Published online Dec 10, 2014. doi: 10.5306/wjco.v5.i5.874
Revised: April 3, 2014
Accepted: April 17, 2014
Published online: December 10, 2014
Processing time: 347 Days and 11.9 Hours
Breast cancer (BC) is the most common cancer among women worldwide. The aetiology and carcinogenesis of BC are not clearly defined, although genetic, hormonal, lifestyle and environmental risk factors have been established. The most common treatment for BC includes breast-conserving surgery followed by a standard radiotherapy (RT) regimen. However, radiation hypersensitivity and the occurrence of RT-induced toxicity in normal tissue may affect patients’ treatment. The role of DNA repair in cancer has been extensively investigated, and an impaired DNA damage response may increase the risk of BC and individual radiosensitivity. Single nucleotide polymorphisms (SNPs) in DNA repair genes may alter protein function and modulate DNA repair efficiency, influencing the development of various cancers, including BC. SNPs in DNA repair genes have also been studied as potential predictive factors for the risk of RT-induced side effects. Here, we review the literature on the association between SNPs in base excision repair (BER) genes and BC risk. We focused on X-ray repair cross complementing group 1 (XRCC1), which plays a key role in BER, and on 8-oxoguanine DNA glycosylase 1, apurinic/apyrimidinic endonuclease 1 and poly (ADP-ribose) polymerase-1, which encode three important BER enzymes that interact with XRCC1. Although no association between SNPs and radiation toxicity has been validated thus far, we also report published studies on XRCC1 SNPs and variants in other BER genes and RT-induced side effects in BC patients, emphasising that large well-designed studies are needed to determine the genetic components of individual radiosensitivity.
Core tip: Single nucleotide polymorphisms (SNPs) in DNA repair genes may modulate DNA repair efficiency, influencing the development of various cancers, including breast cancer (BC). SNPs in DNA repair genes have also been studied as predictors for the risk of radiotherapy-induced side effects. We reviewed the literature on the association between SNPs in base excision repair (BER) genes and BC risk. We focused on X-ray repair cross complementing group 1, 8-oxoguanine DNA glycosylase 1, apurinic/apyrimidinic endonuclease 1 and poly (ADP-ribose) polymerase-1, which encode four important BER proteins. We also report published studies on SNPs in BER genes and individual radiosensitivity in BC patients.
- Citation: Patrono C, Sterpone S, Testa A, Cozzi R. Polymorphisms in base excision repair genes: Breast cancer risk and individual radiosensitivity. World J Clin Oncol 2014; 5(5): 874-882
- URL: https://www.wjgnet.com/2218-4333/full/v5/i5/874.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i5.874
Female breast cancer (BC) is the most prevalent malignancy worldwide, with 5.2 million cases diagnosed between 2004 and 2008[1], and it is the primary cause of cancer-related death among women (http://globocan.iarc.fr).
Breast cancer is considered a multifactorial disease, and its occurrence is related to genetic, reproductive, environmental and lifestyle factors.
Breast cancer susceptibility has a complex genetic basis, which has been widely investigated over the past 20 years. Linkage analysis and a candidate gene approach have been used to identify susceptibility loci for BC with high and moderate penetrance. Rare mutations, principally in genes involved in DNA repair such as BRCA1, BRCA2, TP53, ATM, CHEK2, PALB2 and BRIP1, are associated with a greatly increased risk of BC[2].
Case-control and, in recent years, genome-wide association studies (GWAS) have identified more than 70 low-penetrance common variants [single nucleotide polymorphisms (SNPs)] associated with BC risk. Although many of the associations identified in GWAS are localised in non-coding regions of the genome, which are most likely involved in gene expression regulation, some common patterns have been identified among the BC susceptibility loci. Low-penetrance BC loci are related to mammary gland development, the DNA repair pathway, growth factors, the cell cycle, differentiation and apoptosis[3].
Common SNPs in DNA repair genes have been extensively investigated in candidate-gene and case-control association studies in the context of breast carcinoma and other types of cancer. SNPs in such genes can affect the efficiency of the DNA repair machinery and contribute to genomic instability and cancer development[4].
Among the different DNA repair pathways, base excision repair (BER) is responsible for the repair of bases damaged by the effects of X-rays, reactive oxygen radicals and alkylating agents. Many epidemiological studies have investigated the association between common variants in BER genes and human cancer, including breast cancer[5]. Furthermore, DNA repair genes, including BER genes, have been extensively examined in several epidemiological studies to determine their association with radiation-induced toxicity in cancer patients undergoing radiotherapy (RT)[6]. In fact, inter-individual differences in response to therapeutic radiation exposure have been observed, and this variability may be influenced by genetic factors affecting DNA repair efficiency[7]. The side effects induced by RT in normal tissue largely depend on the capacity of cells to repair DNA damage caused by irradiation.
Here, we survey association studies on the most common variants in BER genes, evaluating their role in BC susceptibility and in the risk of developing adverse reactions after RT.
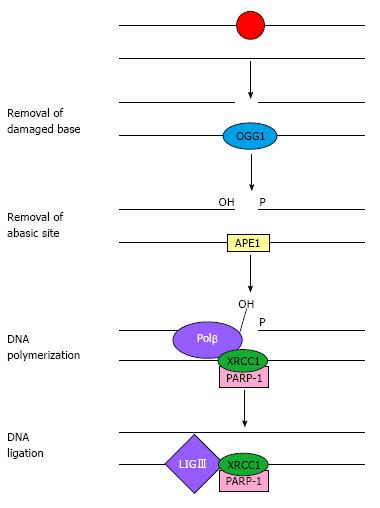
The BER pathway is the primary mechanism that protects cells from oxidative DNA damage; it acts on small DNA lesions or modified bases, where it removes and replaces the damaged base[4]. This process starts with the release of the damaged base by base-specific DNA glycosylases (e.g., the oxidised base 8-oxoguanine is excised by 8-oxoguanine DNA glycosylase), followed by the cleavage of the sugar-phosphate chain, excision of the apurinic/apyrimidinic (AP) site by endonuclease action, DNA synthesis and ligation (Figure 1). This pathway is referred to as short-patch BER and results in the replacement of the AP site via the incorporation of a single nucleotide. In contrast, the long-patch BER pathway produces a repair tract of at least two nucleotides[8,9].
Enzymes involved in BER include 8-oxoguanine DNA glycosylase 1 (OGG1); AP endonuclease 1 (APE1 or APEX1), which excises the abasic residue; poly (ADP-ribose) polymerase-1 (PARP-1), which binds DNA-containing strand breaks; polynucleotide kinase; DNA polymerase-β (Polβ); and DNA ligase III (LIGIII), which completes the restoration phase.
Another protein that plays a central role in BER is X-ray repair cross-complementing group 1 (XRCC1), which is a scaffold protein that coordinates both the initial and late steps of abasic site restoration via multiple protein-protein interactions[10]. XRCC1 stabilises OGG1 on an AP site present in double-stranded DNA until APE1 is able to bind to the DNA, allowing the transfer of the DNA substrate from the DNA glycosylase product to the AP endonuclease. Polβ is then recruited by its interactions with APE1 and XRCC1. The binding of XRCC1 with PARP-1 also plays a role in BER. Moreover, XRCC1 is essential for the stabilisation of LIGIII.
Interactions between XRCC1 and its BER partners are mediated by different domains of the protein, which introduces the possibility that all four proteins, namely XRCC1, Polβ, PARP-1 and LIGIII, could form a single complex[11]. Some of the interactions between BER proteins are illustrated in Figure 1.
Many polymorphisms have been identified in genes encoding proteins involved in BER and in other DNA repair pathways[12]. In particular, many epidemiological studies have been performed to evaluate the association between XRCC1 SNPs and different types of cancer[13].
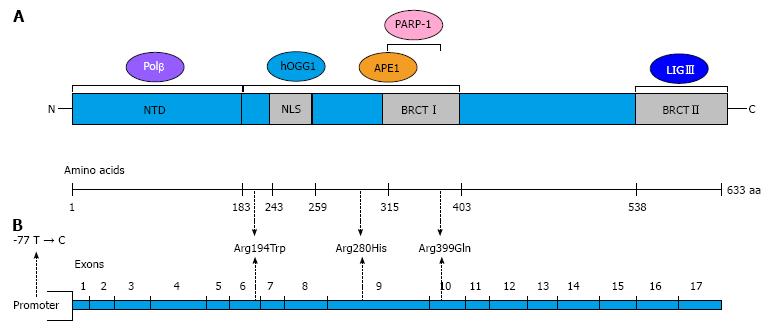
The human XRCC1 gene maps to chromosome 19q13.2 and encodes a scaffold protein of 633 amino acids that plays a major role in the BER pathway and is also involved in other DNA repair mechanisms, such as single-strand break repair and non-homologous end joining. XRCC1 interacts with several BER proteins such as DNA polymerase β, APE1, OGG1, PARP-1 and LIGIII[14]. A schematic representation of XRCC1 and its interactions is shown in Figure 2A.
Among the many SNPs found in the XRCC1 gene, three polymorphisms resulting in non-conservative amino acid substitutions have been identified: Arg194Trp (rs1799782), Arg280His (rs25489) and Arg399Gln (rs25487)[15]. Another XRCC1 variant located in the 5’-untranslated region (5’UTR), -77 T > C (rs3213245), was identified in 2004[16]. The structure of the XRCC1 gene and the localisation of the most common SNPs are illustrated in Figure 2B. XRCC1 variants could affect the function of the protein and impair DNA repair efficiency. In particular, the XRCC1 Arg399Gln variant has been the subject of many case-control studies to investigate its possible association with breast cancer risk. The published data on this association, which are often contradictory, have been collected in several meta-analyses[17-20] (Table 1). Huang et al[17] showed that the Arg399Gln SNP is associated with a trend of increased breast cancer risk using both dominant and recessive models. In ethnic subgroups, and considering the recessive model, this polymorphism is associated with BC risk in Asians and Africans, although it is weakly related with breast cancer in Caucasians. The association between the Arg399Gln variant and BC risk was confirmed in Asian and African populations[19], whereas other studies confirmed these data only among Asians[18,20]. These findings suggest that the role of the XRCC1 Arg399Gln SNP as a risk factor for breast cancer may differ between Caucasian and Asian populations.
| Ref. | XRCC1 SNPs | Number of studies analysed | Result |
| Huang et al[17] | Arg399Gln | 37 | The 399Gln variant allele is associated with an increased risk of BC |
| Arg194Trp | 18 | ||
| Arg280His | 8 | ||
| Li et al[18] | Arg399Gln | 40 | The recessive effect of the 399Gln variant allele increases the risk of BC (significant only in Asians) |
| Arg194Trp | 21 | ||
| Arg280His | 9 | ||
| Wu et al[19] | Arg399Gln | 44 | This SNP is associated with increased BC risk in Asians and Africans |
| Saadat et al[20] | Arg399Gln | 36 | This SNP is associated with increased BC risk in Asians |
| Yi et al[13] | Arg399Gln | 54 | This SNP is associated with increased risk of BC in Asians and Indians |
| XRCC1 haplotypes | |||
| Saadat[21] | Arg399Gln | 10 | The Arg194-Gln399 haplotype is associated with increased BC risk in Asians |
| Arg194Trp |
A very recent meta-analysis of 297 case-control studies evaluated the association between the XRCC1 Arg399Gln polymorphism and overall cancer risk[13]; a significantly increased cancer risk was found in all genetic models. In stratified analyses by cancer type, significantly enhanced breast cancer risk was observed in Asians and particularly in Indians.
Some case-control studies assessing the association between XRCC1 haplotype and susceptibility to breast cancer were collected in a meta-analysis[21]. Haplotypes for the most common non-synonymous XRCC1 SNPs (Arg194Trp and Arg399Gln) were considered, and the analysis showed a slight increased risk of BC associated with the Arg194-Gln399 haplotype in comparison with the Arg194-Arg399 haplotype. In the stratified analysis according to geographic location, the Arg194-Gln399 haplotype was significantly associated with breast cancer risk among individuals from Asian countries, while no association has been found between the XRCC1 haplotype and BC susceptibility in Caucasian populations.
Thus far, a few XRCC1 haplotype analyses have included the -77 T > C SNP in the 5’UTR[22-24]. Two studies[22,24], one performed on French and the other performed on Chinese BC patients and controls, considered XRCC1 SNPs at position -77 and at codons 194, 280 and 399. Brem et al[22] found that the haplotype carrying the variant allele at codon 280 and the wild-type (wt) alleles at the other positions was associated with an increased risk of BC, although the association was not significant. In contrast, Liu et al[24] observed a significantly higher BC risk for the haplotype containing the variant allele at position -77 and the wt alleles at other positions.
In a case-control study on Caucasian BC patients, Sterpone et al[23] performed a haplotype analysis based on XRCC1 genotypes at position -77 and at codons 194 and 399. The haplotype containing the wt allele at position 194 and the variant alleles at other positions was significantly associated with a higher BC risk in this study.
The human OGG1 gene is located on chromosome 3p26.2 and encodes the key enzyme in the repair of 8-oxoguanine, which is one of the most common products generated by exposure to reactive oxygen species[25]. Among the SNPs in OGG1, the most studied variant is the functional substitution Ser326Cys (rs1052133), which is located in exon 7 of the OGG1 gene and causes an amino acid change (from serine to cysteine). Comparative functional analysis via a complementation assay of a defective Escherichia coli mutant revealed that the repair activity of the 326Ser protein was higher than that of the 326Cys protein[26]. Therefore, this OGG1 polymorphism may result in changes in DNA repair activity in human cells.
Many studies that have highlighted the association between the OGG1 Ser326Cys SNP and breast cancer risk have produced conflicting results. To clarify the findings, several meta-analyses have been performed (Table 2)[27-29]. Yuan et al[27] collected data published from 2003 to 2008 and evaluated the association between the OGG1 Ser326Cys SNP and BC onset according to menopausal status and ethnicity by performing a stratified analysis. This meta-analysis suggested that the 326Cys allele had a significant protective effect against BC in European women, both in the additive (326Cys allele vs 326Ser allele) and dominant (Cys/Cys + Cys/Ser vs Ser/Ser) genetic models. No significant association was found between this SNP and menopausal status or other ethnicities. These results were discussed by Ding et al[28], who combined all of the studies on European populations and performed a new meta-analysis, which indicated a lack of association between the OGG1 326Cys allele and BC risk for this ethnicity. This result was confirmed by the meta-analysis performed by Gu et al[29], who identified 11 case-control studies published from 2003 to 2009; however, they did not observe any significant association between Ser326Cys SNP and BC risk, even in the analyses stratified by ethnicity, source of controls and menopausal status. An additional case-control study not included in the aforementioned meta-analysis also showed a lack of association between the OGG1 Ser326Cys variant and BC susceptibility[30].
| Ref. | BER gene | SNPs | Number of studies analysed | Result |
| Yuan et al[27] | OGG1 | Ser326Cys | 10 | This SNP is significantly associated with a protective effect against BC in European subjects (additive and dominant model) |
| Ding et al[28] | OGG1 | Ser326Cys | 4 | There was a lack of association between this SNP and BC risk in a European population |
| Gu et al[29] | OGG1 | Ser326Cys | 11 | There was a lack of association between this SNP and BC risk |
| Wei et al[33] | OGG1 | Ser326Cys | 12 | This SNP did not have a significant effect on BC |
| Wu et al[35] | PARP-1 (ADPRT) | Val762Ala | 6 | There was no association between this SNP and BC (all genetic models) |
| Wu et al[35] | APE1 | Asp148Glu | 5 | There was no association between this SNP and BC (all genetic models) |
Nevertheless, a recent study in a Korean population[31] revealed that different SNPs in BER genes (including the OGG1 Ser326Cys and rs2072668 SNPs) function in combination to increase the risk of breast cancer. Another recent study showed that OGG1 326Cys was significantly associated with an increased BC risk in specific subgroups of Chinese Han women (younger than 55 years, premenopausal, triple-negative or p53-positive)[32].
The Ser326Cys SNP was also significantly associated with overall cancer risk in a more recent meta-analysis[33], and it showed a stronger association with lung cancer risk.
Therefore, there is evidence that the OGG1 Ser326Cys polymorphism is associated with cancer risk, and in particular that it may be a low-penetrance susceptibility factor for lung cancer. Moreover, the Ser326Cys variation could interact with other factors such as age, triple-negative status and p53-positive status, thereby influencing breast cancer carcinogenesis.
PARP-1, also referred to as ADP ribosyl transferase, and APE1 are two of the most important enzymes in the BER pathway.
The human PARP-1 gene is localised to chromosome 1q41-42 and encodes a nuclear protein that specifically recognises and binds DNA strand breaks. PARP-1 also recruits other BER proteins, including the XRCC1-LIGIII complex, to facilitate the core BER reaction.
APE1 initiates the restoration step of the BER pathway by hydrolysing the 5’-phosphodiester bond of the AP site[5]. The human APE1 gene maps to chromosome 14q12[34].
Several studies have assessed the association between common polymorphisms (PARP-1 Val762Ala-rs1136410 and APE1 Asp148Glu-rs3136820) in these two BER genes and BC risk, although inconclusive results were obtained. A meta-analysis of the literature, updated to 2011, was performed to obtain a more accurate estimate of this association[35]. In total, 8 studies were included in the meta-analysis (Table 2). No association between PARP-1 Val762Ala and breast cancer risk was found in any of the genetic models. Additionally, there was no association between BC risk and APE1 Asp148Glu considering all genetic models; therefore, the analysis suggests that these two polymorphisms are not associated with BC susceptibility.
Two additional papers on APE1 SNPs and breast cancer risk were recently published[36,31]. A case-control study performed on a Chinese population reported no association between Asp148Glu and BC susceptibility, as indicated by the above-cited meta-analysis. In contrast, a significant association between a SNP in the APE1 promoter (-656 T > G) and decreased BC risk was found, suggesting that this polymorphism may influence breast cancer occurrence[36]. In a study by Kim et al[31], the association between APE1 Asp148Glu and two OGG1 SNPs, including OGG1 Ser326Cys, and BC risk was evaluated. Whereas APE1 Asp148Glu was weakly associated with BC risk, a combined analysis including the two BER genes revealed a significant effect on breast cancer occurrence, suggesting the importance of assessing a combination of SNPs in different genes and gene haplotypes for the prediction of BC risk.
Breast-conserving surgery followed by a standard RT regimen is the most common treatment for breast cancer. However, therapeutic exposure to IR can induce adverse reactions in normal tissue. These reactions show considerable variation among individuals, suggesting the involvement of genetic factors. Because IR hypersensitivity may lead to interruption of therapy, in the last several years, significant effort has been devoted to the identification of molecular factors that could increase radiotherapy-induced side effects.
SNPs in genes involved in processes such as DNA repair, cell-cycle control, apoptosis, cellular antioxidant defence and cytokine production may influence the individual radioresponse. Several experimental approaches, such as candidate SNP association studies and GWAS, are being used to investigate the genetic basis of normal tissue radiosensitivity[37]. Radiogenomics studies are most numerous in breast cancer patients treated with RT and are aimed at identifying SNP profiles that can be used to select radiosensitive patients. In particular, several studies on BC patients evaluating the association between the risk of acute and late skin reactions to RT and SNPs in DNA repair genes (especially XRCC1) were performed; however, these studies yielded conflicting results. Eleven studies on XRCC1 SNPs, mainly involving Caucasian patients, were collected in a recent meta-analysis[38].
In these studies, the severity of acute and/or late side effects of RT was assessed according to various evaluation criteria, including the Common Terminology Criteria for Adverse Events (CTCAE) (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf), the criteria proposed by the Radiation Therapy Oncology Group (RTOG) and European Organization for Research and Treatment of Cancer (EORTC)[39], the Late Effects of Normal Tissue-Subjective Objective Management Analytical (LENT/SOMA)[40] and the Common Toxicity Criteria of the United States National Institutes of Health (NIH) (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcmanual_v4_10-4-99.pdf#search=‘‘ctc’’).
To evaluate acute RT side effects in BC patients, the following clinical skin reactions within the radiation field of the breast were documented during treatment: erythema, epilation, desquamation and decreased sweating in addition to more severe morbidities such as edema, ulceration, haemorrhage and necrosis. Common late radiation effects (i.e., effects that first occur at least 90 d after the initiation of RT) include fibrosis, telangiectasia and atrophy.
Both early and late normal tissue reactions are graded on a 5-point ordinal scale (0 indicating absence of a radiation effect and 5 indicating an effect leading to death). The grade of toxicity induced by radiotherapy was assessed according to different evaluation criteria, with high-grade toxicity considered as grade ≥ 2 (according to CTCAE, RTOG-EORTC and LENT/SOMA) or grade ≥ 2c (according to the NIH).
The results of the meta-analysis by Xie et al[38] are summarised in Table 3. No significant association between the XRCC1 Arg399Gln SNP and IR-induced toxicity was observed in the overall analysis. Nevertheless, stratified analyses showed that the Arg399Gln SNP was predictive of side effects in some subgroups of BC patients. For example, carriers of the XRCC1 399Gln allele were at higher risk of RT-induced side effects in studies using high-quality genotyping methods, in studies with mixed treatment regimens or when studies on only late toxicity were excluded. On the contrary, the XRCC1 Arg280His variant allele had a protective effect against RT-induced toxicity only in BC patients treated with radiotherapy alone. XRCC1 Arg194Trp and -77 T > C did not have any predictive value.
| Ref. | XRCC1 SNPs | Number of studies analysed | Result |
| Xie et al[38] | Arg399Gln | 8 | The 399Gln variant allele is associated with a higher risk of RT-induced toxicity (only in some subgroups of BC patients) |
| Arg194Trp | 6 | No predictive value was found for this SNP | |
| -77 T > C | 4 | No predictive value was found for this SNP | |
| Arg280His | 4 | The 280His variant allele is protective against RT-induced toxicity (in BC patients treated with RT only) |
This analysis indicated that large well-designed studies are needed to more clearly establish the predictive value of XRCC1 variants and SNPs in other DNA repair genes for radiation-induced side effects. The choice of genotyping method and the selection of well-characterised patient cohorts should be carefully considered.
Few studies investigating the association between SNPs in other BER genes and the risk of adverse reactions to RT in BC patients are available in the literature. Concerning the APE1 Asp148Glu SNP, the 148Glu allele was found to have a protective effect against the development of acute toxicity to radiation in Caucasian BC patients; however, this effect was only observed in the normal-weight subgroup of patients[41]. Moreover, the authors observed that the APE1 148Glu and XRCC1 399Gln alleles exerted a combined protective effect. No association between APE1 SNPs and late complications in normal tissue after radiotherapy has been identified thus far for BC[42,43].
The association of DNA repair SNPs with late RT effects in normal tissue has also been investigated in prostate cancer patients, although the radiogenomics studies reported thus far present interpretive difficulties because of numerous confounding factors[37]. A GWAS was recently performed to identify SNPs associated with the development of erectile dysfunction following RT for prostate cancer[44]. Twelve candidate SNPs identified in this study were associated with cellular functions such as adhesion, signalling and hormone metabolism rather than DNA damage repair. Therefore, the involvement of DNA repair SNPs in the radioresponse after RT for prostate cancer remains an open question.
Few studies have examined patient populations with tumours other than carcinomas of the breast or prostate, although significant acute and late toxicities are frequent in patients with squamous carcinomas of the head and neck who are treated with RT. The association of DNA repair SNPs with effects on normal tissue in the head and neck and in other types of cancers should be more extensively explored[37].
The attempts made thus far to validate the published data on genotype and radiation toxicity did not confirm a clinically relevant predictive value for any published SNP[45]. Currently, it remains controversial whether SNPs could significantly influence the risk of complications in normal tissue[46].
In the last decade, there has been increasing interest in identifying associations between SNPs in DNA repair genes and susceptibility to various cancers, including BC[4,5]. In this context, BER gene polymorphisms have been extensively investigated; however, their association with BC has not been clearly defined. The Arg399Gln SNP, which is the most common variant in the XRCC1 gene, showed an overall weak association with BC risk that became stronger only for some ethnicities. In general, SNPs may contribute to the genetic risk for BC, although their effect is usually only slightly statistically significant. In some studies, SNP-SNP interactions have been examined to evaluate epistatic effects contributing to BC[47]. SNPs in different DNA repair pathways, or in other pathways related to DNA metabolism, were selected, and specific SNP pairs showed a statistical association with BC risk. Significant trends in BC risk were also observed in association with an increasing number of risk alleles in different DNA repair genes[48,23]. Concerning BER genes, XRCC1 SNPs and haplotypes play an important role because they result in amino acid substitutions, which may affect the interaction of the protein with the other BER enzymes and alter DNA repair efficiency. Studies on the interaction between SNPs in BER genes should be encouraged because although a single SNP may have a negligible effect, interactions between different SNPs in genes of the BER pathway could significantly affect cancer risk.
Studies on the interactions between SNPs in genes of different DNA damage signalling and repair pathways should also be performed. Newly available techniques, principally GWAS, will help to explain the role of moderate-risk alleles and common lower-penetrance alleles in sporadic and familial BC risk.
Furthermore, gene-environment interactions should be investigated to elucidate the complex mechanism underlying BC carcinogenesis.
Similarly, it has been demonstrated that SNPs in BER genes (particularly APE1 and XRCC1) may contribute to IR hypersensitivity. Until now, no association between SNPs and late toxicity has been confirmed, either for BC or for prostate cancer[38]. Therefore, large, well-designed studies are needed to obtain more robust results.
Although the analysis of gene polymorphisms for the individualisation of cancer therapy is not yet widespread in routine clinical practice, understanding the genetic components of individual radiosensitivity remains an important goal. To properly assess the value of pre-treatment genotyping approaches, prospective collection of genomic DNA from patients enrolled in clinical trials should be planned to develop personalised radiotherapy protocols for both sensitive and resistant patients.
The establishment of gene polymorphism databases will significantly contribute to these tasks, and meta-analyses that collect a large amount of data will permit faster access to scientific results.
P- Reviewer: Fernandez-Santander A, Lian H, Rameshwar P, Shao R S- Editor: Wen LL L- Editor: A E- Editor: Liu SQ
| 1. | Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1226] [Article Influence: 94.3] [Reference Citation Analysis (0)] |
| 2. | Bogdanova N, Helbig S, Dörk T. Hereditary breast cancer: ever more pieces to the polygenic puzzle. Hered Cancer Clin Pract. 2013;11:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 3. | Ghoussaini M, Pharoah PD, Easton DF. Inherited genetic susceptibility to breast cancer: the beginning of the end or the end of the beginning? Am J Pathol. 2013;183:1038-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 4. | Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513-1530. [PubMed] |
| 5. | Karahalil B, Bohr VA, Wilson DM. Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum Exp Toxicol. 2012;31:981-1005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 6. | Bartsch H, Dally H, Popanda O, Risch A, Schmezer P. Genetic risk profiles for cancer susceptibility and therapy response. Recent Results Cancer Res. 2007;174:19-36. [PubMed] |
| 7. | Andreassen CN, Alsner J, Overgaard M, Overgaard J. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother Oncol. 2003;69:127-135. [PubMed] |
| 8. | Costa S, Medeiros R, Schmitt F. DNA signalling/repair genetic polymorphisms and breast cancer risk: a review. Applied Cancer Res. 2005;25:161-180. |
| 9. | Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 419] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 10. | Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J. 2001;20:6530-6539. [PubMed] |
| 11. | Marsin S, Vidal AE, Sossou M, Ménissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem. 2003;278:44068-44074. [PubMed] |
| 12. | Mohrenweiser HW, Xi T, Vázquez-Matías J, Jones IM. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol Biomarkers Prev. 2002;11:1054-1064. [PubMed] |
| 13. | Yi L, Xiao-Feng H, Yun-Tao L, Hao L, Ye S, Song-Tao Q. Association between the XRCC1 Arg399Gln polymorphism and risk of cancer: evidence from 297 case-control studies. PLoS One. 2013;8:e78071. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Sterpone S, Cozzi R. Influence of XRCC1 Genetic Polymorphisms on Ionizing Radiation-Induced DNA Damage and Repair. J Nucleic Acids. 2010;2010:pii: 780369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604-608. [PubMed] |
| 16. | Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378-4384. [PubMed] |
| 17. | Huang Y, Li L, Yu L. XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms in breast cancer risk: a meta-analysis. Mutagenesis. 2009;24:331-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 18. | Li H, Ha TC, Tai BC. XRCC1 gene polymorphisms and breast cancer risk in different populations: a meta-analysis. Breast. 2009;18:183-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Wu K, Su D, Lin K, Luo J, Au WW. XRCC1 Arg399Gln gene polymorphism and breast cancer risk: a meta-analysis based on case-control studies. Asian Pac J Cancer Prev. 2011;12:2237-2243. [PubMed] |
| 20. | Saadat M, Ansari-Lari M. Polymorphism of XRCC1 (at codon 399) and susceptibility to breast cancer, a meta-analysis of the literatures. Breast Cancer Res Treat. 2009;115:137-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Saadat M. Haplotype analysis of XRCC1 (at codons 194 and 399) and susceptibility to breast cancer, a meta-analysis of the literatures. Breast Cancer Res Treat. 2010;124:785-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Brem R, Cox DG, Chapot B, Moullan N, Romestaing P, Gérard JP, Pisani P, Hall J. The XRCC1 -77T->C variant: haplotypes, breast cancer risk, response to radiotherapy and the cellular response to DNA damage. Carcinogenesis. 2006;27:2469-2474. [PubMed] |
| 23. | Sterpone S, Mastellone V, Padua L, Novelli F, Patrono C, Cornetta T, Giammarino D, Donato V, Testa A, Cozzi R. Single-nucleotide polymorphisms in BER and HRR genes, XRCC1 haplotypes and breast cancer risk in Caucasian women. J Cancer Res Clin Oncol. 2010;136:631-636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Liu L, Yuan P, Liu L, Wu C, Zhang X, Guo H, Zhong R, Xu Y, Wu J, Duan S. A functional -77T>C polymorphism in XRCC1 is associated with risk of breast cancer. Breast Cancer Res Treat. 2011;125:479-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597-609. [PubMed] |
| 26. | Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H, Nohmi T, Kasai H, Yokota J. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219-3225. [PubMed] |
| 27. | Yuan W, Xu L, Feng Y, Yang Y, Chen W, Wang J, Pang D, Li D. The hOGG1 Ser326Cys polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:835-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Ding DP, Zhang Y, He XF. Lack of association between hOGG1 Ser326Cys polymorphism and breast cancer susceptibility in European population. Breast Cancer Res Treat. 2011;129:1023-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Gu D, Wang M, Zhang Z, Chen J. Lack of association between the hOGG1 Ser326Cys polymorphism and breast cancer risk: evidence from 11 case-control studies. Breast Cancer Res Treat. 2010;122:527-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Roberts MR, Shields PG, Ambrosone CB, Nie J, Marian C, Krishnan SS, Goerlitz DS, Modali R, Seddon M, Lehman T. Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis. 2011;32:1223-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Kim KY, Han W, Noh DY, Kang D, Kwack K. Impact of genetic polymorphisms in base excision repair genes on the risk of breast cancer in a Korean population. Gene. 2013;532:192-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Xie H, Xia K, Rong H, Chen X. Genetic polymorphism in hOGG1 is associated with triple-negative breast cancer risk in Chinese Han women. Breast. 2013;22:707-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 33. | Wei B, Zhou Y, Xu Z, Xi B, Cheng H, Ruan J, Zhu M, Hu Q, Wang Q, Wang Z. The effect of hOGG1 Ser326Cys polymorphism on cancer risk: evidence from a meta-analysis. PLoS One. 2011;6:e27545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284-1289. [PubMed] |
| 35. | Wu B, Liu HL, Zhang S, Dong XR, Wu G. Lack of an association between two BER gene polymorphisms and breast cancer risk: a meta-analysis. PLoS One. 2012;7:e50857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Kang H, Dai Z, Ma X, Ma L, Jin Y, Liu X, Wang X. A genetic variant in the promoter of APE1 gene (-656 T>G) is associated with breast cancer risk and progression in a Chinese population. Gene. 2013;531:97-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Parliament MB, Murray D. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Semin Radiat Oncol. 2010;20:232-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Xie XX, Ouyang SY, Jin HK, Wang H, Zhou JM, Hu BQ. Predictive value of Xrcc1 gene polymorphisms for side effects in patients undergoing whole breast radiotherapy: a meta-analysis. Asian Pac J Cancer Prev. 2012;13:6121-6128. [PubMed] |
| 39. | Cox JD, Stetz J, Pajak TF. Toxicity criteria of the Radiation Therapy Oncology Group (RTOG) and the European Organization for Research and Treatment of Cancer (EORTC). Int J Radiat Oncol Biol Phys. 1995;31:1341-1346. [PubMed] |
| 40. | Pavy JJ, Denekamp J, Letschert J, Littbrand B, Mornex F, Bernier J, Gonzales-Gonzales D, Horiot JC, Bolla M, Bartelink H. EORTC Late Effects Working Group. Late effects toxicity scoring: the SOMA scale. Radiother Oncol. 1995;35:11-15. [PubMed] |
| 41. | Chang-Claude J, Popanda O, Tan XL, Kropp S, Helmbold I, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Schmezer P. Association between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11:4802-4809. [PubMed] |
| 42. | Giotopoulos G, Symonds RP, Foweraker K, Griffin M, Peat I, Osman A, Plumb M. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer. 2007;96:1001-1007. [PubMed] |
| 43. | Chang-Claude J, Ambrosone CB, Lilla C, Kropp S, Helmbold I, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Schmezer P. Genetic polymorphisms in DNA repair and damage response genes and late normal tissue complications of radiotherapy for breast cancer. Br J Cancer. 2009;100:1680-1686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 44. | Kerns SL, Stock R, Stone N, Buckstein M, Shao Y, Campbell C, Rath L, De Ruysscher D, Lammering G, Hixson R. A 2-stage genome-wide association study to identify single nucleotide polymorphisms associated with development of erectile dysfunction following radiation therapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2013;85:e21-e28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 45. | Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, Wilkinson JS, Tyrer J, Misra V, Platte R. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13:65-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 46. | Andreassen CN, Dikomey E, Parliament M, West CM. Will SNPs be useful predictors of normal tissue radiosensitivity in the future? Radiother Oncol. 2012;105:283-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 47. | Sapkota Y, Mackey JR, Lai R, Franco-Villalobos C, Lupichuk S, Robson PJ, Kopciuk K, Cass CE, Yasui Y, Damaraju S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS One. 2013;8:e64896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132-2138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 142] [Article Influence: 8.4] [Reference Citation Analysis (0)] |