INTRODUCTION
Autoimmune diseases (AID), which jointly affect 5% to 10% of the general population in developed countries, are the third leading cause of morbidity. Onset of AID occurs from childhood to late adulthood and disproportionately affects women at all ages. AID can affect persons of all racial, ethnic, and socioeconomic groups, although the impact of racial background varies among AID. Over 80 different AID have been characterized and they are defined by the presence of autoreactive lymphocytes (T and B) and autoantibodies. Since the 1950s, AID have been divided into two classes: organ-specific AID in which the immune-mediated injuries are localized to a single organ or tissue, such as the pancreas in type 1 diabetes (T1D); and non-organ-specific (systemic) AID in which the immune reactions are directed against many different organs and tissues resulting in widespread injury, such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and primary Sjögren’s syndrome (pSS)[1]. No definitive diagnostic markers exist for most AID and classification criteria (but not diagnostic criteria) were introduced to circumvent this difficulty in order to categorize the patients into groups. However, classification criteria suffer from many limitations, one of which is a lack of sensitivity since, in some cases, it takes one to two decades to verify the status relative to the criteria. In fact, AID should be viewed as an “immunological disease continuum” driven first by a deregulation of the immune system and leading, in the end, to a classic AID[2]. Another important limitation of this classification is related to the therapeutic responses, which can be extremely variable from one patient to another, especially when highly specific new biotherapy treatments are used, thus suggesting that a specific AID can result from several pathways.
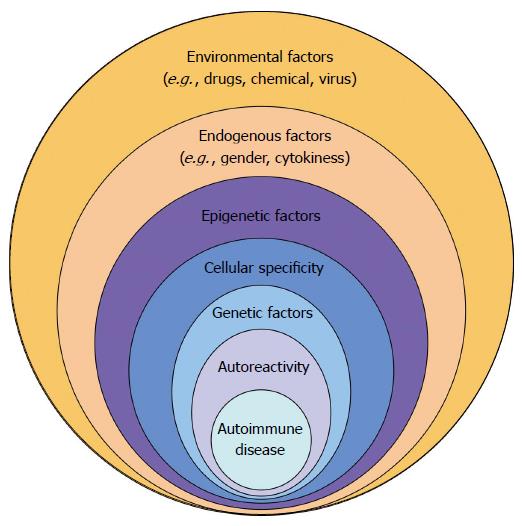
At the beginning of 2015, the ENCODE program (Encyclopedia of DNA elements) has shown the close connection between epigenetic factors, genetic factors and the different immune cell subsets[3]. Indeed, Farh et al[3] demonstrated that susceptibility genes associated with AID were concentrated, not at the level of protein-coding genes, but rather in non-coding regions (90%). These new regions were characterized and it was revealed that they were the major target of epigenetic modifications sensitive to endogenous factors, and were, in particular, specific to one or more immune cell subsets. As a consequence (Figure 1), the development of AID can be viewed as a multi-step process that involves environmental and endogenous factors, both leading to deregulation of the epigenetic machinery, which in turn specifically affects the immune system and/or the target organs. The cellular impact of the epigenetic dysregulation can be amplified in the case of genetic mutations. As a consequence autoreactive lymphocytes and autoantibodies are produced leading subsequently to the development of the AID.
Figure 1 Multiple factors are necessary to promote autoimmune diseases in genetically predisposed individuals.
ENVIRONMENTAL FACTORS
Several lines of evidence strongly support a critical and pathogenic role for environmental factors in AID development. An elegant way in which to evaluate the environmental factor hypothesis consists of determining the disease concordance rate (CR) between monozygotic twins (MZ). Applied to several AID, the use of MZ twins has been decisive in characterizing those AID with high environmental components, such as systemic sclerosis (SSc; CR 5%) and RA (CR 5%-20%), compared to AID with high genetic components, such as Crohn’s disease (CD; CR 75%-85%) and psoriasis (CR 40%-65%). In between, SLE (CR 24%-57%), pSS (CR 15%-25%), T1D (CR 30%-50%) have an intermediate CR, indicating a scenario in which genetics and environmental factors are equally involved[4].
MZ twin studies discordant for AID have further demonstrated the importance of epigenetic modifications in the affected twin. Applied to SLE, studies have revealed that peripheral blood leukocyte CpG methylation profiles are demethylated in MZ twins discordant for SLE[5]. Differences in DNA methylation between unaffected and affected twins were also reported using peripheral lymphocytes from SSc and primary biliary cirrhosis patients[6,7], in T cells from psoriasis patients[8], and in monocytes from T1D patients[9]. Such observations have to be interpreted in relation to the observations that DNA methylation patterns and histone acetylation profiles diverge with age in healthy MZ, with the greatest differences in those who have distinctly different lifestyles[10]. Direct evidence has been provided previously that drugs, UV light, cigarette smoking as well as chemicals can induce important epigenetic changes[11].
ENDOGENOUS FACTORS
Targeting the proinflammatory cytokines of the innate immune system, such as TNF-alpha and IL-6, has been very effective in management of AID. T cell and B cell directed therapies, such as the anti-CD20 monoclonal antibody (mAb) B-cell-depleting agent rituximab, are also effective[12,13]. Treating patients with biotherapy has powerful influence on the epigenetic machinery. In RA, treating patients with anti-TNF-alpha mAb therapy was shown to increase the histone acetyltransferase/histone deacetylase (HAT/HDAC) ratio through the control of HDAC1, whereas rituximab increased nuclear activity of both HAT and HDAC[14]. Similarly, we have observed that treating pSS patients with rituximab restores global DNA methylation in salivary gland epithelial cells[15], and that using anti-IL6 receptor mAb (itolizumab) repairs the defective DNMT1 pathway and DNA methylation in SLE B cells[16].
EPIGENETICS
The pioneering argument supporting epigenetic dysregulation in AID was provided by analyzing the effects of two drugs, procainamide, used to treat cardiac arrhythmias, and hydralazine, used to treat hypertension, which are associated with a 5% and 20% incidence risk for developing drug-induced lupus, respectively. It has been demonstrated for both drugs that they interfere with CpG DNA methylation directly (procainamide) by inhibiting DNMT1, the main DNA methyl transferase, or indirectly (hydralazine) by blocking the MEK/Erk activation pathway that controls DNMT1 expression[17]. Another argument to consider with regards to epigenetic dysregulation is related to the abnormal detection of retrotransposons in AID[18,19]. Retrotransposons, which comprise 45% of the genome, are known to affect the human genome in different ways: first by generating insertions or modifications into protein-coding regions that can result in genetic diseases; second by introducing long-range gene regulatory elements (enhancer, repressor/silencer, insulator); third by producing transposon-derived proteins with antigenic and/or immunosuppressive functions; and, in some cases, by promoting fusion proteins[20,21]. One example is related to the human T cell leukaemia related endogenous retrovirus (HRES-1) that is inserted in the long arm of chromosome 1 at position 1q42. HRES-1 expression is controlled at the epigenetic level by DNA methylation[22] and an association between SLE and HRES-1 polymorphisms has been described[23]. When expressed, HRES-1 has the capacity to produce a p38gag protein that can, in turn, induce the development of Abs as observed in 52% of patients with SLE in contrast to 3.6% in healthy donors[24].
CELLULAR SPECIFICITY
Defects in CpG DNA methylation are usually seen in AID with the particularity that distinct cell-types are involved. In pSS, DNA demethylation involves epithelial cells with a reduction of DNMT1 and an increase of Gadd45alpha[15]. In SLE, T cells, B cells and monocytes are demethylated due to a decrease in the enzymatic activity of DNMT1 and DNMT3a, and an increase in the enzymatic activity of Gadd45alpha and MBD4[25]. In RA, synoviocytes are demethylated which is associated with a reduction in DNMT1, MBD2 and the methyl donor S-adenosylmethionine (SAM). In contrast, DNA hypermethylation has been described in endothelial cells from SSc patients and keratinocytes from patients with psoriasis. Changes in histone modifications are also detected in AID. Global H3 and H4 hypoacetylation and hyper H3k9 trimethylation, plus a negative correlation of H3 acetylation with the disease activity, characterize CD4+ T cells from SLE patients. In synovial cells from RA patients, HDAC1 overexpression has been demonstrated.
GENETICS
The first revelation of strong associations between AID and genetic factors was established with the human leukocyte antigen (HLA) region. The HLA super-locus is present in chromosomal position 6p21, spanning approximately 4 megabases, and contains 132 protein-coding genes composed partly of the classic HLA class I (HLA-A, -B, and -C) and class II (HLA-DP, -DQ, and -DR) genes which are involved in antigen processing and presentation to CD8 and CD4 T cells, respectively. Furthermore, the HLA locus is characterized by an exceptionally high degree of polymorphism, and most of the HLA variants characterized to date have relative associations with specific AID when present.
Moreover, with the development of the genome-wide association study project, up to one hundred non-HLA genetic associations were characterized in AID[26]. The list is not exhaustive as next-generation sequencing technologies contributed to the characterization of rare single nucleotide polymorphisms (SNPs) and additional SNPs, copy number variations and microsatellites associated with AID. Non-HLA gene associations are not disease specific and odds ratios (OR) are usually modest (1.1 to 1.8) in contrast to the HLA genes (OR, usually > 2). Another important point is the distribution of the AID-associated causal genetic factors that are present within protein coding regions (5%), splice junctions (0.2%), promoters (8%), 3’ untranslated region (UTR, 3%) and, last but not least, within cell-specific long range gene-regulatory sequences (60%)[3].
AUTOREACTIVITY
Lymphocyte differentiation starts in the bone marrow from hematopoietic multipotent stem cells, and specific epigenetic signatures accompany each stage. Thus, coordinated transcriptional regulation by DNA methylation/demethylation and histone post-translational modifications (e.g., H3K4me3, H3K27me3) is necessary for cytokine polarization in T helper cells, CD8 cytotoxic T cell differentiation, the selection of regulatory T cells and possibly B cell autoreactivity via rearrangement of the antigen receptor gene[27,28]. Indeed, blocking DNA methylation with specific inhibitors in activated B- and T-cells leads to the emergence of autoreactive lymphocytes and development of a SLE/pSS-like disease when passively transferred to normal mice[29,30]. The implication of DNA methylation in the control of lymphocyte autoreactivity is reinforced by the analysis of lymphocytes in the immunodeficiency centrometric region instability and facial anomalies syndrome, which is characterized by a non-functional DNMT3b[31]. In these patients, there is an absence of mature T cells and accumulation of immature B cells with an autoreactive B cell receptor.
THERAPEUTIC POSSIBILITIES
The beneficial effect of HDAC inhibitors (HDACi) has been demonstrated in several autoimmune models. When used in the spontaneous lupus mouse model MRL/lpr/lpr, trichostatin A, a known inhibitor of HDACs, leads to a decrease in renal disease by modulating cytokines, but the autoAb production was not affected[32]. HDACi ameliorates other autoimmune animal models, including: the rat collagen-induced arthritis; the multiple sclerosis model, experimental autoimmune encephalomyelitis; and the T1D model. In addition, it is suspected that epigenetically modified chromatin complexes represent an important immunogenic stimulus leading to autoantibody production[33].
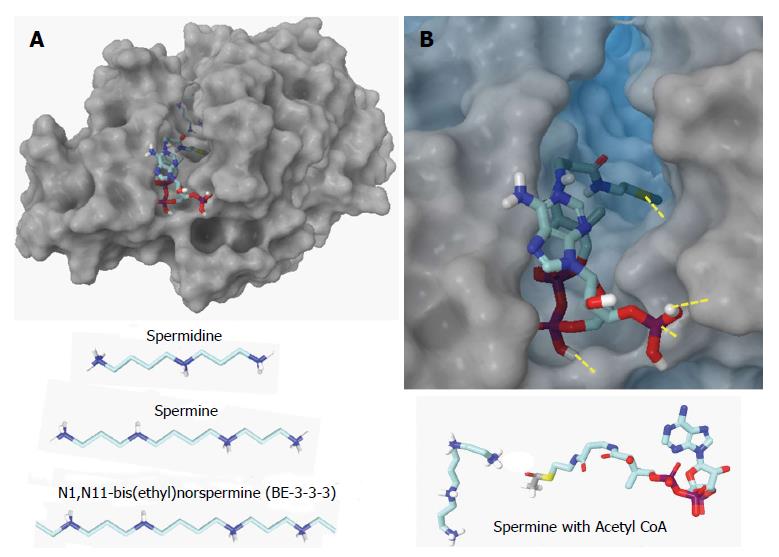
Another recently identified therapeutic set of epigenetic targets in AID is spermidine/spermine N1-acetyltransferase 1 (SAT1) involved in the polyamine pathway which can entail excessive consumption of the cell’s methyl donor, SAM, by the S-adenosylmethionine decarboxylase (AMD1)[34,35]. Both SAT1 and AMD1 are elevated in synovial fibroblasts from RA patients while, at the same time, DNA methyltransferase (DNMT1) and global DNA methylation are decreased[36]. Blocking SAT1 activity with specific siRNAs restores DNA methylation[37]. Therefore, development of specific SAT1 inhibitors is emerging as a new therapeutic approach. Computational tools can greatly accelerate drug discovery targeting SAT1 since crystal structures of wild type human SAT1 are available along with known inhibitors as references, such as N1, N11-bis(ethyl)norspermine (BE-3-3-3) with fairly well understood mechanisms of inhibition (Figure 2).
Figure 2 Human wild type spermidine/spermine N1-acetyltransferase.
Spermidine/spermine N1-acetyltransferase (SAT1) functions as a dimer with two channels in which the acetylation reaction occurs. A: Top – SAT1 with spermine and acetyl CoA docked in one channel. Bottom – the normal substrates, spermidine and spermine, are shown along with a known inhibitor, BE-3-3-3; B: Top - Close-up of acetyl CoA docked in the channel. Yellow dashes denote hydrogen bonds. The substrate will enter the other end of the channel and bind. Bottom – Spermine and acetyl CoA bind in close proximity in SAT1 so that the acetyl group can transfer to the substrate at which point CoA is released and the acetylspermine has reduced affinity so it releases. Structure of SAT1[38] is 2B4D.pdb from the Protein Data Bank (http://www.rcsb.org).
To date, among the five epigenetic drugs approved by the food and drug administration, only one, tofacitinib, a Jak1/2 inhibitor that controls histone phosphorylation, has been proven efficacious in RA. Several Jak inhibitors are currently being tested in AID, and more than 100 epigenetic drugs are in various stages of development. However, the main limitations of these drugs in chronic diseases such as AID are related to the relative lack of specificity, the lack of cellular specificity, the limited activity, as well as the risk of major side effects. As a consequence, new epigenetic medications need to circumvent these limitations. Furthermore, the development of epigenetic biomarkers may also help screen high-risk populations before the onset of the disease for prevention, and to provide a rationale to select responder patients that can benefit from epigenetic drugs.
CONCLUSION
The arguments presented here indicate that epigenetic changes precede AID and confer risk for AID suggesting a strong argument for epigenetic causality in genetically predisposed individuals. As a consequence, better understanding of the pathways leading to epigenetic deregulation would undoubtedly have benefits for prevention, follow-up and treatment of AID.
ACKNOWLEDGMENTS
We are also grateful to Simone Forest and Geneviève Michel for their help in typing the paper.
P- Reviewer: Martinez-Lostao L, Yamakawa M S- Editor: Ji FF L- Editor: A E- Editor: Jiao XK