Revised: February 15, 2014
Accepted: June 18, 2014
Published online: August 6, 2014
Processing time: 358 Days and 6.1 Hours
AIM: To investigate the subcellular localization and the function of mouse transducin β-like 3 (Tbl3).
METHODS: The coding sequence of mouse Tbl3 was cloned from the cDNAs of a promyelocyte cell line by reverse transcription-polymerase chain reaction. Fusion constructs of Tbl3 and enhanced green fluorescent protein (EGFP) were transfected into fibroblasts and examined by fluorescence microscopy to reveal the subcellular localization of tbl3. To search for nucleolar targeting sequences, scanning deletions of Tbl3-EGFP were constructed and transfected into fibroblasts. To explore the possible function of Tbl3, small hairpin RNAs (shRNAs) were used to knock down endogenous Tbl3 in mouse promyelocytes and fibroblasts. The effects of Tbl3 knockdown on ribosomal RNA (rRNAs) synthesis or processing were studied by labeling cells with 5,6-3H-uridine followed by a chase with fresh medium for various periods. Total RNAs were purified from treated cells and subjected to gel electrophoresis and Northern analysis. Ribosome profiling by sucrose gradient centrifugation was used to compare the amounts of 40S and 60S ribosome subunits as well as the 80S monosome. The impact of Tbl3 knockdown on cell growth and proliferation was examined by growth curves and colony assays.
RESULTS: The largest open reading frame of mouse Tbl3 encodes a protein of 801 amino acids (AA) with an apparent molecular weight of 89-90 kilodalton. It contains thirteen WD40 repeats (an ancient protein-protein interaction motif) and a carboxyl terminus that is highly homologous to the corresponding region of the yeast nucleolar protein, utp13. Virtually nothing is known about the biological function of Tbl3. All cell lines surveyed expressed Tbl3 and the level of expression correlated roughly with cell proliferation and/or biosynthetic activity. Using Tbl3-EGFP fusion constructs we obtained the first direct evidence that Tbl3 is targeted to the nucleoli in mammalian cells. However, no previously described nucleolar targeting sequences were found in Tbl3, suggesting that the WD40 motif and/or other topological features are responsible for nucleolar targeting. Partial knockdown (by 50%-70%) of mouse Tbl3 by shRNA had no discernable effects on the processing of the 47S pre-ribosomal RNA (pre-rRNA) or the steady-state levels of the mature 28S, 18S and 5.8S rRNAs but consistently increased the expression level of the 47S pre-rRNA by two to four folds. The results of the current study corroborated the previous finding that there was no detectable rRNA processing defects in zebra fish embryos with homozygous deletions of zebra fish Tbl3. As ribosome production consumes the bulk of cellular energy and biosynthetic precursors, dysregulation of pre-rRNA synthesis can have negative effects on cell growth, proliferation and differentiation. Indeed, partial knockdown of Tbl3 in promyelocytes severely impaired their proliferation. The inhibitory effect of Tbl3 knockdown was also observed in fibroblasts, resulting in an 80% reduction in colony formation. Taken together, these results indicate that Tbl3 is a newly recognized nucleolar protein with regulatory roles at very early stages of ribosome biogenesis, perhaps at the level of rRNA gene transcription.
CONCLUSION: Tbl3 is a newly recognized nucleolar protein with important regulatory roles in ribosome biogenesis.
Core tip: The mouse gene transducin β-like 3 (Tbl3) encodes a protein with thirteen WD40 protein-protein interaction motifs and is the mammalian homologue of yeast utp13. Virtually nothing is known about the function of tbl3. In this report, we provide the first direct evidence that Tbl3 is targeted to the nucleoli and plays an important role in regulating the synthesis of the 47S pre-ribosomal RNA, i.e., at very early stages of ribosome biogenesis. This activity has never been described before and sets Tbl3 apart from all other known nucleolar proteins. TBL3 may provide an attractive target for anti-neoplastic therapy.
-
Citation: Wang J, Tsai S.
Tbl3 encodes a WD40 nucleolar protein with regulatory roles in ribosome biogenesis. World J Hematol 2014; 3(3): 93-104 - URL: https://www.wjgnet.com/2218-6204/full/v3/i3/93.htm
- DOI: https://dx.doi.org/10.5315/wjh.v3.i3.93
Ribosomes are essential for protein synthesis. Defects in ribosome biogenesis or “ribosomopathy” underlie several devastating hematological disorders such as the Diamond-Blackfan anemia, Shwachman-Bodian-Diamond syndrome, dyskeratosis congenita and the 5q- myelodysplastic syndrome[1,2].
The production of ribosomes is a highly coordinated, multi-step process that starts in the nucleoli, the organelles that form around the “nucleolar organizers” on the long arms of acrocentric chromosomes (human chromosomes 13, 14, 15, 21 and 22)[3]. The nucleolar organizers consist of 400 or so rRNA genes (rDNAs). The production of ribosomes begins with the transcription of the rDNAs by RNA polymerase I (RNA Pol I), which synthesizes a long primary transcript, the 47S pre-rRNA, that is then covalently modified (e.g., 2’-O-ribose methylation and pseudouridylation) and processed into mature 28S, 18S and 5.8S rRNAs. The rRNAs are assembled into pre-ribosomes with about eighty ribosomal proteins plus the 5S rRNA, which is separately transcribed by RNA polymerase III outside the nucleolus. A large number (> 150) of small nucleolar RNAs and nonribosomal proteins participate in the post-transcriptional modification, processing and assembly of rRNAs to produce pre-ribosomes, which undergo further maturation to become nascent 40S and 60S ribosomal subunits before being exported into the cytoplasm. Most steps of rRNA processing occur co-transcriptionally with pre-rRNA synthesis[4-7]. As the production of ribosomes consumes the bulk of cellular energy[8], defective or dysregulated ribosome production can affect many aspects of cellular physiology especially cell proliferation.
Due to the relative simplicity and the ease of genetic manipulations in the yeasts, considerably more has been learned about the functions of non-ribosomal nucleolar proteins in yeasts than in mammalian cells. Still, the functions of many yeast nonribosomal nucleolar proteins remain unknown. In mammals, the total number of non-ribosomal proteins involved in ribosome biogenesis is even larger, reflecting the greater complexities of eukaryotic rRNA modification, processing, ribosome assembly, transport and other nucleolar events. Recent proteomic analyses of purified human nucleoli listed over 200 to 700 nonribosomal nucleolar proteins[3,9-11]. The exact functions of many of these proteins are unknown.
In this report, we describe the initial functional characterization of murine transducin-β-like 3 (Tbl3). At least four proteins have been described as “transducin-β-like” based on DNA sequence analysis, including transducin-β-like 1 (Tbl1), transducin-β-like related 1 (Tblr1), transducin-β-like 2 (Tbl2) and tbl3. tbl1 and tblr1 have been shown to be transcription co-regulators (co-repressors) in the signal transduction pathway of retinoic acid receptor-α[12-15]. Tbl2 is a putative protein associated with the Williams-Beuren syndrome based on gene mapping but its function is entirely unknown[16]. Very little is known about tbl3[17]. A literature search yielded 8 publications that mentioned Tbl3. Of these, one is a gene mapping study and two are disease-gene association studies[18-20]. Three are proteomic studies[9-11]. The seventh describes the composition of the co-repressor complex associated with the retinal photoreceptor-specific nuclear receptor (PNR)[21]. The most recent report is from the authors and collaborators and focuses on the phenotypic characterization of a zebra fish mutant, ceylon (cey) with homozygous deletions of Tbl3 (in addition to four other uncharacterized genes)[22]. The cey mutant embryos have normal tissue specification but the sizes of some organs or cell populations such eye, pancreas, T cells and erythrocytes are markedly diminished due to reduced cellular proliferation, which in turn is attributed to cell cycle slowing as a result of Tbl3 deficiency. The proliferative defect of the cey mutant becomes apparent 3-4 d post fertilization (dpf). All mutant embryos die by 10-14 dpf due to depletion of maternally derived Tbl3 mRNA in the embryos[22]. The embryonic lethality of Tbl3 deletion highlights its importance in tissue/organ development. However neither the site of Tbl3 expression nor the mechanism of cell cycle slowing was elucidated. Here, we provide direct experimental evidence that mouse Tbl3 is targeted to the nucleoli where it plays an important role(s) in regulating the production of the 47S pre-rRNA.
The full-length coding sequence of mouse Tbl3 was amplified from oligo d(T)-primed cDNAs of MPRO (Mouse Promyelocyte) cells[23], cloned in frame into the HindIII/SalI sites of the pEGFP C3 (for N-terminal fusion with EGFP) and pEGFP N1 vector (for C-terminal fusion)(Clontech). The truncation mutants of Tbl3 (del 1-632, del 1-400, del 632-801, del 401-632) were generated by polymerase chain reaction (PCR) amplification of the desired coding sequences and cloned into the HindIII/SalI sites of pEGFP N1 or C3 vector. The N-terminal deletion mutants (del 1-10, del 1-12, del 1-16 del 1-20) and C-terminal deletion mutants (del 777-801, del 753-776, del 733-752, del 713-732, del 693-712, del 673-692, del 653-672, del 633-652) were also generated using PCR-based deletion methods. All plasmids were sequenced to ensure accuracy. DNAs were purified using a plasmid DNA purification kit (Qiagen) and further purified by phenol-chloroform extraction and ethanol precipitation.
The short hairpin RNA or shRNA sequences for mouse Tbl3 were designed using the shRNA Sequence Designer (Clontech). The sequences of the two shRNAs we used in the current study are: 5’ccggTGCCAAGGATCAGAGCATAttcaagaga-TATGCTCTGATCCTTGGCAttttttg and 5’ccggTGGCCATTACCTCTTCTGT-ttcaagagaACAGAAGAGGTAATGGCCAttttttg (uppercases denote Tbl3-derived sequences). Synthetic oligonucleotides were annealed and cloned into the AgeI/EcoRI sites of the pMKO 1p vector (referred to as pMKO hereafter)[24]. pMKO contains a puromycin resistance gene expression cassette and confers puromycin resistance in transfected mammalian cells. Both constructs provided specific knockdown but the first shRNA exhibited a stronger knockdown effect and was used in most experiments. Either pMKO or pMKO-Luc shRNA (Luc stands for luciferase) was used as the negative control in all knockdown experiments with similar results.
NIH 3T3 cells were transfected with the pEGFP N1 or C3 fusion constructs by calcium phosphate method. MPRO cells were maintained in Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco BRL) and the conditioned medium of BHK/HM-5 cell line as a source of murine granulocyte-macrophage colony stimulating factor (10% vol/vol)[23]. MPRO cells were transfected with 10 μg of pMKO (or pMKO-Luc shRNA) vs pMKO-Tbl3 shRNA DNAs per 2.5 × 106 cells by electroporation using Gene Pulser Xcell (Bio-Rad). Transfected cells were selected with puromycin (0.375 μg/mL)(Sigma) for 8-10 d to establish stable transfectants.
LAP-3 fibroblasts[25] were transfected in triplicates with 5 μg of pMKO (or pMKO-Luc shRNA) vs pMKO-Tbl3 shRNA by the Lipofectamine (Life Technologies) method in parallel to minimize variation in toxicity and transfection efficiency. After 24-48 h, transfected cells were detached by trypsin/EDTA and subcultured at 1:10-20 ratios in new 60-mm tissue culture dishes and selected with puromycin (1.5 μg/mL) for 5-10 d. Colonies were fixed and stained in situ with Coomassie Stain (BioRad).
Total cellular RNAs were extracted from the same starting numbers of MPRO cells stably transfected with pMKO (or pMKO-Luc shRNA) or pMKO-Tbl3 shRNA in parallel using an RNeasy kit (Qiagen). Total RNAs extracted from equal numbers of starting cells were electrophoresed in 1% formaldehyde-agarose gels, blotted onto Hybond-N (Amersham) and hybridized with a randomly primed, 32P-labled Tbl3 probe in RapidHyb buffer (Amersham) at 70 °C for 3 h, followed by two washings with 2 × standard sodium citrate buffer (SSC) plus 0.1% sodium dodecyl sulfate (SDS) at room temperature for 15 min. The final wash was done with 0.1 × SSC plus 0.1% SDS at 56 °C for 15 min.
To study the synthesis and processing of pre-rRNA, equal numbers of stably transfected MPRO/pMKO (or pMKO-Luc shRNA) and MPRO/pMKO-Tbl3 shRNA were labeled with 2.5 μCi/mL of 5,6-3H-uridine (Perkin Elmer) for 30 min at 37 °C, washed with phosphate buffered saline (PBS) and chased for 0-180 min at 37 °C in nonradioactive media. Total RNAs were purified using an RNeasy kit. In some experiments, Trizol agent was used to recover the 5.8S rRNA quantitatively. Total RNAs from equal numbers of starting cells were electropheresed in 1% formaldehyde-agarose gels and blotted onto Hybond-N. For fluorography, Northern blots were spray-coated with En3Hance Spray (Perkin Elmer) and exposed to X-ray films directly without covering at -80 °C for 1-14 d. The fluor graphs were analyzed using a BioRad Gel Doc XR+.
For ribosome profiling, equal numbers of MPRO/pMKO (or MPRO/pMKO-Luc shRNA) and MPRO/pMKO-Tbl3 shRNA were washed with PBS and pelleted at 4 °C and re-suspended in polysome lysis buffer containing 100 mmol/L KCl, 5 mmol/L MgCl2, 20 mmol/L HEPES (pH 7.4), 1% NP-40, 1 mmol/L dithiothreitol, heparin sodium (200 μg/mL), phenylmethylsulfonyl fluoride (1.0 mmol/L) (Sigma) and RNasin (100 unit/mL)(Promega). Cells were homogenized using a 1-ml syringe with a 25-gauge needle for eight times and centrifuged at 8000 g for 10 min at 4 °C. The supernatant was layered on a 10%-45% (wt/vol) sucrose density gradient made in the polysome gradient buffer (100 mmol/L KCl, 5 mmol/L MgCl2, 20 mmol/L HEPES, pH 7.4) at 38000 g for 3 h at 4 °C in a Beckman SW55Ti rotor and analyzed using a UA-6 Absorbance Detector (Isco). Of note, no cycloheximide was added to the lysates or sucrose gradients to allow the polysomes to dissociate into a single peak of 80S monosomes (i.e., 40S plus 60S subunits) to facilitate the comparison of the total amounts of ribosomes.
NIH3T3 cells expressing Tbl3-EGFP fusion protein were lysed in RIPA buffer. Lysates were denatured and electrophoresed in a 4%-12% denaturing SDS PAGE (NuPage; InVitrogen) along with Magic Mark XP Western Protein Standard (Life Technologies), blotted onto Immobilon-P (Millipore), probed with a rat monoclonal anti-GFP antibody (Pierce), followed by biotinylated goat anti-rat Igs (Pierce) and streptavidin-conjugated horseradish peroxidase (Amersham) and visualized by enhanced chemiluminescence.
The cDNA of the largest open reading frame of mouse Tbl3 was cloned from the MPRO[23] cell line by RT-PCR. It contains 2,406 nucleotides (nt) and encodes a protein of 801 aa. Sequence alignments show that it is highly homologous to human TBL3, Schizosaccharomyces pombe utp13, and Saccharomyces cerevisiae utp13 (Figure 1). Thirteen WD40 repeats (also known as beta-transducin repeats) are present in mouse tbl3. The WD40 repeat consists of the consensus aa sequence [X6-94-(GH-X23-41-WD)] and is found in many proteins involved in signal transduction, rRNA processing, gene regulation, vesicular trafficking and cytoskeletal assembly[26,27]. Crystallography studies reveal that the WD40 repeats cluster together to form a β-propeller structure, which serves as a rigid platform for multiple protein-protein interactions[27]. A second region of homology is found in the C-terminus of mouse Tbl3 and yeast utp13. The conserved sequence in the C-terminus has not been found in any other protein. It is likely that this region contains a functional domain(s) unique to mouse Tbl3 and yeast utp13.
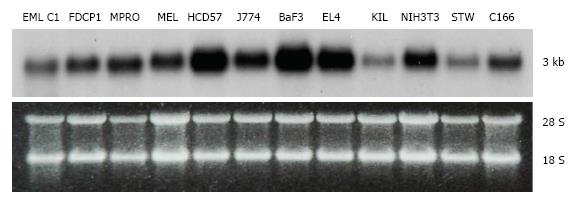
Tbl3 is expressed in all cell lines and tissues that we have examined. In all cases, there is only one band of Tbl3 mRNA measuring approximately 3 kb in length in each cell type (Figure 2). The level of Tbl3 mRNA varies significantly from one cell type to another and roughly correlates with the proliferative or biosynthetic activity of the cells. Among the cell lines surveyed, erythroleukemia (HCD57)[28], macrophage (J774), B-cell lymphoma (BaF3), T-cell lymphoma (EL4) and NIH3T3 fibroblasts express the highest levels of Tbl3. In normal tissues, hepatocytes, myocardium, testes, hematopoietic progenitors, CD8+ T cells and CD14+ monocytes express 3-5 times more Tbl3 than the average tissue (not shown).
To study the subcellular localization of mouse tbl3, we constructed an expression vector expressing mouse Tbl3 as a fusion protein with EGFP in its C-terminus. The vector was transfected into NIH3T3 cells (or CV-1 or 293 cells). Fluorescence microscopy revealed that the overwhelming majority of the fusion protein was targeted to the nucleoli with a very small fraction appearing in the nucleoplasm (Figure 3A-E). The same result was obtained when EGFP was fused to the N-terminus of Tbl3 (not shown). The small nucleoplasmic pool (in a very fine punctate pattern at high magnifications) may represent genuine extra-nucleolar distribution of Tbl3 or an overexpression artefact. Western blot analysis using a monoclonal anti-GFP antibody yielded a single band of tbl3-EGFP fusion protein of approximately 116 kD (Figure 3F). Thus, the deduced apparent molecular weight of mouse Tbl3 is 89-90 kDa.
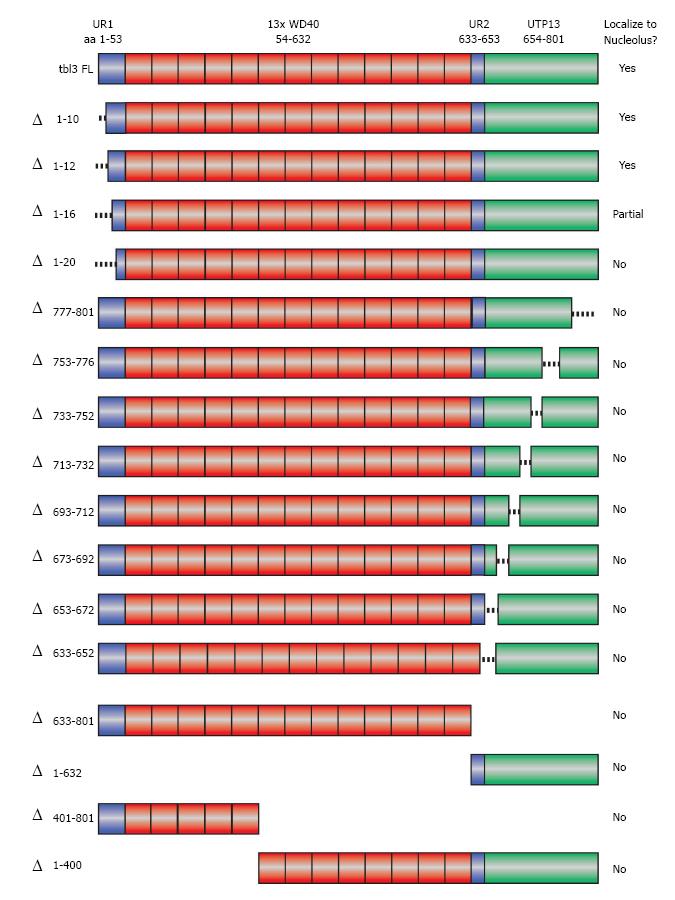
No previously reported nucleolar targeting motifs have been identified in Tbl3. To determine the aa sequence required for nucleolar localization of tbl3, we constructed a series of scanning deletion mutants of Tbl3 as EGFP fusion proteins (Figure 4). These constructs were transfected into NIH3T3 fibroblasts and examined by fluorescence microscopy. Among the constructs examined, only the full-length Tbl3 and those with very short (10-16 aa) N-terminal deletions could localize to the nucleoli. The remaining constructs yielded a diffuse pattern throughout the cytoplasm and nucleoplasm (but largely excluded from the nucleoli), a pattern that is indistinguishable from that of EGFP per se. This result suggests that the overall topology and/or the WD40 repeats rather than a unique localization signal are responsible for the nucleolar localization of tbl3.
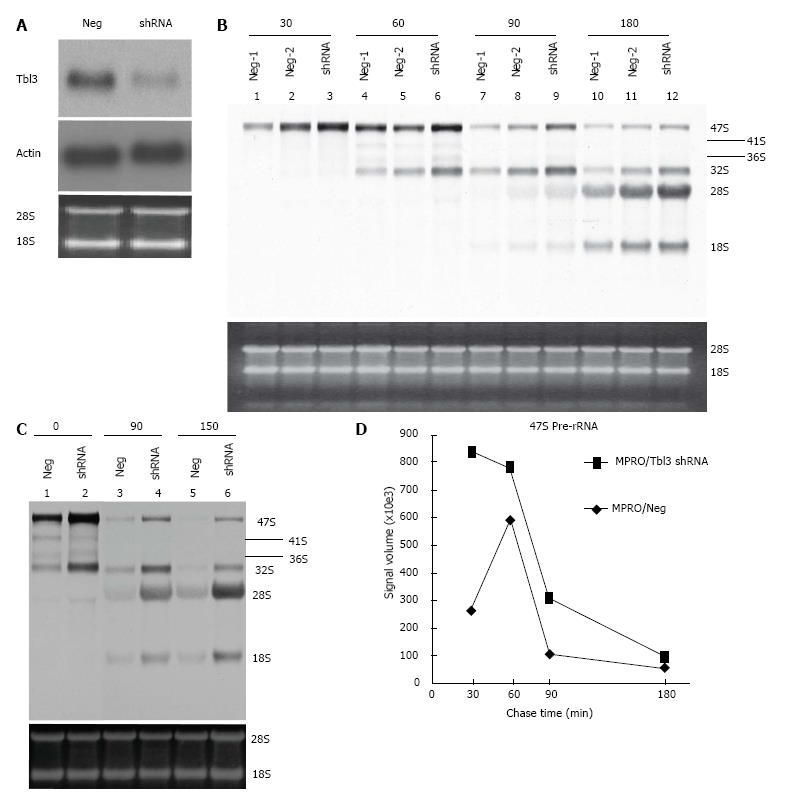
To study the function of Tbl3, we used the MPRO murine promyelocyte cell line as a model system. MPRO was established from a normal mouse marrow[23]. The differentiation of MPRO is reversibly blocked at the promyelocyte stage. DNA array and proteomic studies have shown that MPRO, unlike most transformed leukemia cell lines, closely reflects the physiology of normal promyelocytes[29]. To investigate the function of Tbl3 in MPRO, we trasnsfected the MRPO cells with either pMKO (or pMKO-Luc shRNA; negative controls) or pMKO-Tbl3 shRNA (1 or 2) vector and selected with puromycin to obtain stable transfectants. To avoid natural selection of variants with growth advantages, only low-passage (< 6) stable transfectants were used. Northern blot analyses demonstrated that the levels of Tbl3 mRNA were knocked down by approximately 50%-70% in the pMKO-Tbl3 shRNA transfectants while the levels of Tbl3 mRNA remained unchanged in the pMKO transfectants (Figure 5A, upper panel). The levels of housekeeping genes such as β-actin were unaffected by pMKO-Tbl3 shRNA (Figure 5A, middle panel).
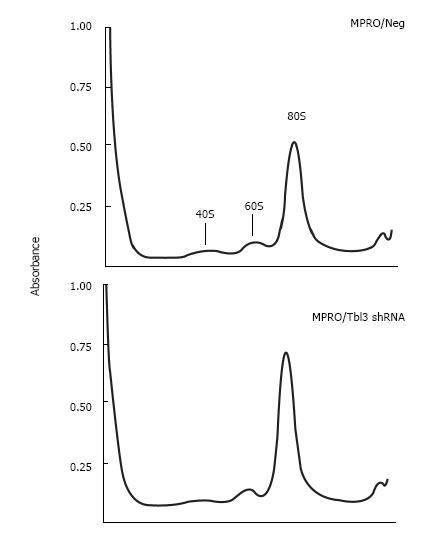
To investigate the effects of Tbl3 knockdown on newly transcribed pre-rRNA and its subsequent processing, we performed 3H-uridine pulse-chase experiments in the MPRO transfectants. 3H-uridine is phosphorylated after entering the cells and incorporated into newly transcribed RNAs, most of which are rRNAs. There was no detectable difference in the amounts of steady-state 28S and 18S rRNAs as revealed by ethidium bromide staining (Figure 5B and C, lower panels) and spectrophotometry. There was also no difference in the steady-state levels of the 5.8S rRNA on prolonged exposure (not shown). Intriguingly the levels of 3H-uridine-labeled, newly transcribed 47S pre-rRNA were consistently increased by 2 to 4 fold in pMKO-Tbl3 shRNA transfectants (Figure 5B and C, upper panels). Furthermore, there was no obvious delay or defect in the processing of the 47S pre-rRNA as evidenced by similar rates of disappearance in the negative control and shRNA groups (Figure 5D) and by the proportional and contemporaneous appearance of the 3H-uridine-labeled 28S and 18S rRNAs (and 5.8S rRNA; not shown) and the 41S, 36S and 32S processing intermediates (Figure 5B and C, upper panels). The key bands in lane 10 (Neg-1) and lane 12 (shRNA) of Figure 5B were further analyzed and the signal volume of the 28S rRNA was assigned a value of 1.00 to facilitate comparison. The calculated ratios of 3H-labeled 32S:28S:18S rRNAs are 0.47:1.00:0.66 for the negative control group and 0.28:1.00:0.65 for the shRNA group. This result is consistent with the interpretation that there is no obvious delay in processing. Ribosome profiling by sucrose gradient centrifugation revealed that there was also no significant difference in the amount or ratio of the 40S and 60S ribosomal subunits and the 80S monosomes in MPRO/pMKO (or MPRO/pMKO-Luc shRNA; negative controls) vs MPRO/pMKO-Tbl3 shRNA cells (Figure 6). This is consistent with the previous finding that the steady-state levels of 28S, 18S and 5.8S rRNA were not affected by Tbl3 knockdown (Figure 5B and C, lower panels). Together, the results of 3H-uridine pulse-chase and ribosome sucrose gradient centrifugation indicate that Tbl3 knockdown leads to increased levels of the 47S pre-rRNA but has no detectable effects on the processing of pre-rRNAs or the amount of steady-state 28S, 18S, 5.8S rRNAs or the amounts of the 40S and 60S subunits or the 80S monosomes.
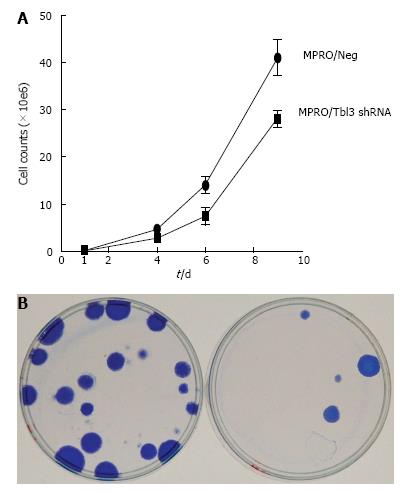
During the course of the study we noticed that the growth of newly established MPRO/pMKO-Tbl3 shRNA transfectants was very slow compared with MPRO/pMKO (or MPRO/pMKO-Luc shRNA) or the parental MPRO. Calculation based on the time it took for the same number of newly established MPRO/pMKO-Tbl3 shRNA stable transfectants to reach the same population size as MPRO/pMKO indicates that the doubling time of MPRO/pMKO-Tbl3 shRNA was 3-4 times longer than that of MPRO/pMKO in newly established transfectants. As the population expanded from a few transfectants to approximately 106 cells, the doubling time of MPRO/pMKO-Tbl3 shRNA became progressively shorter, apparently due to the selection or outgrowth of variants that were less affected by the knockdown effects and hence enjoyed a higher proliferative rate. Nevertheless, the growth rate of MPRO/pMKO-Tbl3 shRNA was still slower compared with MPRO/pMKO in the 2-3 wk after the initial expansion following transfection and puromycin election (Figure 7A).
To further examine the effects of Tbl3 knockdown on cellular proliferation, we used the LAP3 fibroblast cell line as the model system. The LAP3 fibroblast cell line is derived from NIH3T3 (transformed embryonic cells) but is substantially “weakened” compared with the parental line[25]. Theoretical considerations and empiric experience indicate that it is particularly suitable for uncovering defects in ribosome biogenesis[25]. LAP3 cells were transfected with pMKO (or pMKO-Luc shRNA; negative controls) vs pMKO-Tbl3 shRNA (1 or 2) in triplicates in parallel and allowed to form macroscopic colonies in the presence of puromycin to select for stable integrants. The colonies were stained with Coomassie blue in situ to allow direct comparison. As shown in Figure 7B, the pMKO-Tbl3 shRNA-transfected group showed a great reduction (by about 80%) in colony numbers, indicating that Tbl3 knockdown also has deleterious effects on fibroblast cell growth and proliferation.
In this report we provide direct evidence that mouse Tbl3 is targeted to the nucleoli (Figure 3A-D). This finding implicates Tbl3 in the ribosome biogenesis pathway and/or other nucleolar events. The results of targeting studies using a series of scanning deletion mutants of Tbl3 suggest that no particular nucleolar targeting sequence is involved (Figure 4). Thus, Tbl3 likely associates with other proteins in the nucleoli via its WD40 protein-protein interaction motifs and/or other topological features and this forms the basis for its nucleolar localization.
To explore the function of mammalian Tbl3, we used the shRNA approach to achieve a partial knockdown. In retrospect, a partial rather than complete knockdown is desirable since a complete knockdown is likely incompatible with cellular survival based on what we have learned from our cell line studies and from the cey zebra fish embryos[22]. First, we looked at the processing of the 28S, 18S and 5.8S rRNAs. Then, we focused on the newly synthesized 47S pre-rRNA by pulse-labeling cells with 3H-uridine. The latter approach allowed us to focus on newly synthesized pre-rRNA and minimized the contribution of post-transcriptional processing. As shown in Figure 5B and C (bottom panels), Tbl3 knockdown had no discernable effect on the steady-state levels of the mature 28S, 18S and 5.8S rRNAs. Instead, it consistently increased the level of newly synthesized 47S pre-rRNA by two to four folds (Figure 5B and C, upper panels and Figure 5D). The higher levels of newly synthesized 47S pre-rRNA could result, a priori, from increased synthesis or decreased processing of the 47S pre-rRNA or both. If the elevated level of 47S pre-rRNA resulted from decreased processing, then we expect to see a slower rate of disappearance of the 3H-labeled 47S pre-rRNA or a higher 47S to 28S (or 47S to 18S or 47S to 58S) ratio or the ratio of 28S to any processing intermediate in cells with Tbl3 knockdown. However, our results showed that rates of disappearance of 47S pre-rRNA were very similar (Figure 5D) and the various ratios remained very similar in MPRO/pMKO-Tbl3 shRNA and in MPRO/pMKO (negative control) at all time points examined (Figure 5B and C, upper panels). This finding argues against a processing defect that would alter the size or quantity of the 28S, 18S and 5.8S rRNAs and raises the possibility that Tbl3 knockdown primarily increased the synthesis of the 47S pre-rRNA. This interpretation is consistent with the previous conclusion that higher levels of steady-state 47S pre-rRNA reflect higher rates of rDNA transcription in a given cell type[30].
What could be the potential mechanisms by which Tbl3 regulates the synthesis of the 47S pre-rRNA? As Tbl3 is the mammalian homologue of yeast utp13, a review of the data on yeast utp13 may shed some light. Both mouse Tbl3 and yeast utp13 contain thirteen WD40 protein-protein interaction repeats and a conserved region in the C-terminus that is unique to both Tbl3 and utp13. The presence of many WD40 repeats in Tbl3 and utp13 indicates that they very likely form a complex or complexes with other proteins. Indeed, proteomic studies indicate that yeast utp13 forms a primary subcomplex, the so-called “utp-B” complex, with five other nucleolar proteins, namely utp 6, utp 18, utp 21, dip2 and pwp2[31]. This primary subcomplex in turn associates with other subcomplexes such as “UTP-A” and “UTP-C” complexes as well as additional nucleolar proteins to form a 90S megacomplex, which is known as the “small subunit (SSU) processome”. The SSU is assembled co-transcriptionally at the 5’ end of the newly transcribed pre-rRNA[31]. The SSU megacomplex is visible on electron microscopy as a terminal knob on the leading end of the elongating pre-rRNA still attached to the rDNA chromatin in yeast nucleoli[32]. By analogy, mammalian Tbl3 may similarly form a megacomplex via its multiple WD40 motifs with other proteins involved in the synthesis and/or processing of pre-rRNA and associates co-transcriptionally with the 5’ end of the pre-rRNA. Given the observation that Tbl3 knockdown increases the production of 47S pre-rRNA, we hypothesize that Tbl3 is an important component of a putative pathway that normally provides feedback inhibition to the rDNA transcription machinery to coordinate the synthesis of the 47S pre-rRNA with the subsequent processing of rRNAs and ribosome assembly. Without coordination or feedback regulation, the rate of pre-RNA synthesis may become out of sync with that of processing and assembly and this mismatch may cause errors or waste in ribosome biogenesis[33,34].
Since the production of rRNAs and ribosomes consumes the lion share of cellular energy and biosynthetic precursors, over production of rRNAs inevitably will deprive cells of the energy and biosynthetic precursors needed for other physiologic processes and result in decreased cellular proliferation. Thus the consumption or siphoning away of cellular energy and biosynthetic precursors in overproduction of pre-rRNA may explain at least in part the inhibitory effect of Tbl3 knockdown on cellular proliferation in mammalian cells (Figure 7A and B). As Tbl3 contains thirteen WD40 repeats it is possible that Tbl3 interacts with other nuclear or nucleolar proteins directly or indirectly involved in cell cycle regulation or DNA synthesis in addition to its interaction with components of SSU. The previously described detection of Tbl3 in the co-repressor complex of PNR in retina may well represent such an extra-nucleolar role[21]. However, we believe that the main role of Tbl3 is in the nucleoli as most (approximately 99%) if not all Tbl3 protein is found in that organelle (Figure 3A-D).
As we mentioned to earlier, complete depletion of yeast utp13 results in severe 18S rRNA processing defects and lethality in yeasts[32]. Considering the fact that Tbl3 is the mammalian homologue of yeast utp13, it is surprising that no rRNA processing defect was detected in the current study of mouse cells with partial Tbl3 knockdown (Figure 5). Similarly, there were no detectable defects in rRNA processing in the Tbl3-/Tbl3- (cey) zebra fish embryos (supplemental Figure 6 of ref. 22). How can one reconcile these differences? The simplest explanation is that yeast utp13 and mammalian Tbl3 (or zebra fish tbl3) have evolved divergently in functionality. It is also quite possible that additional or redundant factors or mechanisms have evolved in mammals (or zebra fish) for rRNA processing such that a deficiency of Tbl3 alone has no discernible impact on rRNA processing. Still another possibility is that Tbl3 has more than one function and while a total deficiency is required to completely block the rRNA processing function, a partial deficiency is sufficient to interfere with the regulation of pre-rRNA synthesis.
The clinical importance of ribosomes is underscored by the fact that defects in ribosomes or ribosome biogenesis are the causes of several bone marrow failure syndromes such as the Diamond-Blackfan anemia (defective erythropoiesis due to rRNA processing defects caused by mutations in ribosomal protein genes RPS17 or RPS19 or RPS24)[35] and Shwachman-Bodian-Diamond syndrome (bone marrow failure, exocrine pancreas insufficiency, skeletal abnormalities and disposition to myelodysplastic syndrome and acute myeloid leukemia; caused by uncoupling of GTP hydrolysis from eIF6 release on the 60S ribosome subunit)[36]. More recently, a subtype of myelodysplastic syndrome, the so-called “5q- syndrome”, has been experimentally phenocopied by RNAi-mediated haploinsufficiency of RPS14[37]. The propensity of ribosomopathy disorders to afflict hematopoiesis probably reflects the sensitivity of the highly proliferative hematopoietic progenitors to any disturbances in DNA or protein synthesis. Given the pronounced inhibitory effect of partial Tbl3 knockdown on cellular proliferation in the current study, it is tempting to speculate that a loss of function mutation or deletion of human TBL3 may also lead to a bone marrow failure-like syndrome. Furthermore, TBL3 may provide an attractive target for anti-tumor therapy.
The authors would like to thank Drs. Lester Lau and Dmitri Pestov for providing the LAP-3 fibroblasts.
Transducin β-like 3 (tbl3) encodes a protein with thirteen WD40 protein-protein interaction motifs. Virtually nothing is known about the function of Tbl3 including its subcellular localization and the general nature of its function. This report describes the first direct evidence that Tbl3 is targeted to the nucleoli and plays an important role in regulating the synthesis of the 47S pre-ribosomal RNA (pre-rRNA), i.e., the first step in the production of ribosomes, which are indispensable for protein synthesis and hence the survival and normal functions of all cells.
This article addresses a sizable gap in the understanding of the very early stage of the production of ribosomes. Understanding the function(s) of Tbl3 may lead to the development of new antibiotics and anti-tumor drugs.
Tbl3 is a newly identified nucleolar protein with a previously unrecognized regulatory function in the very early stage of ribosome production.
Given the pronounced inhibitory effect of Tbl3 knockdown on cellular proliferation as demonstrated in the current study, the authors suspect that a loss of function mutation or deletion of human TBL3 may also lead to a bone marrow failure-like syndrome or other developmental defects. Furthermore, TBL3 may provide a good target for anti-cancer therapy by interfering with the production of ribosomes at a very early stage.
Ribosomopathy: defects in ribosome production and/or function including the synthesis, modification and processing of ribosomal RNA and assembly of ribosomes. Bone marrow failure syndromes: diseases caused by proliferative and/or differentiation defects in the production of blood cells by bone marrow stem cells.
Authors provided the direct evidence that murine Tbl3 protein is targeted to compartment of nucleoli and plays an important role in synthesis of 47S pre-rRNA. Thus, this protein is important for regulation of ribosome biosynthesis. Manuscript is well written and new information about localization of Tbl3 protein in nucleolus is very interesting from the complex view on proteome of nucleolus.
P- Reviewer: Bartova E, Komada M, Loreni F S- Editor: Gou SX L- Editor: A E- Editor: Lu YJ
| 1. | Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196-3205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 558] [Cited by in RCA: 613] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 2. | Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583-4588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 3. | Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1095] [Cited by in RCA: 1217] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 4. | Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673-683. [PubMed] |
| 5. | Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 383] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 6. | Lapeyre B. Conserved ribosomal RNA modification and their putative roles in ribosome biogenesis and translation. Topics Curr Genetics. 2005;12:263-284. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Raska I, Koberna K, Malínský J, Fidlerová H, Masata M. The nucleolus and transcription of ribosomal genes. Biol Cell. 2004;96:579-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1404] [Cited by in RCA: 1449] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 9. | Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100-4109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 374] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 10. | Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 760] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 11. | Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 913] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 12. | Bassi MT, Ramesar RS, Caciotti B, Winship IM, De Grandi A, Riboni M, Townes PL, Beighton P, Ballabio A, Borsani G. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Genet. 1999;64:1604-1616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 343] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 14. | Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 357] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 15. | Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 445] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 16. | Pérez Jurado LA, Wang YK, Francke U, Cruces J. TBL2, a novel transducin family member in the WBS deletion: characterization of the complete sequence, genomic structure, transcriptional variants and the mouse ortholog. Cytogenet Cell Genet. 1999;86:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 494] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 18. | Weinstat-Saslow DL, Germino GG, Somlo S, Reeders ST. A transducin-like gene maps to the autosomal dominant polycystic kidney disease gene region. Genomics. 1993;18:709-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 19. | Yoshida T, Kato K, Yokoi K, Oguri M, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nozawa Y. Association of genetic variants with hemorrhagic stroke in Japanese individuals. Int J Mol Med. 2010;25:649-656. [PubMed] |
| 20. | Yoshida T, Kato K, Yokoi K, Oguri M, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nozawa Y. Association of gene polymorphisms with chronic kidney disease in Japanese individuals. Int J Mol Med. 2009;24:539-547. [PubMed] |
| 21. | Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26:764-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Hutchinson SA, Tooke-Locke E, Wang J, Tsai S, Katz T, Trede NS. Tbl3 regulates cell cycle length during zebrafish development. Dev Biol. 2012;368:261-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Tsai S, Collins SJ. A dominant negative retinoic acid receptor blocks neutrophil differentiation at the promyelocyte stage. Proc Natl Acad Sci USA. 1993;90:7153-7157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 167] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 1221] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 25. | Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516-5528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 154] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 26. | Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1085] [Cited by in RCA: 1134] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 27. | Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 895] [Cited by in RCA: 926] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 28. | Ruscetti SK, Janesch NJ, Chakraborti A, Sawyer ST, Hankins WD. Friend spleen focus-forming virus induces factor independence in an erythropoietin-dependent erythroleukemia cell line. J Virol. 1990;64:1057-1062. [PubMed] |
| 29. | Lian Z, Wang L, Yamaga S, Bonds W, Beazer-Barclay Y, Kluger Y, Gerstein M, Newburger PE, Berliner N, Weissman SM. Genomic and proteomic analysis of the myeloid differentiation program. Blood. 2001;98:513-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Cui C, Tseng H. Estimation of ribosomal RNA transcription rate in situ. Biotechniques. 2004;36:134-138. [PubMed] |
| 31. | Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27:5414-5429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 142] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 545] [Cited by in RCA: 558] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 33. | Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 427] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 34. | Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506-2517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 198] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 35. | Choesmel V, Fribourg S, Aguissa-Touré AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17:1253-1263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 36. | Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, González Fernández A, Simpson P, D’Santos CS, Arends MJ. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917-929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 230] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 37. | Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 756] [Cited by in RCA: 674] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 38. | Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831-2841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 220] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 39. | DeHart SL, Heikens MJ, Tsai S. Jagged2 promotes the development of natural killer cells and the establishment of functional natural killer cell lines. Blood. 2005;105:3521-3527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |