Published online Jun 18, 2023. doi: 10.5312/wjo.v14.i6.387
Peer-review started: February 15, 2023
First decision: March 24, 2023
Revised: April 6, 2023
Accepted: May 6, 2023
Article in press: May 6, 2023
Published online: June 18, 2023
Processing time: 123 Days and 15 Hours
Artificial intelligence and deep learning have shown promising results in medical imaging and interpreting radiographs. Moreover, medical community shows a gaining interest in automating routine diagnostics issues and orthopedic measure
To verify the accuracy of automated patellar height assessment using deep learning-based bone segmentation and detection approach on high resolution radiographs.
218 Lateral knee radiographs were included in the analysis. 82 radiographs were utilized for training and 10 other radiographs for validation of a U-Net neural network to achieve required Dice score. 92 other radiographs were used for automatic (U-Net) and manual measurements of the patellar height, quantified by Caton-Deschamps (CD) and Blackburne-Peel (BP) indexes. The detection of required bones regions on high-resolution images was done using a You Only Look Once (YOLO) neural network. The agreement between manual and auto
Proximal tibia and patella was segmented with accuracy 95.9% (Dice score) by U-Net neural network on lateral knee subimages automatically detected by the YOLO network (mean Average Precision mAP greater than 0.96). The mean values of CD and BP indexes calculated by orthopedic surgeons (R#1 and R#2) was 0.93 (± 0.19) and 0.89 (± 0.19) for CD and 0.80 (± 0.17) and 0.78 (± 0.17) for BP. Automatic measurements performed by our algorithm for CD and BP indexes were 0.92 (± 0.21) and 0.75 (± 0.19), respectively. Excellent agreement between the orthopedic surgeons’ measurements and results of the algorithm has been achieved (ICC > 0.75, SEM < 0.014).
Automatic patellar height assessment can be achieved on high-resolution radiographs with the required accuracy. Determining patellar end-points and the joint line-fitting to the proximal tibia joint surface allows for accurate CD and BP index calculations. The obtained results indicate that this approach can be valuable tool in a medical practice.
Core Tip: This study presents an accurate method for automatic assessment of patellar height on high-resolution lateral knee radiographs. First, You Only Look Once neural network is used to detect patellar and proximal tibial region. Next, U-Net neural network is utilized to segment bones of the detected region. Then, the Caton-Deschamps and Blackburne-Peel indexes are calculated upon patellar end-points and joint line fitted to proximal tibia joint surface. Experimental results show that our approach has the potential to be used as a pre- and postoperative assessment tool.
- Citation: Kwolek K, Grzelecki D, Kwolek K, Marczak D, Kowalczewski J, Tyrakowski M. Automated patellar height assessment on high-resolution radiographs with a novel deep learning-based approach. World J Orthop 2023; 14(6): 387-398
- URL: https://www.wjgnet.com/2218-5836/full/v14/i6/387.htm
- DOI: https://dx.doi.org/10.5312/wjo.v14.i6.387
Patellofemoral joint (PFJ) disorders are common structural and functional problems that may cause pain and instability leading to joint degeneration. The knee osteoarthritis (OA) is one of the most common diseases and its incidence is increasing due to the aging and obesity[1]. Overall, the treatment costs related to knee OA are substantial. Recent developments in medical informatics and artificial intelligence have led to an increasing number of studies on medical imaging of knee OA, but most of the research is focused on the tibiofemoral joint[2]. However PFJ, a third articulation has received insufficient attention. The assessment of patellar height (PH) is a fundamental parameter in diagnosing PFJ pathologies, selection of appropriate treatment and postoperative evaluation[3]. The standard approach for PH assessment is achieved by measuring ratios on X-ray images in the lateral view[4]. PH abnormalities can be recognized using Insall-Salvati, Blackburne-Peel (BP), and Caton-Deschamps (CD) indexes measured on lateral knee radiographs[5]. Patella Baja (PB) and pseudo-Patella Baja are common complications after total knee replacement (TKR) and related to poor outcomes[6]. From a therapeutic point of view, pseudo-Patella Baja with the elevated femorotibial joint line without shortening the patellar tendon can be recognized with the lower BP and CD indexes. The CD and BP ratios are also more important in the diagnosis of PB after TKR[6].
Recently, a substantial attention to reliability of these parameters have been devoted, among others, due to increasing importance of PH in knee replacement surgery, tibial osteotomy, and anterior cruciate ligament reconstruction[3,7,8]. However, such routine tasks are tedious, time-consuming, and prone to considerable inter-observer and intraobserver variability[9]. In recent years, deep-learning (DL) algorithms have gained popularity in medicine, particularly in different aspects of orthopedics[10-12]. They have been used for image classification[13], fracture detection[14] or osteoarthritis diagnosis[15]. Moreover, DL algorithms have also been developed for automatic segmentation of bones on X-ray images to estimate hallux-valgus angles, axial alignment[16,17] and skeletal metastases[18]. Bone segmentation on X-ray images enables calculation of critical radiological parameters. It is a very difficult task[19], but some work has been done in this area[20,21].
To the best of our knowledge, there are no papers describing the application of neural networks for segmentation of lateral knee radiographs and measurements of patellar indexes on high-resolution radiographs. The method is automatic as it detects the region of interest (ROI) with the patella and proximal tibia region on a high-resolution radiograph, performs bone segmentation on the cropped ROI and then calculates the patellar indexes. The rest of the article is organized as follows. In the next Section we present our dataset for training You Only Look Once (YOLO) neural network, our dataset for training U-Net neural network, training procedure to achieve assumed Dice score and our algorithm for automated calculation of patellar indexes using segmented bones. In the following Section we present experimental results. The last part is devoted to discussion and conclusions.
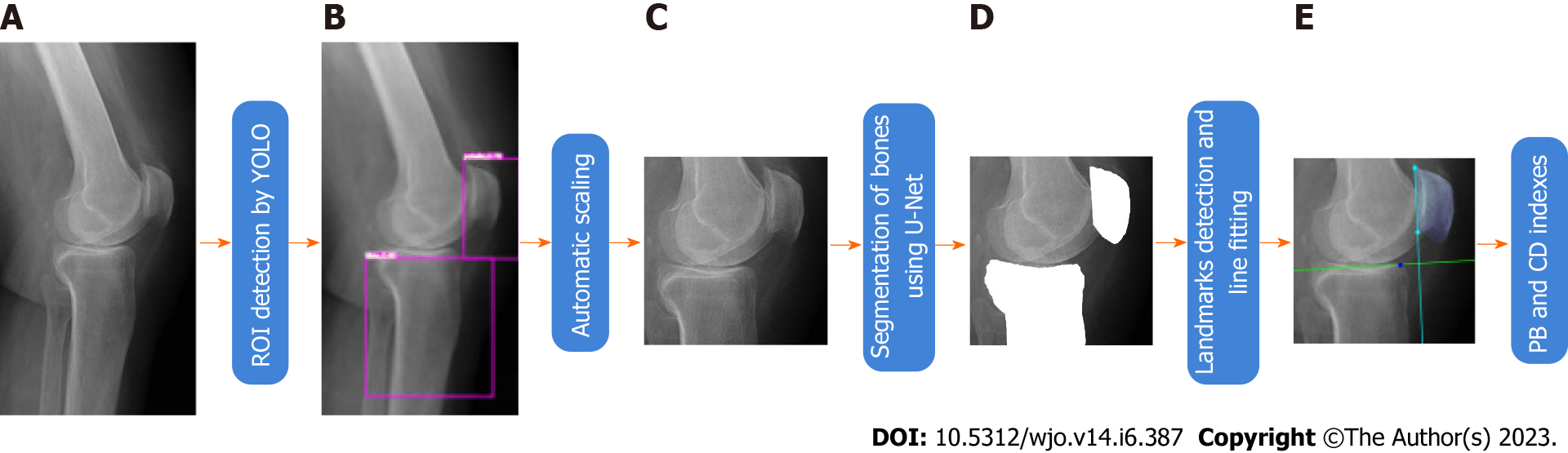
The overall pipeline of our method is shown in Figure 1. In order to detect ROI on radiographs we trained a YOLO neural network[22]. We have trained a U-Net model for automatic segmentation of bones of interest. The U-Net segmented the patella and tibia bones. On the segmented patella we extracted two landmark points and then fitted a patellar articular joint line to them. A second line has been fitted to the segmented proximal tibial articular surface. These lines have been used to calculate the CD and the BP indexes. The CD and BP indexes estimated in such a way have been compared with indexes calculated by medical doctors. The YOLO and U-Net networks have been trained on data collected and labelled by us. Radiological images were acquired using Shimadzu BR120 X-Ray Stand (Shimadzu Medical Systems, United States) and CDXI Software (Canon, United States).
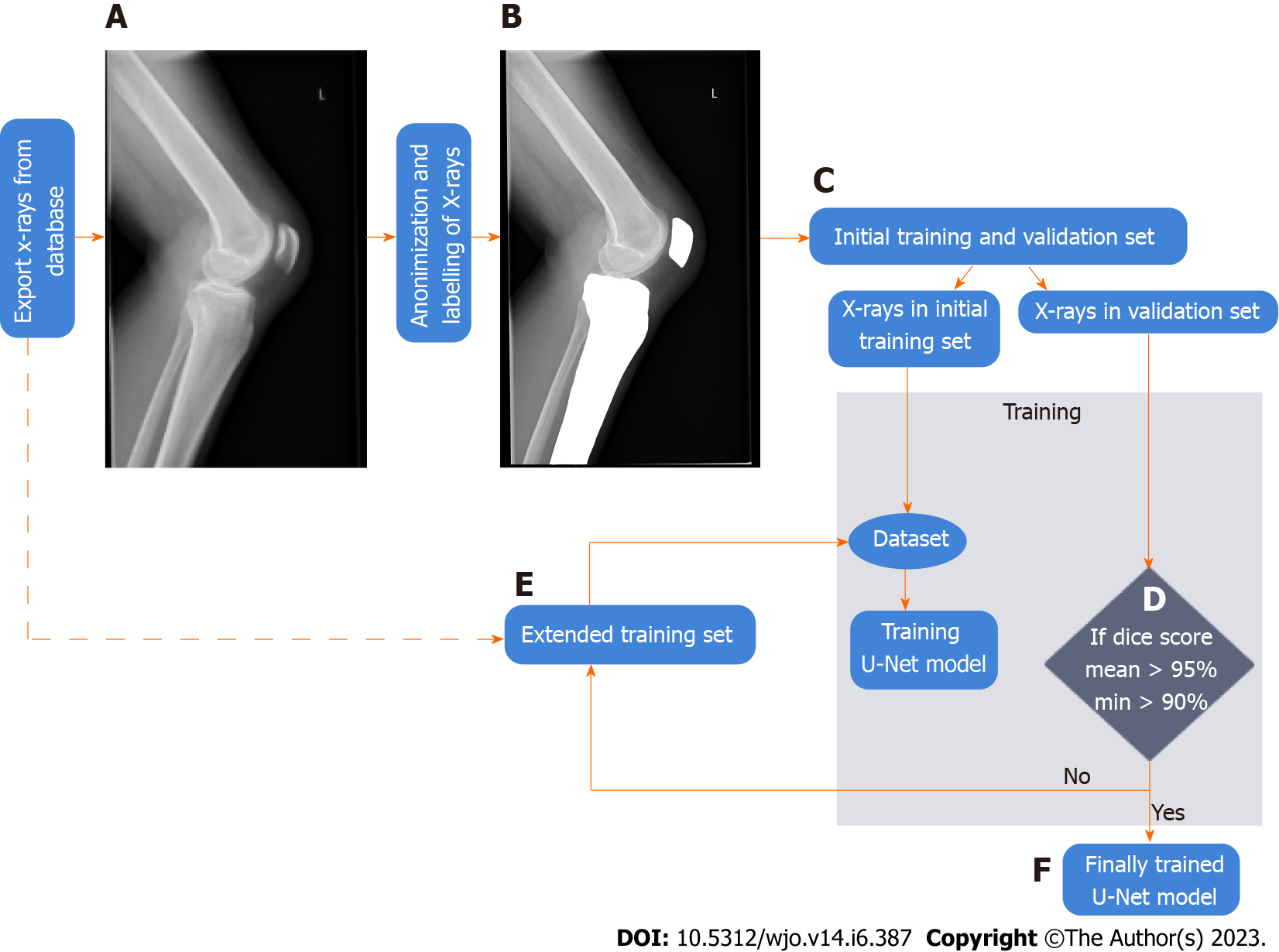
In this research, retrospective analysis of prospectively collected images was conducted on radiographs from an electronic database acquired in our institution. Patients whose radiographs were included in this study were examined in the authors’ institution from January 2019 to April 2021. We collected a dataset for ROI detection and a dataset for bone segmentation. A dataset used for training the YOLO neural network is described below. The flowchart of data collection and methodology of training a U-Net neural network for bone segmentation is depicted in Figure 2 and described in more detail in subsequent paragraphs.
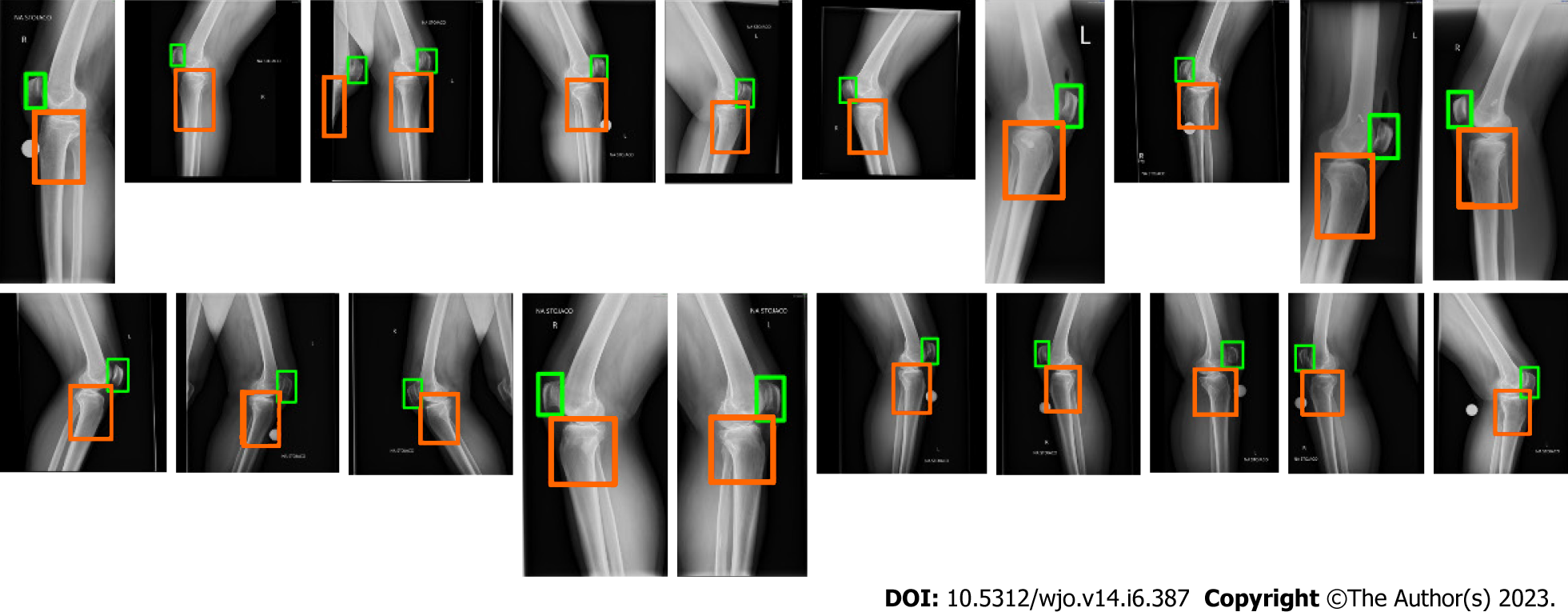
The images in our database are high-resolution images, and only a portion of the radiographs contains pixels belonging to the knee and patella, which are required for calculation of BP and CD indexes. Thus, a YOLO neural network has been used to automatically detect the area of interest[23], i.e., the patella and the proximal tibial artricular surface. One thousand radiographs from our database were selected, anonymized and then manually labeled. On each image we manually determined a rectangle surrounding the patella and a second rectangle surrounding the proximal tibial artricular surface. The YOLO neural network has been trained on 900 images and evaluated on 20 images.
For training a U-Net neural network we randomly selected semiflexed lateral knee radiographs from the electronic database of our institution (stage A in Figure 1 and in Figure 2). The exclusion criteria were: Radiographs performed with improper rotational positioning, grade III or more osteoarthritis in Kellgren-Lawrence scale[24] and/or severe axial knee deformations, visible growth plate, artificial elements distorting the image of the bone outline (e.g., osteosynthesis material or knee prosthesis implant), and heterotopic ossifications around the knee joint. The exported input X-ray images have been anonymized and stored in the lossless .png image format (stage B in Figure 2). Afterward, the bones on radiographs were manually labelled by an orthopedic surgeon (tibia and patella) in Adobe Photoshop ver. 22.0 (Adobe Systems, United States). The labelled images were included in the training and validation set (stage C in Figure 2).
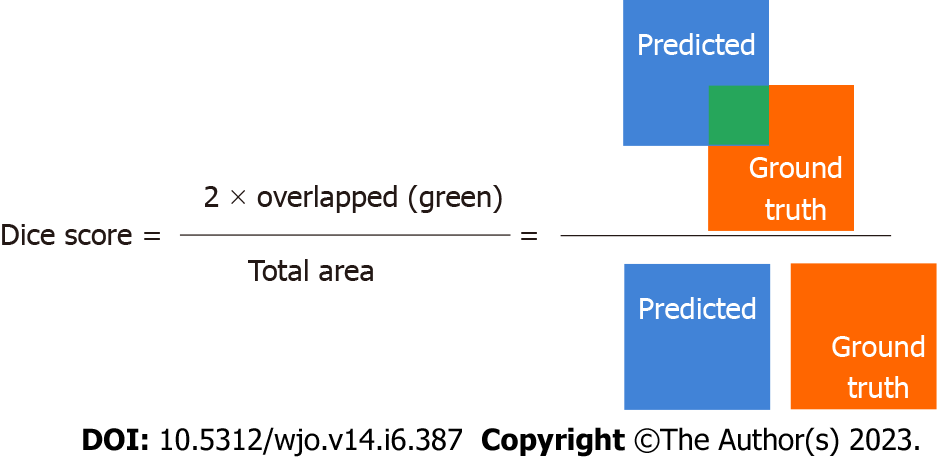
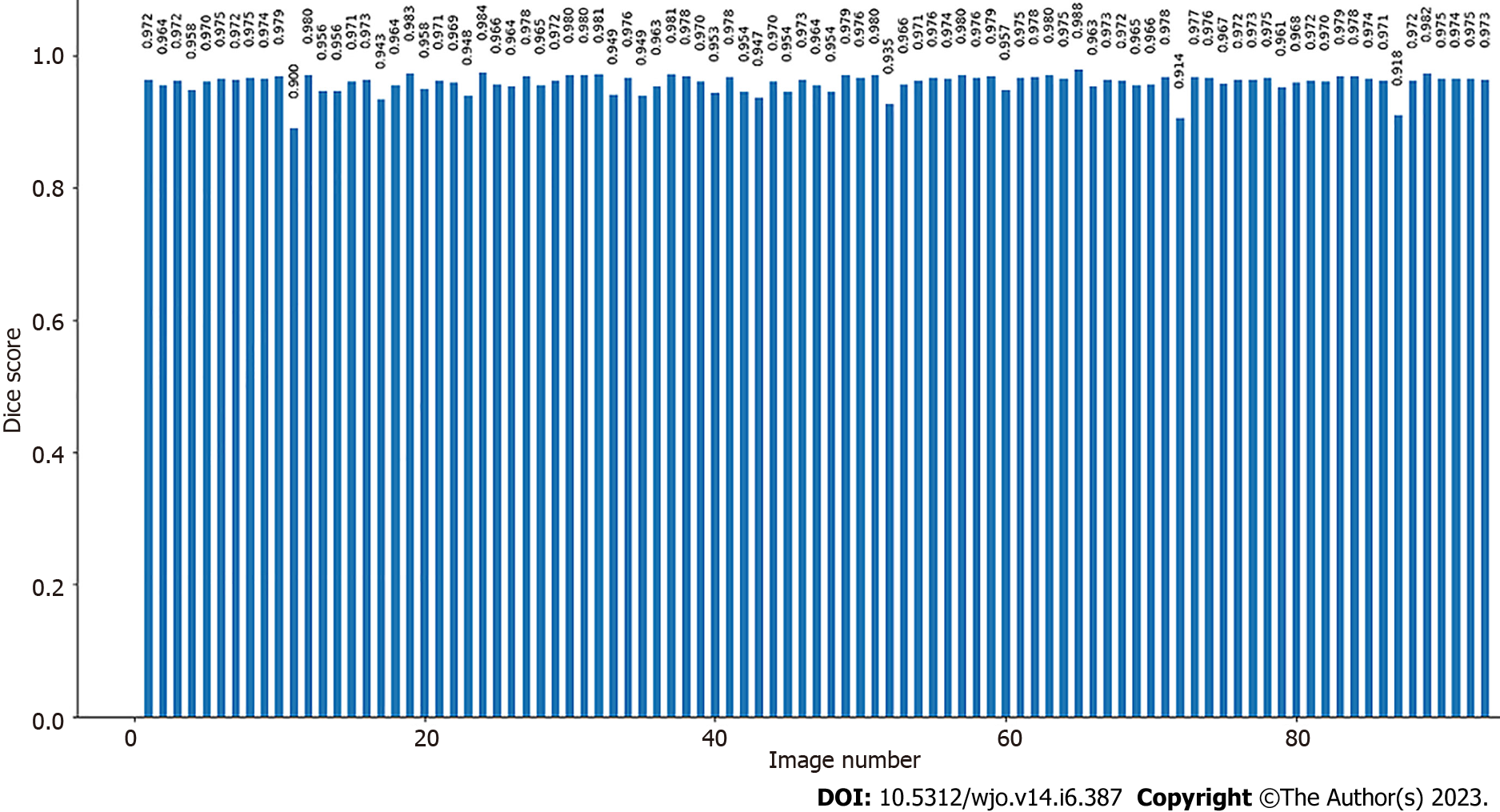
Bone segmentation has been performed using a U-Net[25]. The U-Net has been trained on the training set with verification of the accuracy of automatic segmentation (on validation set). The input images with knee area were automatically cropped on the basis of YOLO detections and then scaled to size 512 × 512 (stage B and C in Figure 1). During training, the resized radiographs were fed to the input of the U-Net, whereas corresponding labeled images were fed to the output of the U-Net. Initially, we manually labelled 50 X-rays of which 10 X-rays were included in the validation set to verify the accuracy of bone segmentation, (stage C in Figure 2). The accuracy was assessed using the Dice score[26] (Figure 3).
The validation set did not change during the training[27]. The U-Net was initially trained on a dataset consisting of 40 X-rays, which was then extended by 10 subjects to achieve the assumed segmentation quality (stage D in Figure 2). After every extension (by 10 radiographs), the U-Net was trained on the extended data from scratch, and the Dice score was calculated (on the validation set). If it was lower than 90%, the extension of the dataset and training of the network were repeated. If Dice score ≥ 90% was achieved, single radiographs were added to the training subset to gain a Dice score ≥ 95% (with minimal Dice score ≥ 90% for each subject (stage E in Figure 2). After the training, we obtained the final U-Net model and final training set (stage F in Figure 2, stage B in Figure 4). The final U-Net model was utilized to segment bones on the test subset. On the segmented bones we determined the characteristic points as well as necessary lines in order to calculate the patellar height and the indexes (stage D and E in Figure 1, stage C in Figure 4). The person responsible for training and evaluating the neural network has not participated in the manual calculation of the patellar indexes and has not seen the results before the final analysis.
The outcomes of the U-Net model (output images with segmented bones - tibia and patella) were used to determine characteristic points and lines (stage E in Figure 1). Articular surface of proximal tibia was automatically determined. Firstly, the anterior corner of the tibial plateau and posterior end-point were determined. Given such boundaries, a line to the points of the proximal tibia joint surface has been fitted. This way the joint line was determined on the basis of many boundary points. An alternative approach would be to determine some landmark points representing articular surface of proximal tibia and then using them to determine the joint line. In contrast to the approach mentioned above, in our approach the joint line is determined more robustly, i.e., on the basis of large number of boundary points segmented by the U-Net. Afterwards, the superior and inferior patellar articular surface endpoints have been determined through simple image analysis, and then used to determine the second line. Finally, these characteristic points and joint lines were automatically used to calculate the CD and the BP[28]. The calculation of the characteristic points and joint lines was done on images with height/with ratio equal to ROI ratio. The novelty of this research is an innovative approach to estimating joint lines based on the line fitting using automatic bone segmentation (more accurate than method based on the characteristic points, c.f. Discussion Section).
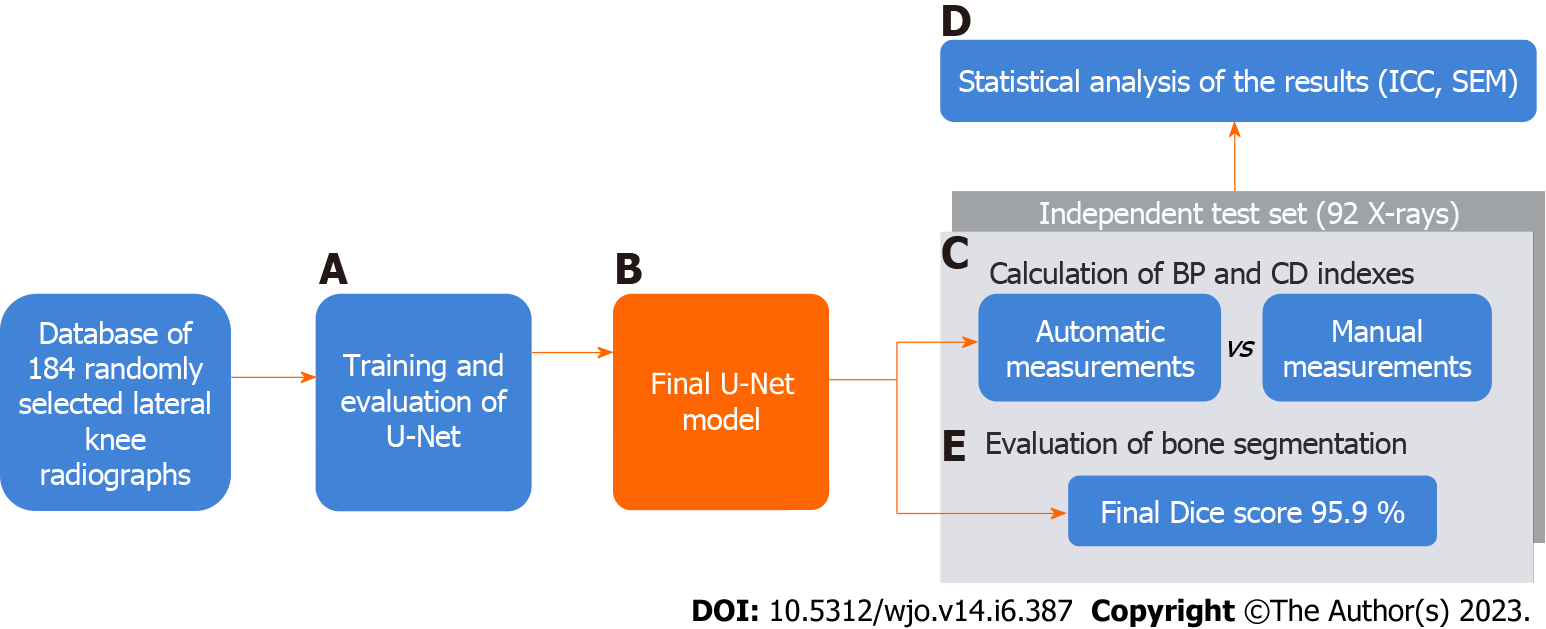
We used 92 randomly selected another lateral knee radiographs, collected from the authors’ institutional electronic database for the automatic and manual calculations of patellar indexes (stage F in Figure 1; stage C in Figure 4). We carefully examined if there is no overlap of patients (or images) in train/valid and test datasets. Like Lee et al[27], we did not create a test set until we had developed the final U-Net model (stage F in Figure 2; stage from B to C in Figure 4). According to Zou et al[29], we assumed a minimal number of subjects to estimate the agreement of the measurements between the two methods as 46.
Automatic and manual measurements were performed on 92 X-rays. The computer-generated results were compared with manual measurements performed independently by two orthopedic surgeons (6 and 38 years of experience, stage C in Figure 4). According to the guidelines[30,31], two orthopedic surgeons performed manual measurements of the CD and BP patellar height indexes. They used Carestream Software ver.12.0 (Carestream Health, United States).
Statistical analysis was performed using Statistica 13.1 Software (Tibco Software, United States) and Microsoft Excel 2019 (Microsoft, United States). The intraclass correlation coefficient (ICC) and the standard error for a single measurement (SEM) were calculated for statistical assessment of reliability and compliance of the measurements stage (D in Figure 4). The ICC of 0.4 demonstrates poor, between 0.4 and 0.75 good, and more than 0.75 indicates excellent reliability of the measurements[32].
The accuracy of bone segmentation (of the final U-Net model) was assessed on the test set to check if the neural network had sufficient generalization capability (whether U-Net could achieve satisfactory results on unknown radiographs). For this evaluation, the X-rays in the test set were manually labelled by the orthopedic surgeon. We compared the Dice scores between the automatically and manually segmented bones (stage E in Figure 4).
Image pre-processing: The labelled images were used for training a U-Net neural network. We designed a U-Net neural network that operates on input images of size 512 × 512 and output images of 512 × 512 with segmented bones. Small holes in automatically segmented bones that can arise from imperfect segmentation were automatically filed.
Each U-Net encoder and decoder contained four layers. It has been trained by optimizing Dice loss in 60 epochs using Adam optimizer, learning rate equal to 0.0001, batch size set to 2. During the training a data augmentation consisting of vertical mirroring of images and scaling of images have been executed. The neural network has been trained on the notebook's GPU (RTX3070 GPU, 6GB RAM). Phyton language was utilized to implement the whole algorithm, the U-Net as well as the YOLO (using Keras API).
At the beginning we evaluated the performance of the trained YOLO in detecting the ROI. We determined mean Average Precision (mAP) on 20 test images. The mAP was equal to 0.962. Figure 5 depicts the detected areas on high-resolution radiographs using YOLO. The rectangles representing the bones of interest were then used to calculate the ROIs.
Finally, 184 randomly selected lateral knee radiographs of the 218 (initially prepared database) were included for the analysis. The mean age of patients was 61.9 (± 17.8) years. There were 65 males (35%) and 119 females (65%). The demographic data of the subgroups of patients are presented in Table 1.
| Total (n = 184) | Training group (n = 82) | Validation group (n = 10) | Testing group (n = 92) | |
| Males/Females | 65/119 | 26/56 | 4/6 | 35/57 |
| Left/Right knee | 99/85 | 47/35 | 3/7 | 49/43 |
| Age (yr) | 61.9 ± 17.83 | 64.0 ± 17.62 | 53.7 ± 11.21 | 61.1 ± 18.37 |
The final training set consisted of 82 X-ray images, as well as 10 X-rays in the validation set (which did not change during the training). To compare the reliability of automatic and manual measurements, 92 randomly selected lateral radiographs of the knee in the test set were used.
The mean value of CD indexes calculated by orthopedic surgeon (R#1 and R#2) was 0.93 (± 0.19) and 0.89 (± 0.19). For BP indexes it was 0.80 (± 0.17) and 0.78 (± 0.17). Automatic measurements performed by the artificial intelligence (AI) for CD index was 0.92 (± 0.21) and for BP index was 0.75 (± 0.19) (Table 2).
| CD index | BP index | |
| Orthopedic surgeon (R#1) | 0.93 ± 0.19 | 0.80 ± 0.17 |
| Orthopedic surgeon (R#2) | 0.89 ± 0.19 | 0.78 ± 0.17 |
| Artificial intelligence | 0.92 ± 0.21 | 0.75 ± 0.19 |
The accuracy of image segmentation performed on the test set was 95.9% (± 1.26). The range of Dice score for individual X-ray images varied from 90% to 97.8% (Figure 6). This result demonstrates that even on a small amount of training data, it is possible to achieve a satisfactory segmentation quality with good generalization on unknown test radiographs that can be used in everyday clinical practice. Our research findings align with Ronneberger et al's conclusions, which pointed out that it is possible to train the U-Net model on very few annotated images[25]. On Google Colab (using CPU) the time needed for the measurement of the patella height on 20 radiographs was equal to about 240 s.
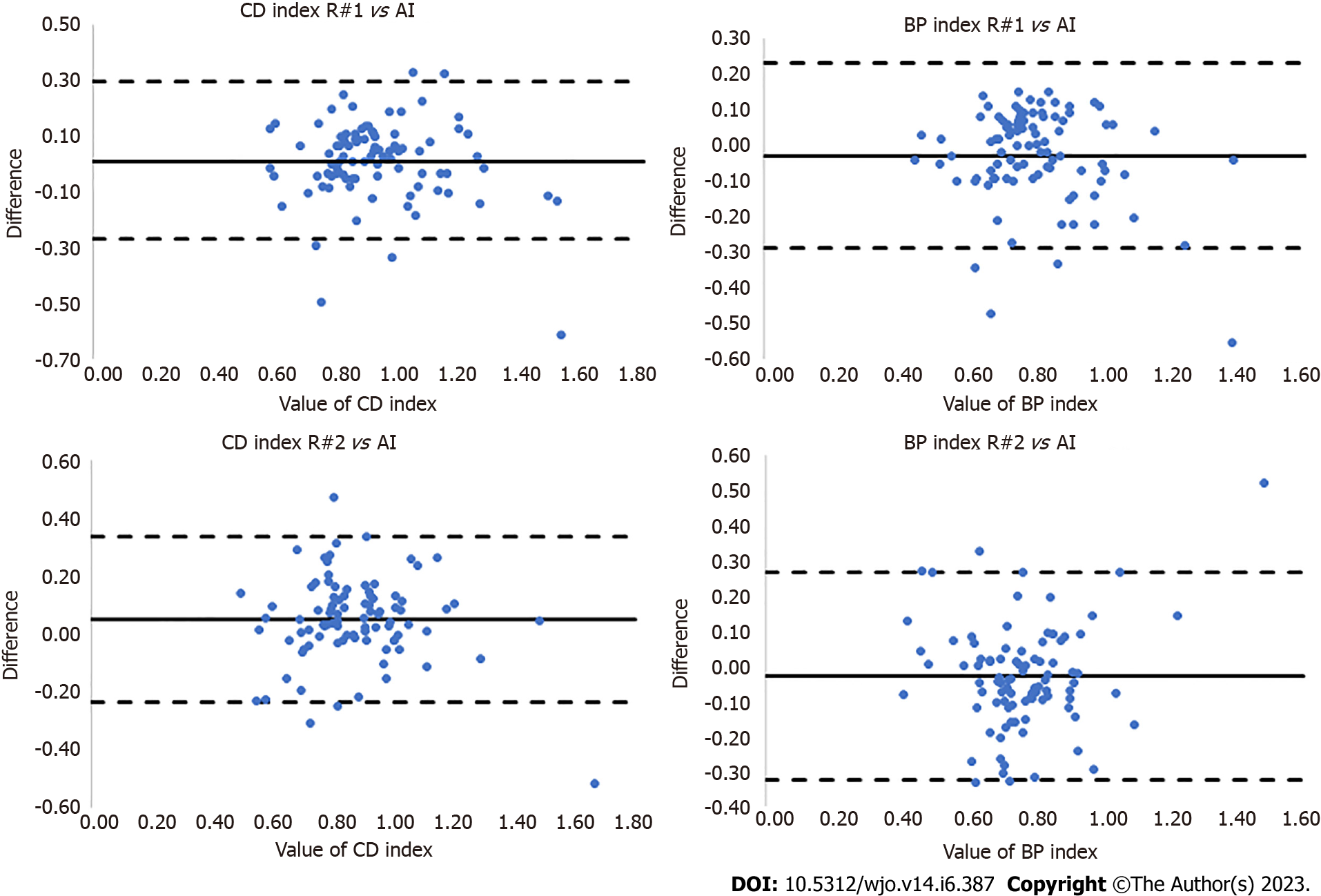
This proposed method uses a U-Net to segment bones making the patellofemoral joint and subsequently measures the Caton-Deshamps and Blackburne-Peel patellar height indexes. Our study revealed that automatic bone segmentation of lateral knee radiographs can be performed with the Dice score of 95.9% (± 1.26). Additionally, a novel approach to automatic estimation of patellar indexes has been proposed. The key points and lines needed for measuring the patellar height indexes were automatically determined based on automatically segmented patella and tibia. Excellent reliability between AI calculations and those performed by orthopedic surgeons for CD [R#1 vs AI: ICC = 0.86 (± 0.38), SEM = 0.015; R#2 vs AI: ICC = 0.80 (± 0.33), SEM = 0.013] and BP [R#1 vs AI: ICC = 0.88 (± 0.38), SEM = 0.015; R#2 vs AI: ICC = 0.79 (± 0.32), SEM = 0.014] were obtained (Table 3, Figure 7).
| ICC | SEM | |
| CD index | ||
| R#1 vs AI | 0.86 ± 0.38 | 0.015 |
| R#2 vs AI | 0.88 ± 0.38 | 0.015 |
| BP index | ||
| R#1 vs AI | 0.80 ± 0.33 | 0.013 |
| R#2 vs AI | 0.79 ± 0.32 | 0.014 |
For comparison, a method for patella segmentation[20] uses the principal component analysis to construct a patella shape model and a dual-optimization approach with genetic algorithm and Active Shape Model to fit the model to the patella boundary. The method has been tested on 20 images. However, this method segments only the patella and requires manually cropped images in which the patella occupies considerable part of the images. In a recently proposed method[21], a convolutional neural network was used to detect landmark points to measure patellar indexes. The detection was performed by a pretrained VGG-16 neural network. It has been trained and evaluated on 916 and 102 images, respectively, with manually annotated landmarks. The method has been evaluated on 400 radiographs. The ICC, Pearson correlation coefficient, mean absolute difference, Root Mean Square and Bland-Altman plots have been calculated to compare this method with manual measurements. However, the method operates on images with the patella and the proximal tibial articular occupying considerable part of the images, i.e. it is not fully automatic as the ROI was manually cropped from the high-resolution x-ray images. Moreover, only radiographs with clear patellar height landmarks have been utilized in experiments.
The method of Ye et al[21] relies on keypoints, which needs far bigger number of annotated radiographs for training the network. In contrast, our work is in line with recent research direction which focuses on training deep models on small datasets. It is worth noting that this is an important aspect as acquiring and labeling medical data is expensive, among others due to privacy concerns. In contrast to keypoint-based methods, our method relies on a large number of boundary points, making it more resistant to outliers and errors (through boundary-aware analysis). Our initial experimental results show that even in the case of training a neural network for keypoints regression on several times larger number of labeled images, the errors are much larger in comparison to the errors of our method. The method is fully automatic as the YOLO detects bones making the patellofemoral joint on high-resolution images, U-Net segments the bones, a line is fitted to the segmented proximal tibial articular surface, and a second line passing through two landmarks detected on the segmented patella are determined. Then they are used to calculate the patellar height indexes. However, good technical execution and sufficient quality of the input images are mandatory to obtain reliable results for automatic measurements. In some specific cases, additional hardware (screws, plates), heterotopic ossifications, large osteophytes that distort the bone outline and oblique view (instead of proper lateral), may slightly reduce quality of bone segmentation, which is the main limitation of this method. We agree with Zheng et al[33], who emphasized that absolute errors may be reduced by increasing the number of subjects in the training group.
Our algorithm for automatic measurements of patellar indexes permits the evaluation of indexes on high data volume. Taking into account that the proposed algorithm allows measurements on high-resolution radiographs, low effort is needed to get patellar indexes, i.e. no manual cropping of knee region is required in contrast to the previous methods. One of the advantage of the proposed method is that relatively small amount of manually labelled bones on images is needed to achieve reliable bone segmentation. Although, higher number of manual annotations for training YOLO responsible for knee detection is needed, the labelling of knee regions can be done in relatively short time. This study has some limitations. Firstly, in the current study, the ROIs that were determined on the basis of YOLO detections were resized to size required by the U-Net, i.e., 512 × 512. This means that the keypoints and lines were determined on images with somewhat smaller resolution than the original radiographs. Secondly, the accuracy of the results depends on: The manual segmentation performed by the researcher during the training phase; the amount and quality of training data, and the architecture of the applied neural network. In current work, the training of neural networks and evaluation of the algorithm was performed on images acquired in a single institution, i.e. our hospital. Thus, further work is needed to collect radiographs from various hospitals to train networks and asses accuracy of the algorithm on radiograms acquired by different devices. Moreover, the input images may have different levels of intensity and quality. These technical aspects should be emphasized and resolved in medical centers that will implement the algorithm for automatic measurement of patellar indexes. In future work we are planning to combine boundary-aware analysis with landmark-based DL measurements. We also plan to extend the U-Net and compare it with recent networks for image segmentation. Additionally images from different hospitals will be used in the research.
The aim of this study was to investigate the reliability of automated patellar height estimation using DL-based bone segmentation and detection on high-resolution images. It showed that reliable automatic patellar height measurements can be achieved on lateral knee radiographs with the accuracy required for the clinical practice. We demonstrated that proximal tibia and patella bones can be segmented precisely (Dice score greater than 95%) by U-Net neural network on knee regions automatically detected by the YOLO network (mean Average Precision mAP greater than 0.96). Determining patellar end-points and the joint line by fitting to points of the proximal tibia joint surface enables calculating the Caton-Deschamps and Blackburne-Peel indexes with very good reliability. Automated measurements are comparable to measurements performed by orthopedic surgeons (SEM greater than 0.75). Experimental results indicate that our approach can be valuable as a pre-operative and potentially as a postoperative assessment tool for big volume data analysis in medical practice.
Recent advancements in artificial intelligence and deep learning have contributed to the development of medical imaging techniques, leading to better interpretation of radiographs. Moreover, there is an increasing interest in automating routine diagnostic activities and orthopedic measurements.
The automation of patellar height assessment using deep learning-based bone segmentation and detection on high-resolution radiographs could provide a valuable tool in medical practice.
The aim of this study was to verify the accuracy of automated patellar height assessment using a U-Net neural network and to determine the agreement between manual and automatic measurements.
Proximal tibia and patella was segmented by U-Net neural network on lateral knee subimages automatically detected by the You Only Look Once (YOLO) network. The patellar height was quantified by Caton-Deschamps and Blackburne-Peel indexes. The interclass correlation coefficient and standard error for single measurement were used to calculate agreement between manual and automatic measure
Proximal tibia and patella were segmented with 95.9% accuracy by the U-Net neural network on lateral knee subimages automatically detected by the YOLO network (mean Average Precision mAP greater than 0.96). Excellent agreement achieved between manual and automatic measurements for both indexes (interclass correlation coefficient > 0.75, SEM < 0.014).
Automatic patellar height assessment can be achieved with high accuracy on high-resolution radiographs. Proximal tibia and patella can be segmented precisely by U-Net neural network on lateral knee subimages automatically detected by the YOLO network. Determining patellar endpoints and fitting the line to the proximal tibia joint surface enables accurate Caton-Deschamps and BP index calculations, making it a valuable tool in medical practice.
Future research can focus on the clinical implementation of this automated method, which has the potential to enhance diagnostic accuracy, reduce human error, and improve patient outcomes.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Orthopedics
Country/Territory of origin: Poland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mahmoud MZ, Saudi Arabia; Mijwil MM, Iraq S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Louati K, Vidal C, Berenbaum F, Sellam J. Association between diabetes mellitus and osteoarthritis: systematic literature review and meta-analysis. RMD Open. 2015;1:e000077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 2. | Kokkotis C, Moustakidis S, Papageorgiou E, Giakas G, Tsaopoulos DE. Machine learning in knee osteoarthritis: A review. Osteoarthr Cartil Open. 2020;2:100069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 3. | Salem KH, Sheth MR. Variables affecting patellar height in patients undergoing primary total knee replacement. Int Orthop. 2021;45:1477-1482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Cabral F, Sousa-Pinto B, Pinto R, Torres J. Patellar Height After Total Knee Arthroplasty: Comparison of 3 Methods. J Arthroplasty. 2017;32:552-557.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Yue RA, Arendt EA, Tompkins MA. Patellar Height Measurements on Radiograph and Magnetic Resonance Imaging in Patellar Instability and Control Patients. J Knee Surg. 2017;30:943-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Xu B, Xu WX, Lu D, Sheng HF, Xu XW, Ding WG. Application of different patella height indices in patients undergoing total knee arthroplasty. J Orthop Surg Res. 2017;12:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 7. | Degnan AJ, Maldjian C, Adam RJ, Fu FH, Di Domenica M. Comparison of Insall-Salvati ratios in children with an acute anterior cruciate ligament tear and a matched control population. AJR Am J Roentgenol. 2015;204:161-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Portner O. High tibial valgus osteotomy: closing, opening or combined? Patellar height as a determining factor. Clin Orthop Relat Res. 2014;472:3432-3440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Picken S, Summers H, Al-Dadah O. Inter- and intra-observer reliability of patellar height measurements in patients with and without patellar instability on plain radiographs and magnetic resonance imaging. Skeletal Radiol. 2022;51:1201-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology. 2018;287:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 254] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 11. | Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci Rep. 2018;8:1727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 286] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 12. | Aggarwal K, Mijwil MM, Grag S, Al-Mistarehi AH, Alomari S, Gök M, Zein Alaabdin AM, Abdulrhman SH. Has the Future Started? The Current Growth of Artificial Intelligence, Machine Learning, and Deep Learning. Iraqi Journal for Computer Science and Mathematics. 3:115-123. [DOI] [Full Text] |
| 13. | Tanzi L, Vezzetti E, Moreno R, Aprato A, Audisio A, Massè A. Hierarchical fracture classification of proximal femur X-Ray images using a multistage Deep Learning approach. Eur J Radiol. 2020;133:109373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 14. | Kuo RYL, Harrison C, Curran TA, Jones B, Freethy A, Cussons D, Stewart M, Collins GS, Furniss D. Artificial Intelligence in Fracture Detection: A Systematic Review and Meta-Analysis. Radiology. 2022;304:50-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 134] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 15. | Tiwari A, Poduval M, Bagaria V. Evaluation of artificial intelligence models for osteoarthritis of the knee using deep learning algorithms for orthopedic radiographs. World J Orthop. 2022;13:603-614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 16. | Kwolek K, Liszka H, Kwolek B, Gadek A. Measuring the Angle of Hallux Valgus Using Segmentation of Bones on X-Ray Images. Artificial Neural Networks and Machine Learning. ICANN 2019. In: Munich, Germany, 11731: 313-325. |
| 17. | Kwolek K, Brychcy A, Kwolek B, Marczynski W. Measuring Lower Limb Alignment and Joint Orientation Using Deep Learning Based Segmentation of Bones. Hybrid Artificial Intelligent Systems. HAIS 2019 In: Leon, Spain 2019; 11734: 514-525. |
| 18. | Lindgren Belal S, Sadik M, Kaboteh R, Enqvist O, Ulén J, Poulsen MH, Simonsen J, Høilund-Carlsen PF, Edenbrandt L, Trägårdh E. Deep learning for segmentation of 49 selected bones in CT scans: First step in automated PET/CT-based 3D quantification of skeletal metastases. Eur J Radiol. 2019;113:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 19. | Wu J, Mahfouz MR. Robust x-ray image segmentation by spectral clustering and active shape model. J Med Imaging (Bellingham). 2016;3:034005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Chen HC, Wu CH, Lin CJ, Liu YH, Sun YN. Automated segmentation for patella from lateral knee X-ray images. Annu Int Conf IEEE Eng Med Biol Soc. 2009;2009:3553-3556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Ye Q, Shen Q, Yang W, Huang S, Jiang Z, He L, Gong X. Development of automatic measurement for patellar height based on deep learning and knee radiographs. Eur Radiol. 2020;30:4974-4984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Floyd C, Ni H, Gunaratne RS, Erban R, Papoian GA. On Stretching, Bending, Shearing, and Twisting of Actin Filaments I: Variational Models. J Chem Theory Comput. 2022;18:4865-4878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 23. | Lawal MO. Tomato detection based on modified YOLOv3 framework. Sci Rep. 2021;11:1447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 24. | Kohn MD, Sassoon AA, Fernando ND. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin Orthop Relat Res. 2016;474:1886-1893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 852] [Cited by in RCA: 844] [Article Influence: 93.8] [Reference Citation Analysis (0)] |
| 25. | Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Proceedings Paper. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 2015; Olaf Ronneberger, Philipp Fischer, Thomas Brox 9351: 234-241. |
| 26. | Baskaran L, Al'Aref SJ, Maliakal G, Lee BC, Xu Z, Choi JW, Lee SE, Sung JM, Lin FY, Dunham S, Mosadegh B, Kim YJ, Gottlieb I, Lee BK, Chun EJ, Cademartiri F, Maffei E, Marques H, Shin S, Choi JH, Chinnaiyan K, Hadamitzky M, Conte E, Andreini D, Pontone G, Budoff MJ, Leipsic JA, Raff GL, Virmani R, Samady H, Stone PH, Berman DS, Narula J, Bax JJ, Chang HJ, Min JK, Shaw LJ. Automatic segmentation of multiple cardiovascular structures from cardiac computed tomography angiography images using deep learning. PLoS One. 2020;15:e0232573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 27. | Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, Lev MH, Do S. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng. 2019;3:173-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 28. | White AE, Otlans PT, Horan DP, Calem DB, Emper WD, Freedman KB, Tjoumakaris FP. Radiologic Measurements in the Assessment of Patellar Instability: A Systematic Review and Meta-analysis. Orthop J Sports Med. 2021;9:2325967121993179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 29. | Zou KH, Warfield SK, Bharatha A, Tempany CM, Kaus MR, Haker SJ, Wells WM 3rd, Jolesz FA, Kikinis R. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11:178-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 985] [Cited by in RCA: 1067] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 30. | Portner O, Pakzad H. The evaluation of patellar height: a simple method. J Bone Joint Surg Am. 2011;93:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 31. | Caton J, Deschamps G, Chambat P, Lerat JL, Dejour H. [Patella infera. Apropos of 128 cases]. Rev Chir Orthop Reparatrice Appar Mot. 1982;68: 317-325. [PubMed] |
| 32. | Zou GY. Sample size formulas for estimating intraclass correlation coefficients with precision and assurance. Stat Med. 2012;31:3972-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 231] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 33. | Zheng Q, Shellikeri S, Huang H, Hwang M, Sze RW. Deep Learning Measurement of Leg Length Discrepancy in Children Based on Radiographs. Radiology. 2020;296:152-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |