Published online Jun 9, 2024. doi: 10.5409/wjcp.v13.i2.90499
Revised: March 27, 2024
Accepted: April 17, 2024
Published online: June 9, 2024
Processing time: 184 Days and 14.8 Hours
Preterm birth is the leading cause of mortality in newborns, with very-low-birth-weight infants usually experiencing several complications. Breast milk is considered the gold standard of nutrition, especially for preterm infants with delayed gut colonization, because it contains beneficial microorganisms, such as Lactobacilli and Bifidobacteria.
To analyze the gut microbiota of breastfed preterm infants with a birth weight of 1500 g or less.
An observational study was performed on preterm infants with up to 36.6 wk of gestation and a birth weight of 1500 g or less, born at the University Hospital Dr. José Eleuterio González at Monterrey, Mexico. A total of 40 preterm neonates were classified into breast milk feeding (BM) and mixed feeding (MF) groups (21 in the BM group and 19 in the MF group), from October 2017 to June 2019. Fecal samples were collected before they were introduced to any feeding type. After full enteral feeding was achieved, the composition of the gut microbiota was analyzed using 16S rRNA gene sequencing. Numerical variables were compared using Student’s t-test or using the Mann–Whitney U test for nonparametric variables. Dominance, evenness, equitability, Margalef’s index, Fisher’s alpha, Chao-1 index, and Shannon’s diversity index were also calculated.
No significant differences were observed at the genus level between the groups. Class comparison indicated higher counts of Alphaproteobacteria and Betaproteobacteria in the initial compared to the final sample of the BM group (P < 0.011). In addition, higher counts of Gammaproteobacteria were detected in the final than in the initial sample (P = 0.040). According to the Margalef index, Fisher’s alpha, and Chao-1 index, a decrease in species richness from the initial to the final sample, regardless of the feeding type, was observed (P < 0.050). The four predominant phyla were Bacteroidetes, Actinobacteria, Firmicutes, and Proteobacteria, with Proteobacteria being the most abundant. However, no significant differences were observed between the initial and final samples at the phylum level.
Breastfeeding is associated with a decrease in Alphaproteobacteria and Betaproteobacteria and an increase of Gammaproteobacteria, contributing to the literature of the gut microbiota structure of very low-birth-weight, preterm.
Core Tip: Gut microbiota in very low-weight preterm infants is characterized by delayed colonization and decreased bacterial species which can lead to complications. In this study, we analyzed it using 16S rRNA gene sequencing in 40 hispanic infants classified into two groups: those receiving breast milk (BM) and those with mixed feeding. A decrease in the counts of Alpha and Betaproteobacteria, with higher counts of bifidobacteria, Bacteroidetes, and Clostridium were observed in the BM group. This study contributes to the literature on the structure of the gut microbial of preterm infants.
- Citation: Sánchez-González SG, Cárdenas-del-Castillo BG, Garza-González E, Padilla-Rivas GR, Rodríguez-Balderrama I, Treviño-Garza C, Montes-Tapia FF, Palacios-Saucedo GC, Gutiérrez-Rodríguez A, de-la-O-Cavazos ME. Gut microbiota in preterm infants receiving breast milk or mixed feeding. World J Clin Pediatr 2024; 13(2): 90499
- URL: https://www.wjgnet.com/2219-2808/full/v13/i2/90499.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v13.i2.90499
Every year, nearly 15 million preterm infants are born worldwide, of whom 1.1 million die because of childbirth complications. Preterm birth is considered the leading cause of mortality in newborns, with very low-birth-weight infants usually experiencing neurological, gastrointestinal, and respiratory complications[1-4].
Several structural and functional changes occur in the neonatal gastrointestinal tract as a result of their diet[5]. Breast milk has various benefits to this highly vulnerable population, such as lower rates of late-onset sepsis, retinopathy, and necrotizing enterocolitis, compared with infants receiving formula[6]. Breast milk contains beneficial microorganisms, such as lactobacilli and bifidobacteria[7,8]. Compared with the multiple aerobic and anaerobic conditions of the microbiota of full-term babies, the microbiota of preterm infants is characterized by delayed gut colonization and decreased bacterial species[9].
Bifidobacteria are the predominant intestinal bacteria for the first three–six days of life in full-term breastfed infants[10]. These bacteria represent almost 90% of the total bacterial count in the intestinal microbiota. The guts of formula-fed newborns contain approximately 20% fewer bifidobacteria than breastfed newborns and increased Bacteroides, Clostridium, and Enterobacteriaceae[10-12].
Kingdom Bacteria comprises nearly 30 phyla. The main phyla present in the human gut microbiota are Firmicutes, which include lactobacilli, and Bacteroidetes (> 90%), followed by Actinobacteria, which include bifidobacteria, Proteobacteria, and microorganisms that can be pathogenic for humans, such as Escherichia, Salmonella, Vibrio, and Helicobacter[13-15]. While an imbalance in the microbiota can affect the health of a newborn, maintaining it as healthy as possible is essential for their well-being[16]. To better understand how breastmilk influences gut microbiota, we conducted this study, in which we analyzed the gut microbiota composition using 16S rRNA gene sequencing in preterm infants weighing under 1500 g classified into breast milk feeding (BM) and mixed feeding (MF) groups[17].
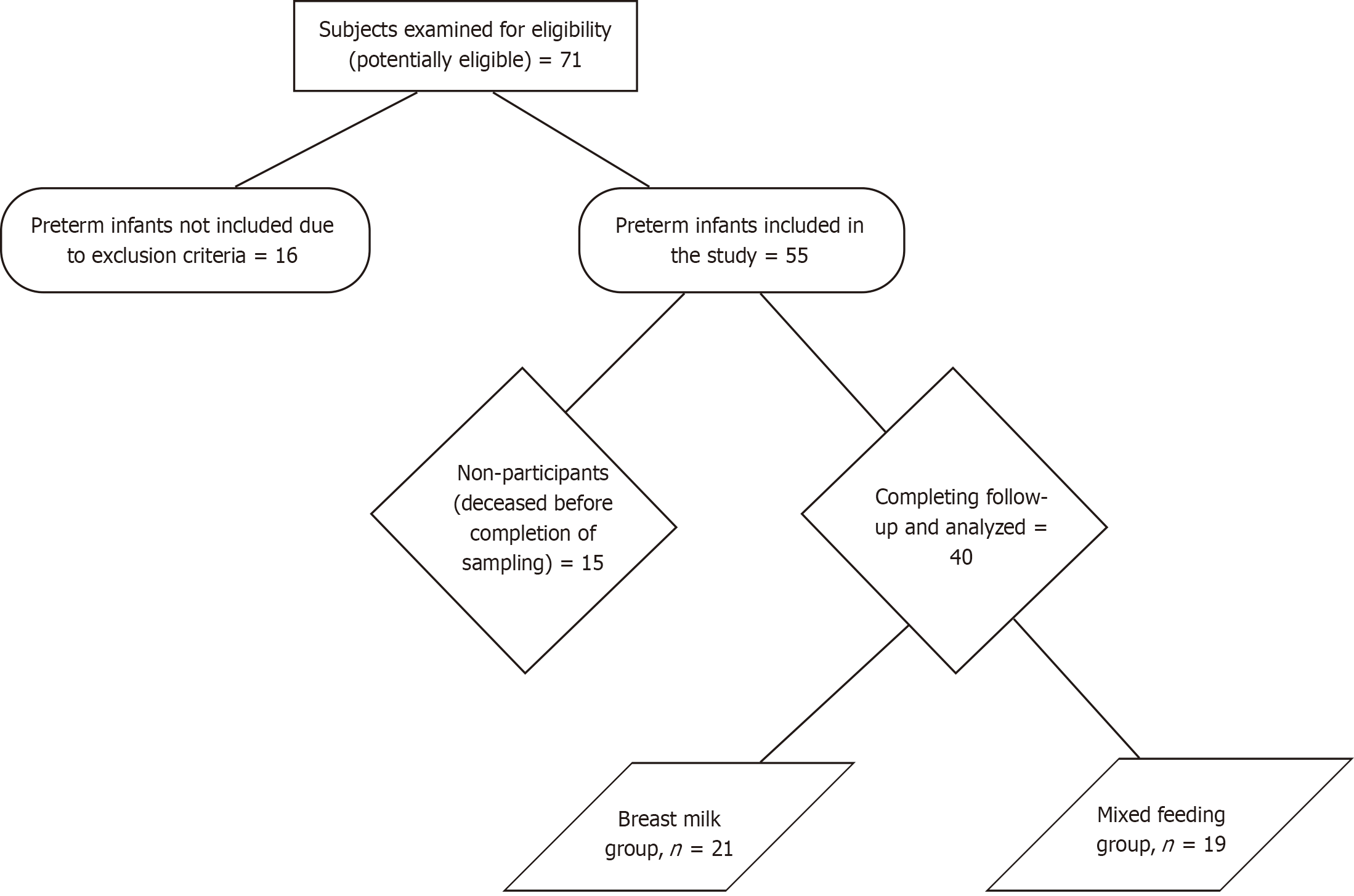
An observational, longitudinal, comparative, and prospective study was performed on preterm infants with up to 36.6 weeks of gestation or less at the time of birth and a birth weight of 1500 g or less[18], born at the University Hospital Dr. José Eleuterio González at Monterrey, northeastern Mexico. Preterm infants were included during the period from October 2017 to June 2019. Infants with congenital gastrointestinal anomalies were excluded. Infants who were transferred to another institution or had incomplete files were also excluded. Number of subjects at each stage of study has been described using a flow diagram (Figure 1).
The mothers of infants in the neonatal intensive care unit (NICU) were encouraged to provide breast milk for their preterm babies. Breastfeeding was typically preferred, and formula (NAN® formula for preterm babies, composed of: 3.4 g of protein/100 kcal, DHA-ARA, Medium chain triglycerides, Whey milk, Skim milk powder, vitamins, lactose and Soy lecithin) was used in case the amount of breast milk was insufficient to meet the needs of the infants. As soon as minimal enteral stimulation could be initiated (decision of the attending neonatologist), feeding was started with a minimal stimulus (0.5 mL/kg/h per day for three days) and then increased to 1 mL/kg/h per day until full enteral feeding was achieved (150 mL/kg/d) in all clinically stable newborns weighing under 1 kg. Newborns weighing over 1 kg were started on enteral feeding (1 mL/kg/h per day) until reaching 150 mL/kg/d. No human donor milk or milk fortifiers were used, and no human milk bank was available at our hospital during the study period. Two independent groups were formed based on the availability of breast milk. One group was exclusively fed with their mother’s BM, and the other group was fed with a mixed diet including their mothers’ breast milk and preterm formula (MF). A total of 40 preterm infants were included, 21 in the BM group and 19 in the MF group.
The study protocol was approved by the Ethics and Research Committee of the School of Medicine, Universidad Autónoma de Nuevo León (registration code PE16-00009). Written informed consent was obtained from the parents of all newborns.
Before any intervention (breast milk administration or mixed feeding), a sample of each infant’s first bowel movement was collected to analyze the gut microbiome. All preterm infants were followed up during their NICU stay, and a second stool sample was obtained for analysis after full enteral feeding was achieved (defined as 150 mL/kg/d) (Table 1). Stool samples were first obtained by trained NICU nurses using a sterile applicator and then placed in a sterile container. Subsequently, all samples were frozen at -70° C and stored for later analysis. A unique identification number was assigned to each sample. In addition, the demographic and clinical characteristics of the newborns were collected, including the type of delivery, feeding type, gender, weeks of gestation, weight, and morbidities at the NICU (Table 1).
| Characteristics | Type of feeding | P value | |
| Breast milk, n = 21 | Mixed, n = 19 | ||
| Gender | |||
| Female | 5 (24) | 9 (47) | 0.113 |
| Male | 16 (76) | 10 (53) | |
| Type of delivery | |||
| Vaginal | 6 (28) | 4 (21) | 0.582 |
| Cesarean section | 15 (71) | 15 (78) | |
| Prematurity category | |||
| Late preterm (34-36.6 gestational weeks) | 0 (0) | 2 (10) | 0.225 |
| Preterm (28-33.6 gestational weeks) | 20 (95) | 15 (79) | |
| Extreme preterm (< 27.6 gestational weeks) | 1 (5) | 2 (10) | |
| Apgar score at 5 min | |||
| 0–3 | 0 (0) | 0 (0) | 0.087 |
| 4–6 | 2 (9) | 2 (10) | |
| 7–10 | 19 (90) | 17 (89) | |
| Gestational weeks, mean ± SD | 29 ± 2 | 30 ± 2 | 0.103 |
| Birth weight (g), mean ± SD | 1,180 ± 217 | 1276 ± 169 | 0.132 |
| Birth height (cm), mean ± SD | 37 ± 3 | 38 ± 2 | 0.338 |
| Intubated at birth | 4 (19) | 5 (26) | 0.581 |
| Days to achieve full enteral feeding, mean ± SD | 18 ± 10 | 15 ± 8 | 0.346 |
| NICU stay (days), mean ± SD | 28 ±19 | 26 ±17 | 0.668 |
| Late-onset sepsis | 5 (24) | 2 (10) | 0.413 |
| Retinopathy | 1 (5) | 0 (0) | 0.997 |
| Necrotizing enterocolitis | 3 (14) | 4 (21) | 0.685 |
| Patent ductus arteriosus | 6 (29) | 5 (26) | 0.994 |
| Respiratory distress syndrome | 18 (86) | 19 (100) | 0.239 |
| Bronchopulmonary displasia | 4 (19) | 3 (16) | 0.991 |
| Intraventricular hemorrhage | 10 (48) | 5 (26) | 0.202 |
| Mortality | 0 (0) | 2 (10) | 0.218 |
Using a formula to compare two proportions for a one-tailed hypothesis with a confidence level of 95%, a statistical power of 90%, and an expected proportion of beneficial bacteria (bifidobacteria) of 90% for the breast-milk-fed group and 40% for the group with mixed feeding[16], the number of participants required was 14 per group. Adjusting this value to 10% of possible losses during follow-up determined that the number of patients to include was 16.
All analyses were performed in MR DNA (Shallowater, TX, United States, (http://www.mrdnalab.com). The semiconserved V4 region of the 16S rRNA gene was amplified using previously described primers[19]. The samples were sequenced using Illumina HiSeq Chemistry (Illumina, Inc., San Diego, CA, United States) following the manufacturer’s protocol.
Q25 sequence data (sequencing base calls with an error rate of less than 0.3%) derived from the sequencing process were processed using a proprietary analysis pipeline (MR DNA). Sequences were depleted of barcodes and primers, and then short sequences (< 200 bp), ambiguous base calls, and homopolymer runs exceeding 6 bp were removed. Operational taxonomic units were defined after singleton sequences were removed and after clustering at 3% divergence (97% similarity). They were then taxonomically classified using BLASTn against a curated National Center for Biotechnology Information database and compiled into counts and percentage files at each taxonomic level (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The statistical methods of this study were reviewed by Neri Alejandro Álvarez-Villalobos, MD, from School of Medicine of Universidad Autónoma de Nuevo León. Descriptive statistical analysis was performed for group comparison between BM and MF. Continuous variables are represented as means ± SD. Percentages and frequencies were used for categorical variables. Numerical variables were compared using Student’s t-test for independent samples with a normal distribution or using the Mann–Whitney U test for nonparametric variables. All analyses were performed using IBM SPSS Statistics version 20 (IBM Corp., Armonk, NY, United States).
Dominance, evenness, equitability, Brillouin’s index, Margalef’s index, Fisher’s alpha, Berger–Parker index, Chao-1 index, Simpson’s index, Menhinick’s index, and Shannon’s diversity index were calculated using the paleontological statistics software PAST (version 4.03)[20].
Statistical comparisons for each sample group were conducted using Kruskal–Wallis pairwise comparisons. Statistical significance was set at P < 0.050.
The microbial community structure was analyzed using weighted UniFrac distance matrices. Principal coordinate analysis plots were used to visualize the data in these matrices, and pairwise analysis of similarities (ANOSIM) was used to determine difference in microbial communities between groups.
A total of 40 preterm neonates weighing less than 1500 g completed follow-up and were analyzed and divided into two groups: 21 in the BM group and 19 in the MF group. All infants were Hispanic, and 65% (26/40) were male. All neonates received antibiotics (ampicillin and aminoglycosides) for the initial week while participating in the study as part of their NICU treatment, however, there was no use of antibiotics before delivery and mothers didn’t present premature rupture of membranes. No significant differences were observed in the demographic characteristics between the two groups. In addition, no differences were observed in the clinical characteristics and outcomes of the infants between groups (Table 1).
After strict quality sequence curation, 3132039 reads were analyzed, and 2940310 reads were pooled. In addition, 2937522 reads identified within the Bacteria and Archaea domains were used for the final microbiota analysis. The mean reads per sample were 36719. For alpha and beta diversity analysis, samples were rarefied by 18000 reads. The data were then multivariate-evaluated to determine the differences between groups.
Data has been deposited in a publicly accessible database: GenBank SRA Accession Number: SRR17156517. BioProject: PRJNA786526 BioSample: SAMN23683919.
No significant differences were observed at the genus level between the BM and MF groups (at both the initial and final time points). A dual hierarchal dendrogram evaluation of taxonomic classification data was constructed to view the data of predominant genera. Samples with similar microbial populations were mathematically clustered, and genera (consortium) were used for clustering. However, no significant difference was observed between the BM and MF groups, given the lack of clustering between groups.
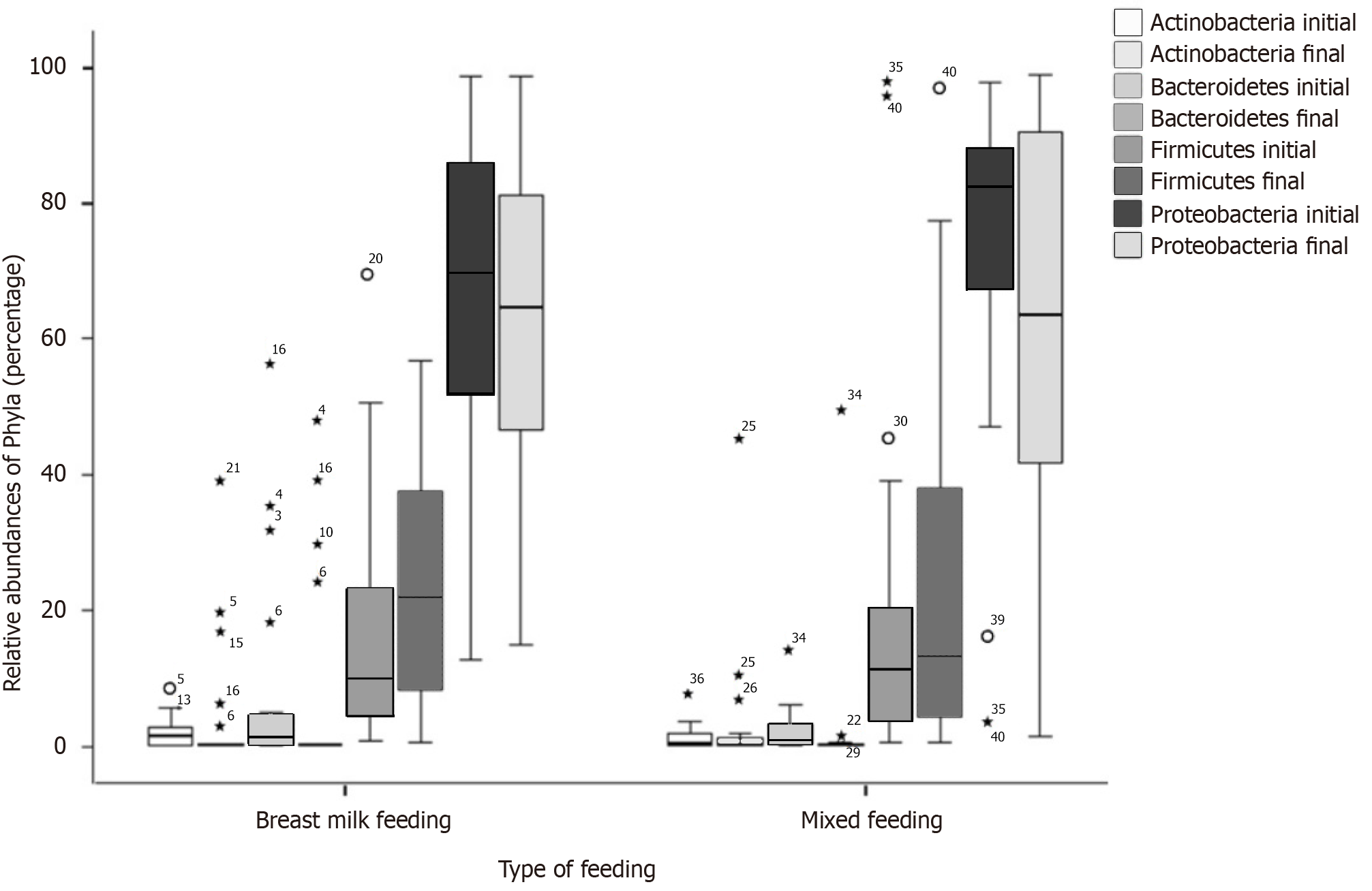
In the BM group, comparisons at the class level indicated higher counts of Alphaproteobacteria and Betaproteobacteria in the initial compared to the final sample (P < 0.011) and higher counts of Gammaproteobacteria in the final than in the initial sample (P = 0.040) (Table 2). The four predominant phyla were Bacteroidetes, Actinobacteria, Firmicutes, and Proteobacteria, with Proteobacteria being the most abundant (Figure 2). No significant differences were observed between the initial and final samples at the phylum level (Table 2).
| Type of feeding | ||||||
| Breast milk, n = 21 | Mixed, n = 19 | |||||
| Initial, mean ± SD | Final, mean ± SD | P value | Initial, mean ± SD | Final, mean ± SD | P value | |
| Class | ||||||
| Bacteroidia | 7.468 ± 15 | 6.835 ± 14 | 0.782 | 1.890 ± 3 | 2.872 ± 11 | 0.612 |
| Flavobacteria | 0.245 ± 10 | 0.008 ± 0.004 | 0.106 | 0.404 ± 1 | 0.005 ± 0.005 | 0.041 |
| Sphingobacteria | 0.030 ± 0.09 | 0.001 ± 0.001 | 0.155 | 0.055 ± 0.1 | 0.002 ± 0.006 | 0.100 |
| Cytophagia | 0.030 ± 0.10 | 0.001 ± 0.002 | 0.208 | 0.0004 ± 0.001 | 0.063 ± 0.268 | 0.320 |
| Actinobacteria | 2.270 ± 2.0 | 4.519 ± 9 | 0.261 | 1.396 ± 2 | 3.520 ± 10 | 0.398 |
| Bacilli | 14.975 ± 17 | 17.794 ± 17 | 0.539 | 19.112 ± 29 | 20.131 ± 26 | 0.875 |
| Clostridia | 1.664 ± 30 | 3.525 ± 6 | 0.177 | 1.517 ± 2 | 5.084 ± 15 | 0.326 |
| Mollicutes | 4.334 ± 17 | 0.054 ± 0.076 | 0.274 | 4.424 ± 19 | 4.607 ± 20 | 0.324 |
| Erysipelotrichia | 0.0001 ± 0.0005 | 0.0006 ± 0.001 | 0.253 | 0.028 ± 0.06 | 0.0002 ± 0.0008 | 0.063 |
| Negativicutes | 1.038 ± 3.0 | 2.440 ± 5 | 0.138 | 0.387 ± 1 | 0.337 ± 0.813 | 0.857 |
| Alphaproteobacteria | 0.989 ± 1.0 | 0.017 ± 0.011 | 0.005 | 0.570 ± 1 | 0.237 ± 0.623 | 0.103 |
| Betaproteobacteria | 23.550 ± 27.0 | 0.643 ± 0.290 | 0.001 | 18.517 ± 28 | 12.789 ± 28 | 0.509 |
| Deltaproteobacteria | 1.585 ± 4.0 | 0.200 ± 0.593 | 0.081 | 0.163 ± 0.4 | 0.103 ± 0.168 | 0.582 |
| Epsilonproteobacteria | 0.130 ± 0.4 | 0.001 ± 0.002 | 0.179 | 0.009 ± 0.02 | 0.0009 ± 0.002 | 0.218 |
| Gammaproteobacteria | 40.831 ± 30.0 | 64.381 ± 24 | 0.004 | 51.063 ± 36 | 50.182 ± 34 | 0.921 |
| Phylum | ||||||
| Bacteroidetes | 7.775 ± 15 | 6.786 ± 14 | 0.667 | 2.350 ± 3 | 2.943 ± 11 | 0.762 |
| Actinobacteria | 2.282 ± 2 | 4.131 ± 9 | 0.373 | 1.396 ± 2 | 3.520 ± 10 | 0.391 |
| Firmicutes | 17.678 ± 19 | 23.761 ± 17 | 0.219 | 21.046 ± 29 | 25.553 ± 28 | 0.558 |
| Proteobacteria | 67.087 ± 24 | 65.243 ± 24 | 0.772 | 70.324 ± 31 | 63.314 ± 31 | 0.403 |
Species diversity was compared between the initial and final samples for both groups. Significant differences were observed in the Margalef, Fisher’s alpha, and Chao-1 indexes in the BM group. Regarding evenness, significant differences were also observed in the Margalef, Fisher’s alpha, Chao-1, and Menhinick’s index in the MF group (P < 0.050) (Table 3).
| Index | Type of feeding | |||||
| Breast milk (n = 21) | Mixed (n = 19) | |||||
| Initial, mean ± SD or median (range) | Final, mean ± SD or median (range) | P value | Initial, mean ± SD or median (range) | Final, mean ± SD or median (range) | P value | |
| Species richness | ||||||
| Margalef’s index | 1.60 ± 0.43 | 1.37 ± 0.27 | 0.042 | 1.53 | 1.31 | 0.0061 |
| Fisher’s alpha | 2.20 ± 0.68 | 1.82 ± 0.41 | 0.037 | 2.05 | 1.71 | 0.0071 |
| Chao-1 index | 8.33 ± 1.96 | 7.29 ± 1.23 | 0.044 | 8 | 7 | 0.0071 |
| Menhinick’s index | 0.9 | 0.7 | 0.079 | 0.8 | 0.7 | 0.0071 |
| Species evenness or dominance | ||||||
| Dominance | 0.63 ± 0.18 | 0.60 ± 0.22 | 0.617 | 0.747 ± 0.153 | 0.699 ± 0.189 | 0.396 |
| Evenness | 0.26 ± 0.09 | 0.29 ± 0.13 | 0.363 | 0.194 ± 0.046 | 0.246 ± 0.080 | 0.020 |
| Brillouin’s index | 0.51 ± 0.26 | 0.53 ± 0.33 | 0.807 | 0.344 ± 0.226 | 0.392 ± 0.265 | 0.551 |
| Simpson’s index | 98.00 | 0.4600 | 0.5891 | 0.253 ± 0.153 | 0.301 ± 0.189 | 0.396 |
| Shannon’s diversity index | 0.68 ± 0.29 | 0.65 ± 0.34 | 0.755 | 0.49 ± 0.250 | 0.51 ± 0.26 | 0.819 |
| Equitability | 0.33 ± 0.15 | 0.34 ± 0.19 | 0.873 | 0.226 ± 0.109 | 0.264 ± 0.141 | 0.355 |
| Berger–Parker index | 0.73 ± 0.16 | 0.67 ± 0.20 | 0.284 | 0.85 | 0.81 | 0.4181 |
The microbial community structure was analyzed using weighted UniFrac distance matrices. However, no significant difference was observed in the phylogenetic assemblage in either group. In addition, no significant difference was observed in the ANOSIM R values (R = 0.007922601, P = 0.242, Q-value = 0.24) between the microbial communities of the BM and MF groups.
There was no significant difference in the structure of the gut microbiota between the two groups after feeding.
Regarding clinical characteristics, even though it is well known that breast milk lowers the rate of late-onset sepsis and necrotizing enterocolitis[6], and we could have expected that the BM group would have this clinical significance compared to the MF group, these differences were not observed in our study possibly due to a no longer-term follow-up.
We analyzed the gut microbiota composition using 16S rRNA gene sequencing in preterm infants, comparing those exclusively fed with BM with those fed with MF. In the BM group, class comparison indicated higher counts of Alphaproteobacteria and Betaproteobacteria in the initial compared to the final sample and higher counts of Gammaproteobacteria in the final than in the initial sample. All three classes (Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria) belong to the phylum Proteobacteria.
The gut microbial community of healthy newborns typically starts with the colonization of facultative anaerobes, followed by the establishment of obligate anaerobic organisms, such as bifidobacteria and Bacteroidetes[7,21]. However, intestinal colonization is delayed in preterm infants, and the microbiome is less diverse, with a lower abundance of bifidobacteria and Bacteroidetes and a greater abundance of Proteobacteria. Multiple factors can influence the immediate colonization of the gastrointestinal tract, especially the type of delivery (vaginal or cesarean section), and maybe gestational weeks, which could explain the differences observed in the relative abundances of classes and phyla in neonates depending on the type of feeding even before the intervention[7,22,23]. Vertical transmission of bacteria from mother to child is one of the most important factors that influence the maturation of microbiota. Different studies describe the impact caused by delivery mode. Cesarean delivery has a great influence on the early colonization of the gut, which is characterized by depletion of Bacteroidetes, unlike vaginally born infants[24].
Most of the patients in our study were born by cesarean section at the decision of the gynecologist. We observed similar results to those described in the literature, with fewer Bacteroidetes in subjects delivered by cesarean section.
In our study, the four predominant phyla were Bacteroidetes, Actinobacteria, Firmicutes, and Proteobacteria, with Proteobacteria being the most abundant. No significant differences were observed between the initial and final samples at the phylum level and none of them suggested a tendency. However, a higher sample size or a long-term study may reflect a significant difference.
Our results indicated no significant differences between groups within the phylum Proteobacteria. However, analysis of specific classes from this phylum revealed a decrease in the counts of Alphaproteobacteria and Betaproteobacteria in preterm infants receiving breast milk. This result is significant because it has been described that the delayed gut microbiota maturation coupled with a Proteobacteria-dominated bacterial composition correlates with necrotizing enterocolitis and late-onset sepsis in preterm infants[25,26].
Higher counts of bifidobacteria, Bacteroidetes, and Clostridium were observed in the BM group compared to those in the MF group. However, the differences observed were not statistically significant. It has been described that bifidobacteria are fortified in the gut microbiome of term infants receiving breast milk compared with formula, thought to be due to the provision of human milk oligosaccharides. Embleton et al[27] also described a higher relative abundance of Bifidobacterium in human milk-fed preterms, although it was described in NICUs who were also using probiotics.
The presence of higher amounts of Bifidobacteria has been described to have benefits in the gut of preterm and is associated with protection from late-onset sepsis[28]. Also, it has been proposed that bifidobacteria produce acetate, which lowers luminal pH and favors the gut barrier function[29,30]. Additionally, this acetate may be converted into butyrate, which is considered an anti-inflammatory molecule[31].
Furthermore, the combination of Bifidobacteria with other probiotics has been associated with benefits for preterm infants. A systematic review that included 63 trials (15712 preterm infants) showed that the combination of one or more Lactobacillus species and 1 or more Bifidobacterium spp had moderate- or high-quality evidence of reduced all cause mortality (OR: 0.56; 95%CI: 0.39-0.80[32].
In our study, no human donor milk or milk fortifiers were used. Furthermore, our findings differ from a similar trial (Asbury et al[33]) that reported lower microbial diversity and lower abundances of Clostridium in preterm infants also receiving an exclusive human milk diet. However, these preterm infants were receiving newly available human-milk-based fortifiers. Clinical practice varies with some NICUs routinely using breast milk fortifiers whereas others never use fortifiers. Studies of the benefits of using fortifiers show there are no high-quality data on longer-term functional outcomes and most used fortifiers are of bovine origin[27].
Alpha diversity is the mean diversity of species in different habitats. It is considered a ubiquitous approach and has several definitions depending on the assumption of species diversity[34]. This means that more than one index of diversity can be used. In this study, we used 11 indices of diversity. Diversity indices are broadly divided into two types: indices that assess species richness (how many types there are) (e.g. Margalef’s index, Menhinick’s index, Chao-1 index, and Fisher’s alpha[35]) and indices that assess species evenness or dominance (how individual organisms are distributed among species) (e.g., Shannon’s diversity index, Brillouin’s index, Simpson’s index, and evenness)[36]. Here, we compared the alpha diversity among the initial and final samples and observed a decrease in species richness from the initial to the final sample regardless of the feeding type (according to the results of Margalef’s index, Fisher’s alpha, and Chao-1 index) (Table 3). Although we expected an increase in species richness, we observed opposite results. Ma et al[37] found that alpha diversity can increase significantly up to six months of age, but in our study neonates were not followed up until that period. Similarly, previous studies have reported that microbial diversity increases with age, indicating a more complex microbiota over time, which could explain why such diversity is not yet observed in our samples[37-40].
The main limitation of this study was that we did not include an exclusively formula-fed group for comparison purposes. However, our NICU protocol was to promote breast milk administration in all premature infants. This means that finding exclusively formula-fed preterm infants would have been challenging, and randomization would have been unethical. Also, the impact of antibiotics administration that all neonates received for the initial week as part of their NICU treatment on the gut represents another limitation. The antibiotic treatment used in our NICU could have especially affected gram-negative bacilli such as Haemophilus influenzae, Klebsiella pneumoniae, Legionella pneumophila, Pseudomonas aeruginosa, E. coli, Proteus mirabilis, Helicobacter pylori, Salmonella enteritidis, Salmonella typhi among others. It is known that antibiotic exposure decreases gut microbiota diversity and changes its composition, allowing a much larger amount of Proteobacteria, which was observed in our study, and impairing quantities of Clostridia and Bifidobacterium[24].
In conclusion, this study contributes to the literature on the structure of the gut microbial community of breastfed, very low-birth-weight, preterm infants. Breastfeeding is associated with a decrease in counts of Alphaproteobacteria and Betaproteobacteria and an increase in Gammaproteobacteria in preterm infants. In addition, regardless of the feeding type, the species richness decreases from the initial to the final sample. Growing evidence in this field suggests that there is potential to create strategies that could help reduce morbidities associated with very low-birth-weight preterms through early-age manipulation of their microbiota[9]. Further studies are required to better understand the factors that may affect the development of newborn microbiota.
We want to thank Sergio Lozano-Rodríguez M.D. and Sergio Antonio Ramírez-Cortinas M.D., for their review and editing of the manuscript. Also, a special acknowledgment to all the NICU and Gastroenterology Service staff who made this work possible.
| 1. | Walani SR. Global burden of preterm birth. Int J Gynaecol Obstet. 2020;150:31-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 509] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 2. | Blencowe H, Cousens S, Chou D, Oestergaard M, Say L, Moller AB, Kinney M, Lawn J; Born Too Soon Preterm Birth Action Group. Born too soon: the global epidemiology of 15 million preterm births. Reprod Health. 2013;10 Suppl 1:S2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1277] [Cited by in RCA: 1358] [Article Influence: 113.2] [Reference Citation Analysis (0)] |
| 3. | Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379:2162-2172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2858] [Cited by in RCA: 3114] [Article Influence: 239.5] [Reference Citation Analysis (0)] |
| 4. | Barfield WD. Public Health Implications of Very Preterm Birth. Clin Perinatol. 2018;45:565-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 5. | Groer MW, Luciano AA, Dishaw LJ, Ashmeade TL, Miller E, Gilbert JA. Development of the preterm infant gut microbiome: a research priority. Microbiome. 2014;2:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 197] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 6. | Walls Castellanos M, Claud EC. The microbiome, guard or threat to infant health. Trends Mol Med. 2021;27:1175-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Cong X, Judge M, Xu W, Diallo A, Janton S, Brownell EA, Maas K, Graf J. Influence of Feeding Type on Gut Microbiome Development in Hospitalized Preterm Infants. Nurs Res. 2017;66:123-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 8. | Layuk N, Sinrang AW, Asad S. Early initiation of breastfeeding and gut microbiota of neonates: A literature review. Medicina Clínica Práctica. 2021;4:10022. [DOI] [Full Text] |
| 9. | Unger S, Stintzi A, Shah P, Mack D, O'Connor DL. Gut microbiota of the very-low-birth-weight infant. Pediatr Res. 2015;77:205-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 10. | Parra-Llorca A, Gormaz M, Alcántara C, Cernada M, Nuñez-Ramiro A, Vento M, Collado MC. Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk. Front Microbiol. 2018;9:1376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 11. | Parm Ü, Metsvaht T, Ilmoja ML, Lutsar I. Gut colonization by aerobic microorganisms is associated with route and type of nutrition in premature neonates. Nutr Res. 2015;35:496-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Zhu B, Zheng S, Lin K, Xu X, Lv L, Zhao Z, Shao J. Effects of Infant Formula Supplemented With Prebiotics and OPO on Infancy Fecal Microbiota: A Pilot Randomized Clinical Trial. Front Cell Infect Microbiol. 2021;11:650407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Collado MC, Cernada M, Neu J, Pérez-Martínez G, Gormaz M, Vento M. Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr Res. 2015;77:726-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 14. | Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, Maas K, Graf J. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PLoS One. 2016;11:e0152751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 174] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 15. | Korpela K, Blakstad EW, Moltu SJ, Strømmen K, Nakstad B, Rønnestad AE, Brække K, Iversen PO, Drevon CA, de Vos W. Intestinal microbiota development and gestational age in preterm neonates. Sci Rep. 2018;8:2453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 194] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 16. | Voreades N, Kozil A, Weir TL. Diet and the development of the human intestinal microbiome. Front Microbiol. 2014;5:494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 325] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 17. | Westaway JAF, Huerlimann R, Miller CM, Kandasamy Y, Norton R, Rudd D. Methods for exploring the faecal microbiome of premature infants: a review. Matern Health Neonatol Perinatol. 2021;7:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Cutland CL, Lackritz EM, Mallett-Moore T, Bardají A, Chandrasekaran R, Lahariya C, Nisar MI, Tapia MD, Pathirana J, Kochhar S, Muñoz FM; Brighton Collaboration Low Birth Weight Working Group. Low birth weight: Case definition & guidelines for data collection, analysis, and presentation of maternal immunization safety data. Vaccine. 2017;35:6492-6500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 271] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 19. | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621-1624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5437] [Cited by in RCA: 5649] [Article Influence: 434.5] [Reference Citation Analysis (0)] |
| 20. | Hammer Ø, Harper DAT, Paul R. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron. 2001;4:1. |
| 21. | Saturio S, Nogacka AM, Alvarado-Jasso GM, Salazar N, de Los Reyes-Gavilán CG, Gueimonde M, Arboleya S. Role of Bifidobacteria on Infant Health. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 22. | Westaway JAF, Huerlimann R, Kandasamy Y, Miller CM, Norton R, Staunton KM, Watson D, Rudd D. The bacterial gut microbiome of probiotic-treated very-preterm infants: changes from admission to discharge. Pediatr Res. 2022;92:142-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 23. | Healy DB, Ryan CA, Ross RP, Stanton C, Dempsey EM. Clinical implications of preterm infant gut microbiome development. Nat Microbiol. 2022;7:22-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 71] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 24. | Beghetti I, Barone M, Brigidi P, Sansavini A, Corvaglia L, Aceti A, Turroni S. Early-life gut microbiota and neurodevelopment in preterm infants: a narrative review. Front Nutr. 2023;10:1241303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 25. | Granger CL, Embleton ND, Palmer JM, Lamb CA, Berrington JE, Stewart CJ. Maternal breastmilk, infant gut microbiome and the impact on preterm infant health. Acta Paediatr. 2021;110:450-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 26. | Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1476] [Cited by in RCA: 2440] [Article Influence: 244.0] [Reference Citation Analysis (0)] |
| 27. | Embleton ND, Sproat T, Uthaya S, Young GR, Garg S, Vasu V, Masi AC, Beck L, Modi N, Stewart CJ, Berrington JE. Effect of an Exclusive Human Milk Diet on the Gut Microbiome in Preterm Infants: A Randomized Clinical Trial. JAMA Netw Open. 2023;6:e231165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 28. | Stewart CJ, Embleton ND, Marrs ECL, Smith DP, Fofanova T, Nelson A, Skeath T, Perry JD, Petrosino JF, Berrington JE, Cummings SP. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome. 2017;5:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 202] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 29. | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1690] [Article Influence: 120.7] [Reference Citation Analysis (0)] |
| 30. | Alcon-Giner C, Dalby MJ, Caim S, Ketskemety J, Shaw A, Sim K, Lawson MAE, Kiu R, Leclaire C, Chalklen L, Kujawska M, Mitra S, Fardus-Reid F, Belteki G, McColl K, Swann JR, Kroll JS, Clarke P, Hall LJ. Microbiota Supplementation with Bifidobacterium and Lactobacillus Modifies the Preterm Infant Gut Microbiota and Metabolome: An Observational Study. Cell Rep Med. 2020;1:100077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 135] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 31. | Rivière A, Gagnon M, Weckx S, Roy D, De Vuyst L. Mutual Cross-Feeding Interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 Explain the Bifidogenic and Butyrogenic Effects of Arabinoxylan Oligosaccharides. Appl Environ Microbiol. 2015;81:7767-7781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 32. | Morgan RL, Preidis GA, Kashyap PC, Weizman AV, Sadeghirad B; McMaster Probiotic, Prebiotic, and Synbiotic Work Group. Probiotics Reduce Mortality and Morbidity in Preterm, Low-Birth-Weight Infants: A Systematic Review and Network Meta-analysis of Randomized Trials. Gastroenterology. 2020;159:467-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 151] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 33. | Asbury MR, Shama S, Sa JY, Bando N, Butcher J, Comelli EM, Copeland JK, Forte V, Kiss A, Sherman PM, Stintzi A, Taibi A, Tomlinson C, Unger S, Wang PW, O'Connor DL; OptiMoM Feeding Group. Human milk nutrient fortifiers alter the developing gastrointestinal microbiota of very-low-birth-weight infants. Cell Host Microbe. 2022;30:1328-1339.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 34. | Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557-578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 399] [Cited by in RCA: 326] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 35. | Benton MJ. The origins of modern biodiversity on land. Philos Trans R Soc Lond B Biol Sci. 2010;365:3667-3679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Kim BR, Shin J, Guevarra R, Lee JH, Kim DW, Seol KH, Kim HB, Isaacson R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J Microbiol Biotechnol. 2017;27:2089-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 549] [Article Influence: 91.5] [Reference Citation Analysis (0)] |
| 37. | Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep. 2020;10:15792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 38. | Toubon G, Butel MJ, Rozé JC, Lepage P, Delannoy J, Ancel PY, Charles MA, Aires J; EPIFLORE Study Group. Very Preterm Children Gut Microbiota Comparison at the Neonatal Period of 1 Month and 3.5 Years of Life. Front Microbiol. 2022;13:919317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 39. | Chen X, Shi Y. Determinants of microbial colonization in the premature gut. Mol Med. 2023;29:90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 40. | Salas AA, Willis KA, Carlo WA, Yi N, Zhang L, Van Der Pol WJ, Younge NE, Lefkowitz EJ, Lal CV. The gut microbiome of extremely preterm infants randomized to the early progression of enteral feeding. Pediatr Res. 2022;92:799-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |