Copyright
©2014 Baishideng Publishing Group Inc.
World J Immunol. Jul 27, 2014; 4(2): 107-115
Published online Jul 27, 2014. doi: 10.5411/wji.v4.i2.107
Published online Jul 27, 2014. doi: 10.5411/wji.v4.i2.107
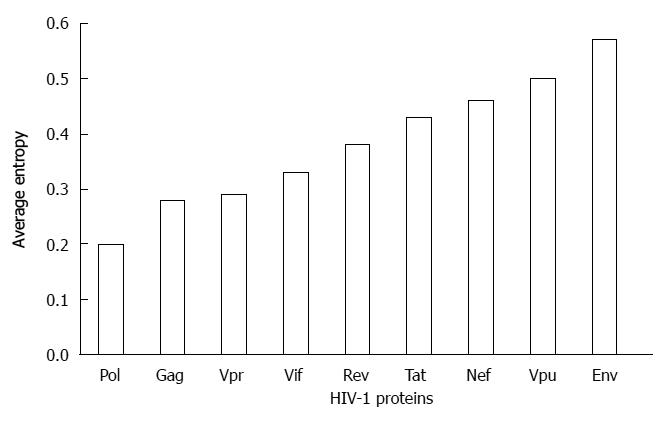
Figure 1 Sequence variability of human immunodeficiency virus type 1 proteins.
The sequence variability of nine proteins of human immunodeficiency virus type 1 (HIV-1) shown in the graph was determined by using Shannon entropy approach[24,90]. The full genome clade B sequences of the individual patients were retrieved from Los Alamos database (n = 544). Vpu: Viral protein U.
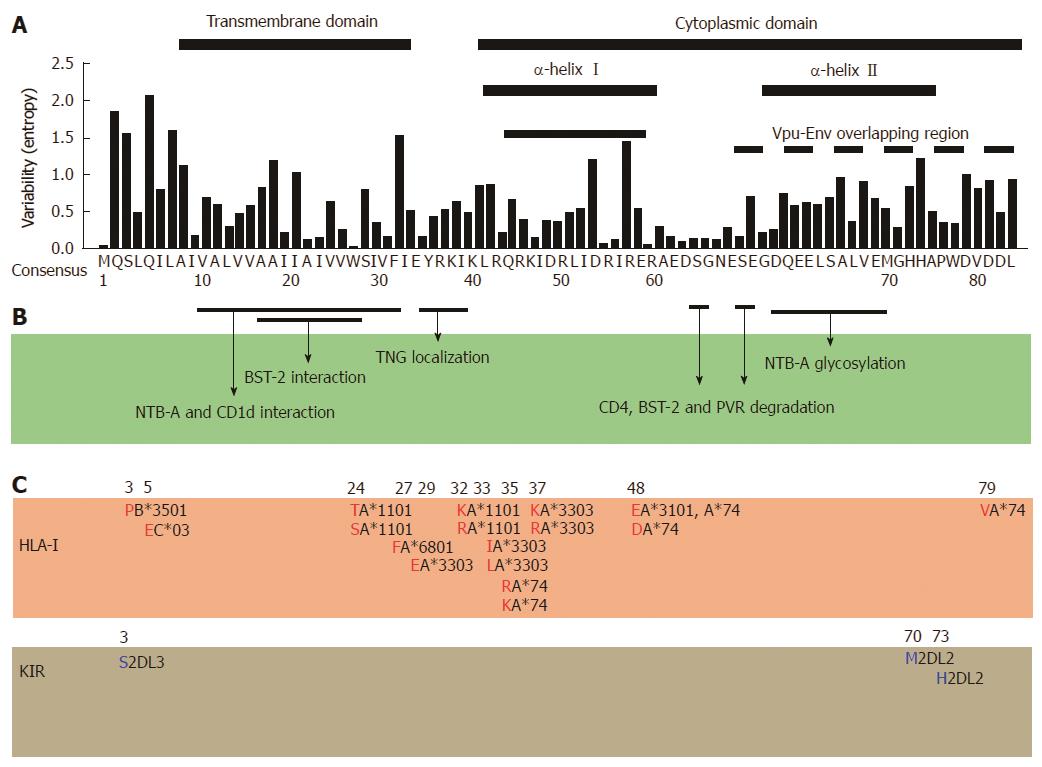
Figure 2 Correlation among amino-acid codon variability, functional regions, and host-mediated immune escape map of human immunodeficiency virus type 1 viral protein U.
A: Amino acid codon variability is assigned to each position of Vpu using Shannon entropy approach[24,90]. Sequences were retrieved from Los Alamos database (n =1139), the consensus subtype B sequence is indicated as a reference; B: Interacting positions and domains responsible for the indicated functions of human immunodeficiency virus type 1 (HIV-1) Vpu are shown[14,62,91]; C: Immune escape map shows amino acid codons and residues (red and blue) associated with HLA-I alleles[20,45,50] and killer-cell immunoglobulin-like receptors (KIR)[44], respectively. The specific alleles are indicated in black adjacent to the amino acid.
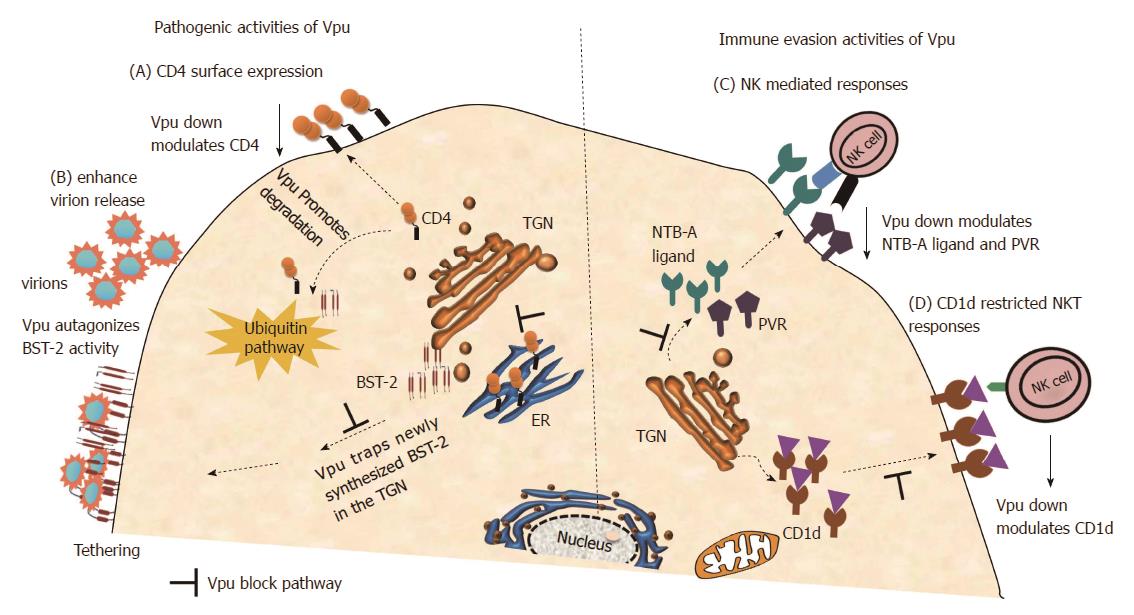
Figure 3 Viral protein U functionality including immune evasion activity.
The schematic representation of the cell illustrates some key functions of viral protein U (Vpu) including immune evasion activities. (A): Panel A illustrates CD4 down regulation by Vpu through degradation in a β-TrCP dependent ubiquitination pathway[11,12,64]; (B): Panel B demonstrates enhancement of virion release by Vpu through antagonizing BST-2, which is achieved through direct interaction with BST-2 which subsequently leads to trapping of BST-2 in the trans-Golgi networks[14-16] and also indicates β-TrCP dependent ubiquitination of BST-2[62,65,66]; (C): Panel C demonstrates Vpu evasion of NK cell recognition through down modulation of NTB-A ligand[70] and PVR[71]; D: Panel D shows down modulation of CD1d from cell surface hence avoid CD1d-restricted NKT cell responses[72,73]. NK: Natural killer; NKT: Natural killer T.
- Citation: Hasan Z, Kamori D, Ueno T. Role of host immune responses in sequence variability of HIV-1 Vpu. World J Immunol 2014; 4(2): 107-115
- URL: https://www.wjgnet.com/2219-2824/full/v4/i2/107.htm
- DOI: https://dx.doi.org/10.5411/wji.v4.i2.107