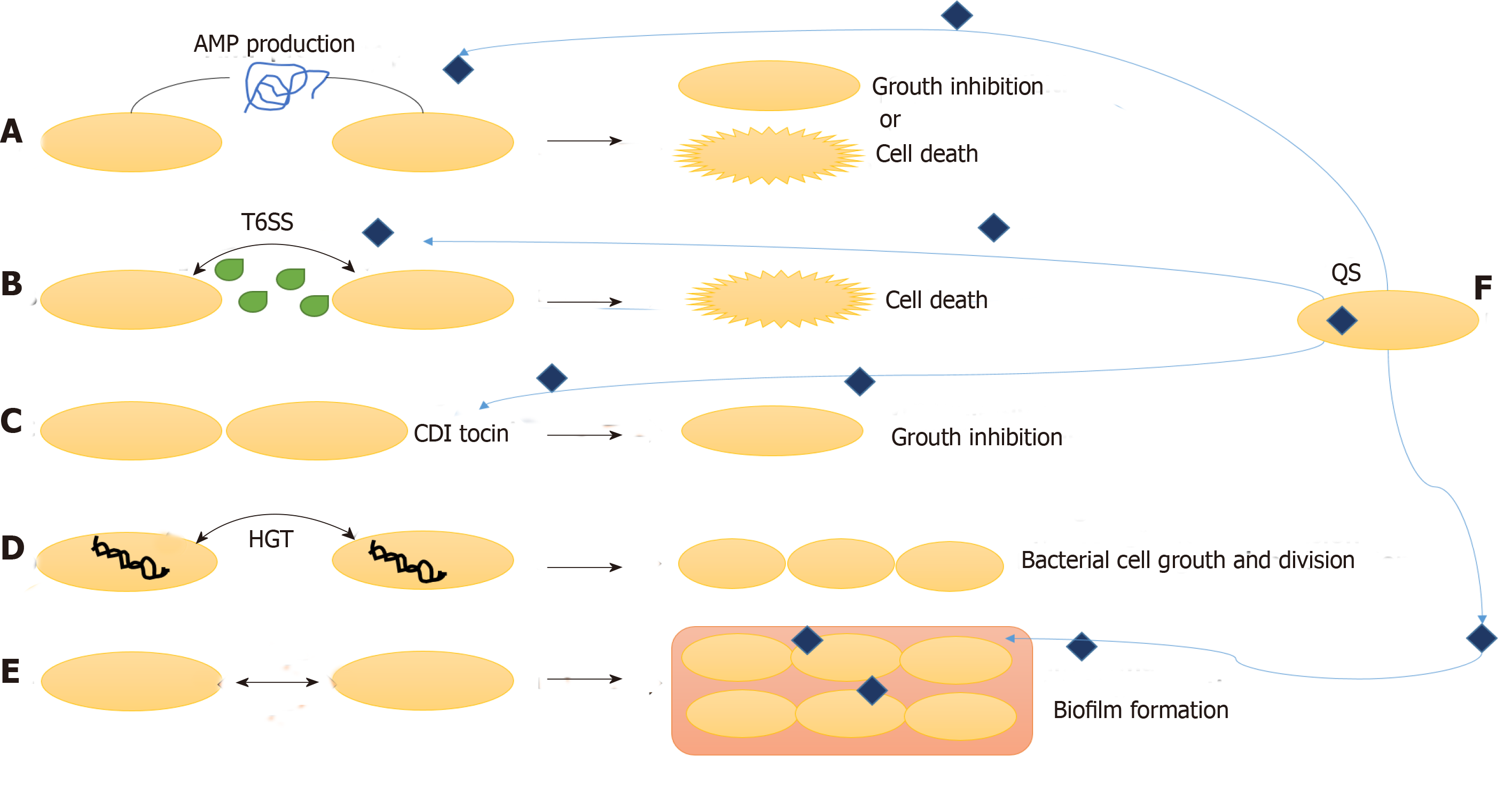
Other than interaction with the host cells, the survival and activity of gut microbiota also depends upon their interaction with the other surrounding microbes. These interactions can be unfriendly or mutualistic, which primarily depends upon the type of microbes. The three main mechanisms by which the bacteria communicate with fellow bacteria around them are: combatting, competing and cooperating (Figure 3).
Gut bacteria combat other microbes by production of antimicrobial peptides (AMP) and specialized secretions. Bacteriocins, colicins and microcins are some AMP that inhibits other microbial invading species without harming the eukaryotic gut cells. Bacteriocins are produced by Gram-positive bacteria, and they fight against other microbes by pore formation in their cell wall. Gram-negative bacteria produce colicins and microcins, which kill other microbes by a variety of strategies such as nuclease activity, inhibition of RNA polymerase, intervention cell wall synthesis and pore formation[41,42]. These bacterial AMPs under the effect of environmental factors control surrounding microbiota composition and concentration.
Other mechanisms of combatting surrounding microbes are contact dependent growth inhibition systems and type VI secretion systems. In contact dependent growth inhibition system, the C terminal end of CdiA protein move in on the target cell and kill it by nuclease activity. Type VI secretion system cells inject toxins in Gram-negative bacteria and eukaryotic cells to combat bacterial competition and cause pathogenesis, respectively[41,42].
One of the essential parameters that governs colonization of microbes and dysbiosis in gut is the competition for carbohydrates. Other than carbohydrates, bacteria also compete for phosphorus, nitrogen, vitamins, trace elements and other vital cofactors. Bacteria encode a variety of transporters for smooth uptake of vitamins and essential cofactors[43,44]. Some bacteria interfere with the uptake of trace elements by other bacteria by secretion of some inhibitors or AMP. For instance, a study by Raffatellu et al[45] showed that Salmonella typhimurium produced lipocalin-2 to inhibit iron uptake by surrounding microbes.
Even after such limiting conditions and competition for food, gut bacteria also communicate and live in cooperation and share by-products. One of the methods of such cooperation is horizontal gene transfer. Horizontal gene transfer is responsible for secretome molecules, which is the most common method to help in production of certain molecules for communal use. For instance, chelators of iron (siderophores), which help in absorption of iron from the surrounding, can be used by any bacteria in the surrounding. In this way, horizontal gene transfer of such genes and public use of these secretions helps the bacterial population to live in cooperation in the gut[46,47].
Formation of biofilms also protects the bacterial population from host immune responses and antimicrobials/antibiotics by secretion of extracellular matrix and binding bacteria together in layers. It prevents the loss of useful secretions and nutrients from a population of microbes living in cooperation with other surrounding microbes. Thick, dense and resistant biofilm formation is very common in IBD patients and is the prime cause of dysbiosis and resistance to treatments/therapies including antibiotics[5,8].
Many cooperative/group behaviors in the gut are governed by the cell-to-cell communication called quorum sensing (QS). The signaling molecules are released in the environment and regulate gene expression in the surrounding communities. QS governs several mechanisms important to survival such as biofilm formation, antibiotic production and expression and release of secretions like type VI secretion systems. Although QS has been known in Lactobacillus species and probiotics, its role in human gut commensal bacteria and pathogens is still not understood well. Further studies are necessary for understanding the microbial communication in both homeostasis and IBD conditions[48-50].
QS occurs by the production and release of extracellular chemical signal molecules called autoinducers by bacteria that are subsequently detected by other bacteria. When a particular threshold concentration of autoinducers is reached, the gene expression is altered in all the bacterial cells residing there. This alteration in gene expression is associated with the variations in cell population density[9]. Several physiological activities such as biofilm formation, virulence, competence, symbiosis, conjugation, antibiotic production, sporulation and motility are controlled by QS communications in bacteria. This communication through autoinducers can take place both within and amongst the bacterial species. Acylated homoserine lactones are produced as autoinducers in Gram-negative bacteria, while Gram-positive bacteria release oligopeptides for communication[9,51].
Gram-negative bacteria V. fischeri demonstrated LuxI/LuxR-type QS. LuxI-like proteins synthesized a particular acylated homoserine lactone signaling molecule, which is an autoinducer. When these homoserine lactone signaling molecules reach a threshold concentration, LuxR-like proteins bind to them and stimulate particular gene transcription[52]. Several other Gram-negative bacteria, including those involved in pathogenesis of IBD regulate gene expression through homologous LuxI/LuxR-type circuit. For instance, in P. aeruginosa, LasI/LasR-RhlI/RhlR virulence system circuit regulates QS. Homoserine lactone signaling autoinducers N-(3-oxododecanoyl)-homoserine lactone and N-(butryl)-homoserine lactone are produced by the action of LasI and RhlI autoinducers. Expression on several virulence factors (lasB, lecA and aprA) in P. aeruginosa is controlled by this circuit[53,54].
Gram-positive bacteria also show QS mediated regulation by secreting peptides as autoinducers. An example of this is Staphylococcus aureus AgrC/AgrA virulence system. An RNA molecule named RNAIII regulates biofilm formation and pathogenicity in S. aureus. The agrBDCA operon regulates the levels of RNAIII. Function of RNAIII is to express several virulence factors in S. aureus[55,56].
In a recent study by Goliska et al[57], pathogenicity of enterococci (Enterococcus faecalis) in IBD was studied. The expression of several genes encoding the virulence factors (gelatinase, extracellular surface protein, cytolysin and hyaluronidase) was studied in IBD patients and control groups. The strains with these virulence genes were also found to have QS genes fsrA-C that regulates the expression of these virulence factors.
Any effective therapeutic strategy to treat IBD should be able to treat all its pathophysiological constituents including inflammation, dysbiosis and leaky gut. The currently available medical treatments of IBD involve three approaches: immune-based therapies, microbiota-based therapies and barrier function-based therapies. Even after these advanced approaches, some patients still require surgical removal of severely affected portions of GI tract[58].
Immune-based therapies directly modulate the immune system to avoid further destruction of commensal microbiota and the gut tissues to relieve inflammation. Furthermore, these therapies also indirectly modulate the mucosal barrier function and the microbiota[58,59]. Immune-based therapy medication includes immunomodulators (e.g., methotrexate, azathioprine), aminosalicylates, corticosteroids, integrin inhibitors and antitumor necrosis factor agents. Several other biologics are being developed to downregulate inflammatory cytokines and their receptors along with rebuilding healthy barrier function[59]. Mesalamine and other salicylates are known to modulate the intestinal microbial composition and decrease microbial adherence and biofilm formation in the gut[39,58].
Microbiota-based therapies are based upon use of antibiotics, probiotics, fecal microbiota transplantation and alteration in diet.
Antibiotics: Antibiotics have been found most effective in CD and only of little use in UC, where it is sometimes used to treat children. Antibiotics such as rifaximin, metronidazole, ciprofloxacin and antimycobacterial agents are being used. These antibiotics decrease the concentration of some pathogens and increase the content of healthy microbiota including Bifidobacteria and F. prausnitzii. Indiscriminate use of antibiotics can lead to severe side effects such as reduction in microbial biodiversity in gut and developing resistance in microbes[60,61].
Probiotics: Probiotics can be defined as the living organism (specific bacterial strains together) that when ingested provides health benefits and protective regulatory activities in the body. Commonly the content of yogurts such as Lactobacilli, Bifidobacteria and Streptococci play the role of probiotics. Probiotics have more efficiently been used in UC and are less effective for CD patients. The most extensively used probiotics for IBD are VSL#3 and E. Coli Nissle 1917. VSL#3 are a combination of eight diverse bacterial strains (four strains of Lactobacilli, three strains of Bifidobacteria and one strain of Streptococcus). These bacteria stimulate the growth of anti-inflammatory bacteria and prevent the growth of pathogenic microbes[62,63].
The use of lactic acid bacteria, which includes Lactobacillus, as probiotics for treatment and prevention of IBD has been proposed in several studies. They are Gram-positive bacteria that normally reside in anaerobic conditions but are facultative aerobes[64,65]. They provide probiotic action by production of bacteriocins, hydrogen peroxide, lactic acid and by forming dense biofilms on gut epithelium, which block all the adhesion sites for attachment of pathogens[65]. Lactobacillus species such as L. casei, L. acidophilus, L. plantarum, L. rhamnosus, L. fermentum, L. amylovorus and L. delbrueckii, etc have been extensively studied for probiotic action against H. pylori, Salmonella, E. coli, Clostridium difficile, Yersinia enterocolitica and Listeria monocytogenes, etc[65,66]. The microbial interactions amongst themselves and with other commensal/pathogenic microbes in the biofilms formed in the gut are regulated by the luxS gene QS mechanisms. Several other proteins that are involved in biofilm formation in different Lactobacillus strains consist of collagen-binding protein, biofilm associated proteins, glycosyl-transferases and mucus-binding protein[66,67]. Most of the Lactobacillus species are bacteriocin producing. A specific class of bacteriocin, called plantaricins are produced by some species such as L. plantarum and L. fermentum. The plantaricin genes which can be identified in Lactobacillus are plnA, plnB, plnC, plnD, plnEF, plnI, plnJ, plnK, plnG, plnN and plantaricin structural genes[68,69].
Fecal microbiota transplantation: This strategy of re-establishing healthy microbiota and treating IBD involves ingestion of healthy donor stool by an ill individual. This has been extensively used with mixed and moderate results in IBD patients. Repetitive infusions are required for sustained results. Keeping in view the chance of transmitting infectious microbes, recent studies suggest the use of artificial stool with limited risk of such infections[70,71].
Alteration in diet: A balanced, high fiber, healthy diet with vegetables, fruits and grains is recommended for IBD patients. Diets with high protein content, excessive red meat and alcohol abuse are discouraged in IBD as it may aggravate or relapse the inflammation. Specific carbohydrate diets that eliminates milk, grains and sugar from the diet has been found effective in some cases of IBD showing an improvement in microbiota diversity[72,73].
This therapy is evolving as the most effective and future approach for treating IBD. If the mucus layer in the gut epithelia is restored, then eventually the immune responses and inflammation can be controlled as well as microbiota diversity can flourish.
Establishing healthy microbiota and inhibition of TNFα and other inflammatory cytokines have recorded an enhanced barrier function in case of IBD. Amino acid, L-glutamine supplements and natural ingredients such as curcumin have been shown to restore tight junctions and hence can be used in treating IBD. Delayed released drugs such as phosphatidylcholine are also being examined extensively as a cure for IBD[39,74-76].
A number of studies with the aim to distinguish between residing microbes in a healthy patient and an IBD patient revealed an overall decline in bacterial diversity in the gut of patients with IBD[25,26]. Clustering of microbial communities was found over inflamed gut surfaces, but the microbial communities may not vary over the healthy gut tissues. Different studies have also reported the presence of pathogenic microbes in gastrointestinal tracts of IBD patients.
Ma et al[77] studied the occurrence of Campylobacter concisus in CD patients colonic biopsy samples and concluded there may be potential involvement of C. concisus in IBD. In another study by Arora et al[78] the risk of C. jejuni infection in UC was established.
Similarly, the presence of several virulence factors such as hyaluronidase, cytolysin and extracellular surface protein in Enterococcus strains isolated from colon tissue samples of children with IBD was observed[57].
There are several studies that link Helicobacter pylori biofilms with gastrointestinal diseases, but it still remains debatable. Mice have developed IBD symptoms in the presence of abnormal immune response and H. pylori infection. No symptoms appeared in germfree conditions that establish single pathogen infection[79]. Halme et al[80] and Luther et al[81] observed contradictory results and demonstrated a protective or inverse relation of Helicobacter towards IBD.
Furthermore, a study by Saebo et al[82] concluded that infection of Yersinia enterocolitica was an activator for IBD. Similarly, Ruckdeschel et al[83] analyzed the impact of virulence factors (such as cytotoxin, invasin and adhesin) of Y. enterocolitica against the action of polymorphonuclear leukocytes.
As far as the role of Listeria monocytogenes in IBD is concerned, studies revealed uncertain outcomes. L. monocytogenes was found proliferating at a higher rate in the colon of patients with IBD than in healthy controls[84]. Listeria forms resistant biofilms on several surfaces including synthetic as well as gut epithelium. The biofilm formation is regulated by QS autoinducer 2 genes luxS and pfs. Chen et al[85] studied the presence of L. monocytogenes in gut biopsies of IBD patients and controls in New Zealand and reported no direct role of L. monocytogenes in causing IBD. Another study by Huijsdens et al[86] also reported a similar result. However, Ooi et al[87] found L. monocytogenes responsible for causing mucosal inflammation in healthy individuals. Another recent study by Miranda-Bautista et al[88] confirmed the presence of L. monocytogenes in patients of CD and concluded that IBD patients are at a serious risk of L. monocytogenes infection.
Numerous studies also demonstrate a relation between Salmonella enterica infection and IBD. It has been reported that IBD and Salmonella infection with coinciding medical and histological symptoms in an elderly woman. This case study reports that colitis may be associated with Salmonella infection[89]. There are various other studies which elucidate the high risk of S. enterica infection in IBD[90-92].
Even though these studies have shown a relationship between pathogens and IBD, a specific pathogenic microbe being responsible for CD or UC is still debatable.
The majority of IBD has been reported in northern Europe and North America, whereas it has been considered rare among Asians and Africans. But, in the past two decades cases of such western diseases have been witnessed in India, including UC and CD, and the number of affected individuals is rising alarmingly[93]. Detailed studies regarding epidemiology and pathogenesis of IBD are lacking in developing countries like India due to neglected health care services, lack of reliable data collection and patient based studies[93,94].
Several studies have reported the possible involvement of specific pathogens in IBD. Verma et al[95] described that different sets of bacteria are responsible for pathogenesis of UC and CD. An increase in Gram-positive Eubacterium and Peptostreptococcus was reported in CD patients but not in UC patients, whereas Campylobacter spp. significantly increased in both CD and UC patients. A study by Patra et al[96] reported the presence of various serogroups of adherent Escherichia coli in rectal biopsies of individuals and associated it with epithelial damage and CD. Banerjee et al[97] reported occurrence of parasitic and viral infections (such as Ankylostoma duodenale, Strongyloides stercoralis, Entamoeba histolytica, etc) in UC patients by analysis of stool samples and rectal biopsies. Iyer et al[98] investigated the relationship between intestinal infections (with Clostridium difficile and other parasites) and UC. They reported that the presence of such infections in UC patients deteriorated the condition and is associated with disease severity. Tripathi et al[99] detected the presence of Salmonella enterica in the stool of 80% of UC patients and concluded an active infection of Salmonella sp. in IBD.
Another recent study examined the gut microbiota profile of vegetarian and non-vegetarian healthy individuals and IBD individuals and colon carcinoma and reported that IBD and colon cancer patients had higher proportions of Bacteroidetes than Firmicutes[100]. Several studies discuss the food-borne infection of Listeria monocytogenes in India. In another study[101] established the expression of virulence gene regulator prfA in L. monocytogenes infecting mammalian host, which helps in formation of resistant biofilms and virulence. Expression of prfA was also studied in co-cultured biofilms with B. subtilis, which made L. monocytogenes less virulent in such biofilms.
Several studies have also been conducted regarding the anti-biofilm nature and probiotic properties of lactic acid bacteria against pathogenic bacteria. Kaur et al[102] reported the action of Lactobacillus spp. against diarrhea causing Vibrio cholerae, which causes high mortality in developing countries like India. Another study examined the antimicrobial action of Lactobacillus spp. from curd and milk against several human infecting pathogens such as Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Klebsiella pneumonia, Salmonella enterica serovar Typhi, Bacillus cereus, Listeria monocytogenes and Shigella flexneri[103]. Furthermore, in some studies it has been demonstrated the antimicrobial action of thirteen Lactobacillus isolates from the gastrointestinal track of broiler chicken against Escherichia coli and pathogenic fungi such as Aspergillus niger, Aspergillus flavus, Penicillium expansum, Penicillium roqueforti and Candida albicans, etc[104-106]. The lactic acid bacteria showed effective inhibition against these pathogens.
The understanding of gut microbiota in terms of composition and its complex interaction in both normal and diseased conditions has been assisted by the use of molecular, metagenomics and meta transcriptomics techniques. Even though several studies (as mentioned above) have shown relationships between pathogens and IBD, a specific pathogenic microbe being responsible for CD or UC is still debatable. Also, detailed research has been done for gut microbiota and IBD, but the molecular basis of their virulence and biofilm formation still remains to be discovered. This has hampered the generation of effective therapeutic agents for IBD that would benefit a high percentage of the world population affected by the disease. Therefore, we need to understand the pathogenesis of IBD and develop strong treatment strategies against it.
Manuscript source: Invited manuscript
Specialty type: Pharmacology and Pharmacy
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bourgoin SG, Huang X, Zhai KF S-Editor: Cui LJ L-Editor: Filipodia E-Editor: Wu YXJ
| 1. | Steed H, Macfarlane GT, Macfarlane S. Prebiotics, synbiotics and inflammatory bowel disease. Mol Nutr Food Res. 2008;52:898-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 2. | Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15:1528-1536. [PubMed] |
| 3. | Reiff C, Kelly D. Inflammatory bowel disease, gut bacteria and probiotic therapy. Int J Med Microbiol. 2010;300:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 4. | Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn's disease and ulcerative colitis - an overview. J Physiol Pharmacol. 2009;60 Suppl 6:61-71. [PubMed] |
| 5. | Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380-3389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 664] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 6. | Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 480] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 7. | Loftus EV. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504-1517. [PubMed] |
| 8. | Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24:50-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 229] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 9. | Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3283] [Cited by in RCA: 2881] [Article Influence: 125.3] [Reference Citation Analysis (0)] |
| 10. | Pirzer U, Schönhaar A, Fleischer B, Hermann E, Meyer zum Büschenfelde KH. Reactivity of infiltrating T lymphocytes with microbial antigens in Crohn's disease. Lancet. 1991;338:1238-1239. [PubMed] |
| 11. | Macpherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365-375. [PubMed] |
| 12. | Orholm M, Munkholm P, Langholz E, Nielsen OH, Sørensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991;324:84-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 411] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 13. | Shanahan F. Inflammatory bowel disease: immunodiagnostics, immunotherapeutics, and ecotherapeutics. Gastroenterology. 2001;120:622-635. [PubMed] |
| 14. | Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1881] [Cited by in RCA: 1789] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 15. | Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182-205. [PubMed] |
| 16. | Rhodes J, Thomas GA. Smoking: good or bad for inflammatory bowel disease? Gastroenterology. 1994;106:807-810. [PubMed] |
| 17. | Koutroubakis I, Manousos ON, Meuwissen SG, Pena AS. Environmental risk factors in inflammatory bowel disease. Hepatogastroenterology. 1996;43:381-393. [PubMed] |
| 18. | Mawdsley JE, Rampton DS. Psychological stress in IBD: new insights into pathogenic and therapeutic implications. Gut. 2005;54:1481-1491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 446] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 19. | Cámara RJ, Ziegler R, Begré S, Schoepfer AM, von Känel R; Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS) group. The role of psychological stress in inflammatory bowel disease: quality assessment of methods of 18 prospective studies and suggestions for future research. Digestion. 2009;80:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 20. | Bitton A, Sewitch MJ, Peppercorn MA, deB Edwardes MD, Shah S, Ransil B, Locke SE. Psychosocial determinants of relapse in ulcerative colitis: a longitudinal study. Am J Gastroenterol. 2003;98:2203-2208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 147] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Mardini HE, Kip KE, Wilson JW. Crohn's disease: a two-year prospective study of the association between psychological distress and disease activity. Dig Dis Sci. 2004;49:492-497. [PubMed] |
| 22. | Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Häsler R, Little RD, Huse K, Ruether A, Balschun T, Wittig M, Elsharawy A, Mayr G, Albrecht M, Prescott NJ, Onnie CM, Fournier H, Keith T, Radelof U, Platzer M, Mathew CG, Stoll M, Krawczak M, Nürnberg P, Schreiber S. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One. 2007;2:e691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 23. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ; NIDDK IBD Genetics Consortium, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E; Belgian-French IBD Consortium; Wellcome Trust Case Control Consortium, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2114] [Cited by in RCA: 2044] [Article Influence: 120.2] [Reference Citation Analysis (0)] |
| 24. | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5700] [Cited by in RCA: 5577] [Article Influence: 278.9] [Reference Citation Analysis (2)] |
| 25. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3419] [Article Influence: 189.9] [Reference Citation Analysis (1)] |
| 26. | Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 319] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 27. | Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis as a prerequisite for IBD. Gut. 2004;53:1057. [PubMed] |
| 28. | Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529-5537. [PubMed] |
| 29. | Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hébuterne X, Hofman P, Darfeuille-Michaud A. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355-1362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 30. | Bibiloni R, Mangold M, Madsen KL, Fedorak RN, Tannock GW. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J Med Microbiol. 2006;55:1141-1149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 163] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 31. | Nibali L, Henderson B, Sadiq ST, Donos N. Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases. J Oral Microbiol. 2014;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 2698] [Article Influence: 168.6] [Reference Citation Analysis (0)] |
| 33. | Merga Y, Campbell BJ, Rhodes JM. Mucosal barrier, bacteria and inflammatory bowel disease: possibilities for therapy. Dig Dis. 2014;32:475-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 34. | Michielan A, D'Incà R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediators Inflamm. 2015;2015:628157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 485] [Article Influence: 48.5] [Reference Citation Analysis (1)] |
| 35. | Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729-1737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 405] [Article Influence: 28.9] [Reference Citation Analysis (1)] |
| 36. | Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 2014;260:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 37. | Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. 2012;30:330-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 38. | Man SM, Kaakoush NO, Mitchell HM. The role of bacteria and pattern-recognition receptors in Crohn's disease. Nat Rev Gastroenterol Hepatol. 2011;8:152-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 39. | Vindigni SM, Zisman TL, Suskind DL, Damman CJ. The intestinal microbiome, barrier function, and immune system in inflammatory bowel disease: a tripartite pathophysiological circuit with implications for new therapeutic directions. Therap Adv Gastroenterol. 2016;9:606-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 145] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 40. | Fernandes P, MacSharry J, Darby T, Fanning A, Shanahan F, Houston A, Brint E. Differential expression of key regulators of Toll-like receptors in ulcerative colitis and Crohn's disease: a role for Tollip and peroxisome proliferator-activated receptor gamma? Clin Exp Immunol. 2016;183:358-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 41. | Braun V, Patzer SI. Intercellular communication by related bacterial protein toxins: colicins, contact-dependent inhibitors, and proteins exported by the type VI secretion system. FEMS Microbiol Lett. 2013;345:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 42. | Rebuffat S. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans. 2012;40:1456-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 43. | Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B₁₂ analogs and compete in the gut. Cell Host Microbe. 2014;15:47-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 210] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 44. | Seth EC, Taga ME. Nutrient cross-feeding in the microbial world. Front Microbiol. 2014;5:350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 201] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 45. | Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 427] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 46. | Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430:1024-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 683] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 47. | Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EP. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol. 2009;19:1683-1691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 170] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 48. | van Hemert S, Meijerink M, Molenaar D, Bron PA, de Vos P, Kleerebezem M, Wells JM, Marco ML. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 2010;10:293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 49. | Dickschat JS. Quorum sensing and bacterial biofilms. Nat Prod Rep. 2010;27:343-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 50. | Lukás F, Gorenc G, Kopecný J. Detection of possible AI-2-mediated quorum sensing system in commensal intestinal bacteria. Folia Microbiol (Praha). 2008;53:221-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 51. | Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101-104. [PubMed] |
| 52. | Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773-781. [PubMed] |
| 53. | Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490-1494. [PubMed] |
| 54. | Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:6424-6428. [PubMed] |
| 55. | Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446-458. [PubMed] |
| 56. | Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569-4577. [PubMed] |
| 57. | Goliska E, Tomusiak A, Gosiewski T, Więcek G, Machul A, Mikołajczyk D, Bulanda M, Heczko PB, Strus M. Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J Gastroenterol. 2013;19:3562-3572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 58. | Andrews CN, Griffiths TA, Kaufman J, Vergnolle N, Surette MG, Rioux KP. Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2011;34:374-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 59. | Peng JC, Shen J, Ran ZH. Novel agents in the future: Therapy beyond anti-TNF agents in inflammatory bowel disease. J Dig Dis. 2014;15:585-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Bejaoui M, Sokol H, Marteau P. Targeting the Microbiome in Inflammatory Bowel Disease: Critical Evaluation of Current Concepts and Moving to New Horizons. Dig Dis. 2015;33 Suppl 1:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 61. | Maccaferri S, Vitali B, Klinder A, Kolida S, Ndagijimana M, Laghi L, Calanni F, Brigidi P, Gibson GR, Costabile A. Rifaximin modulates the colonic microbiota of patients with Crohn's disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother. 2010;65:2556-2565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 142] [Article Influence: 9.5] [Reference Citation Analysis (1)] |
| 62. | Cammarota G, Ianiro G, Cianci R, Bibbò S, Gasbarrini A, Currò D. The involvement of gut microbiota in inflammatory bowel disease pathogenesis: potential for therapy. Pharmacol Ther. 2015;149:191-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 63. | Henker J, Müller S, Laass MW, Schreiner A, Schulze J. Probiotic Escherichia coli Nissle 1917 (EcN) for successful remission maintenance of ulcerative colitis in children and adolescents: an open-label pilot study. Z Gastroenterol. 2008;46:874-875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 64. | Holzapfel WH, Haberer P, Geisen R, Björkroth J, Schillinger U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am J Clin Nutr. 2001;73:365S-373S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 326] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 65. | Figueroa-González I, Quijano G, Ramírez G, Cruz-Guerrero A. Probiotics and prebiotics--perspectives and challenges. J Sci Food Agric. 2011;91:1341-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 66. | Salas-Jara MJ, Ilabaca A, Vega M, García A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms. 2016;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 207] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 67. | Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4645-4652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 261] [Cited by in RCA: 244] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 68. | Ben Omar N, Abriouel H, Keleke S, Sánchez Valenzuela A, Martínez-Cañamero M, Lucas López R, Ortega E, Gálvez A. Bacteriocin-producing Lactobacillus strains isolated from poto poto, a Congolese fermented maize product, and genetic fingerprinting of their plantaricin operons. Int J Food Microbiol. 2008;127:18-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Mokoena MP. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules. 2017;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 225] [Cited by in RCA: 328] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 70. | Borody TJ, Paramsothy S, Agrawal G. Fecal microbiota transplantation: indications, methods, evidence, and future directions. Curr Gastroenterol Rep. 2013;15:337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 189] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 71. | Petrof EO, Khoruts A. From stool transplants to next-generation microbiota therapeutics. Gastroenterology. 2014;146:1573-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 72. | Cohen SA, Gold BD, Oliva S, Lewis J, Stallworth A, Koch B, Eshee L, Mason D. Clinical and mucosal improvement with specific carbohydrate diet in pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2014;59:516-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 73. | Tilg H, Kaser A. Diet and relapsing ulcerative colitis: take off the meat? Gut. 2004;53:1399-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 74. | Karner M, Kocjan A, Stein J, Schreiber S, von Boyen G, Uebel P, Schmidt C, Kupcinskas L, Dina I, Zuelch F, Keilhauer G, Stremmel W. First multicenter study of modified release phosphatidylcholine "LT-02" in ulcerative colitis: a randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol. 2014;109:1041-1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 75. | Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 853] [Article Influence: 60.9] [Reference Citation Analysis (1)] |
| 76. | Wang N, Wang G, Hao J, Ma J, Wang Y, Jiang X, Jiang H. Curcumin ameliorates hydrogen peroxide-induced epithelial barrier disruption by upregulating heme oxygenase-1 expression in human intestinal epithelial cells. Dig Dis Sci. 2012;57:1792-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 77. | Ma R, Liu F, Yap SF, Lee H, Leong RW, Riordan SM, Grimm MC, Zhang L. The Growth and Protein Expression of Inflammatory Bowel Disease-Associated Campylobacter concisus Is Affected by the Derivatives of the Food Additive Fumaric Acid. Front Microbiol. 2018;9:896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 78. | Arora Z, Mukewar S, Wu X, Shen B. Risk factors and clinical implication of superimposed Campylobacter jejuni infection in patients with underlying ulcerative colitis. Gastroenterol Rep (Oxf). 2016;4:287-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 79. | Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126-3131. [PubMed] |
| 80. | Halme L, Rautelin H, Leidenius M, Kosunen TU. Inverse correlation between Helicobacter pylori infection and inflammatory bowel disease. J Clin Pathol. 1996;49:65-67. [PubMed] |
| 81. | Luther J, Dave M, Higgins PD, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis. 2010;16:1077-1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 167] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 82. | Saebo A, Vik E, Lange OJ, Matuszkiewicz L. Inflammatory bowel disease associated with Yersinia enterocolitica O:3 infection. Eur J Intern Med. 2005;16:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 83. | Ruckdeschel K, Roggenkamp A, Schubert S, Heesemann J. Differential contribution of Yersinia enterocolitica virulence factors to evasion of microbicidal action of neutrophils. Infect Immun. 1996;64:724-733. [PubMed] |
| 84. | Chiba M, Fukushima T, Koganei K, Nakamura N, Masamune O. Listeria monocytogenes in the colon in a case of fulminant ulcerative colitis. Scand J Gastroenterol. 1998;33:778-782. [PubMed] |
| 85. | Chen W, Li D, Paulus B, Wilson I, Chadwick VS. Detection of Listeria monocytogenes by polymerase chain reaction in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. J Gastroenterol Hepatol. 2000;15:1145-1150. [PubMed] |
| 86. | Huijsdens XW, Linskens RK, Taspinar H, Meuwissen SG, Vandenbroucke-Grauls CM, Savelkoul PH. Listeria monocytogenes and inflammatory bowel disease: detection of Listeria species in intestinal mucosal biopsies by real-time PCR. Scand J Gastroenterol. 2003;38:332-333. [PubMed] |
| 87. | Ooi ST, Lorber B. Gastroenteritis due to Listeria monocytogenes. Clin Infect Dis. 2005;40:1327-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 147] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 88. | Miranda-Bautista J, Padilla-Suárez C, Bouza E, Muñoz P, Menchén L, Marín-Jiménez I. Listeria monocytogenes infection in inflammatory bowel disease patients: case series and review of the literature. Eur J Gastroenterol Hepatol. 2014;26:1247-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 89. | Nimmons D, Limdi JK. Elderly patients and inflammatory bowel disease. World J Gastrointest Pharmacol Ther. 2016;7:51-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 90. | Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One. 2011;6:e20338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 91. | Gradel KO, Nielsen HL, Schønheyder HC, Ejlertsen T, Kristensen B, Nielsen H. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 316] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 92. | Ternhag A, Törner A, Svensson A, Ekdahl K, Giesecke J. Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis. 2008;14:143-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 93. | Kedia S, Ahuja V. Epidemiology of Inflammatory Bowel Disease in India: The Great Shift East. Inflamm Intest Dis. 2017;2:102-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 94. | Ray G. Inflammatory bowel disease in India - Past, present and future. World J Gastroenterol. 2016;22:8123-8136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 95. | Verma R, Verma AK, Ahuja V, Paul J. Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J Clin Microbiol. 2010;48:4279-4282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 96. | Patra S, Samal SC, Kang G, Pulimood A, Mathan M, Ramakrishna BS. Adherent Escherichia coli in colorectal mucosal biopsies: a histological and ultrastructural evaluation. Indian J Pathol Microbiol. 2012;55:485-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 97. | Banerjee D, Deb R, Dar L, Mirdha BR, Pati SK, Thareja S, Falodia S, Ahuja V. High frequency of parasitic and viral stool pathogens in patients with active ulcerative colitis: report from a tropical country. Scand J Gastroenterol. 2009;44:325-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 98. | Iyer VH, Augustine J, Pulimood AB, Ajjampur SS, Ramakrishna BS. Correlation between coinfection with parasites, cytomegalovirus, and Clostridium difficile and disease severity in patients with ulcerative colitis. Indian J Gastroenterol. 2013;32:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 99. | Tripathi MK, Pratap CB, Dixit VK, Singh TB, Shukla SK, Jain AK, Nath G. Ulcerative Colitis and Its Association with Salmonella Species. Interdiscip Perspect Infect Dis. 2016;2016:5854285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 100. | Bamola VD, Ghosh A, Kapardar RK, Lal B, Cheema S, Sarma P, Chaudhry R. Gut microbial diversity in health and disease: experience of healthy Indian subjects, and colon carcinoma and inflammatory bowel disease patients. Microb Ecol Health Dis. 2017;28:1322447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 101. | Tirumalai PS, Prakash S. Expression of virulence genes by Listeria monocytogenes J0161 in natural environment. Braz J Microbiol. 2012;43:834-843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 102. | Kaur S, Sharma P, Kalia N, Singh J, Kaur S. Anti-biofilm Properties of the Fecal Probiotic Lactobacilli Against Vibrio spp. Front Cell Infect Microbiol. 2018;8:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 103. | Sharma C, Singh BP, Thakur N, Gulati S, Gupta S, Mishra SK, Panwar H. Antibacterial effects of Lactobacillus isolates of curd and human milk origin against food-borne and human pathogens. 3 Biotech. 2017;7:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 104. | Russo P, Arena MP, Fiocco D, Capozzi V, Drider D, Spano G. Lactobacillus plantarum with broad antifungal activity: A promising approach to increase safety and shelf-life of cereal-based products. Int J Food Microbiol. 2017;247:48-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 156] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 105. | Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C, Barrett JC, Cummings FR, Drummond H, Lees CW, Onnie CM, Hanson CE, Blaszczyk K, Inouye M, Ewels P, Ravindrarajah R, Keniry A, Hunt S, Carter M, Watkins N, Ouwehand W, Lewis CM, Cardon L; Wellcome Trust Case Control Consortium, Lobo A, Forbes A, Sanderson J, Jewell DP, Mansfield JC, Deloukas P, Mathew CG, Parkes M, Satsangi J. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 106. | Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1435] [Cited by in RCA: 1388] [Article Influence: 77.1] [Reference Citation Analysis (0)] |