Published online Apr 24, 2025. doi: 10.5306/wjco.v16.i4.104182
Revised: January 11, 2025
Accepted: January 23, 2025
Published online: April 24, 2025
Processing time: 104 Days and 2.2 Hours
Emerging evidence implicates Candida albicans (C. albicans) in human oncogenesis. Notably, studies have supported its involvement in regulating outcomes in co
To investigate the dual role of C. albicans in the development and progression of CRC through metabolite profiling and to establish a prognostic model that integrates the microbial and metabolic interactions in CRC, providing insights into potential therapeutic strategies and clinical outcomes.
A prognostic model integrating C. albicans with CRC was developed, incorporating enrichment analysis, immune infiltration profiling, survival analysis, Mendelian randomization, single-cell sequencing, and spatial transcriptomics. The effects of the C. albicans metabolite mixture on CRC cells were subsequently validated in vitro. The primary metabolite composition was characterized using liquid chromatography-mass spectrometry.
A prognostic model based on five specific mRNA markers, EHD4, LIME1, GADD45B, TIMP1, and FDFT1, was established. The C. albicans metabolite mixture significantly reduced CRC cell viability. Post-treatment analysis revealed a significant decrease in gene expression in HT29 cells, while the expression levels of TIMP1, EHD4, and GADD45B were significantly elevated in HCT116 cells. Conversely, LIME1 expression and that of other CRC cell lines showed reductions. In normal colonic epithelial cells (NCM460), GADD45B, TIMP1, and FDFT1 expression levels were significantly increased, while LIME1 and EHD4 levels were markedly reduced. Following metabolite treatment, the invasive and migratory capabilities of NCM460, HT29, and HCT116 cells were reduced. Quantitative analysis of extracellular ATP post-treatment showed a significant elevation (P < 0.01). The C. albicans metabolite mixture had no effect on reactive oxygen species accumulation in CRC cells but led to a reduction in mitochondrial membrane potential, increased intracellular lipid peroxidation, and induced apoptosis. Metabolomic profiling revealed significant alterations, with 516 metabolites upregulated and 531 downregulated.
This study introduced a novel prognostic model for CRC risk assessment. The findings suggested that the C. albicans metabolite mixture exerted an inhibitory effect on CRC initiation.
Core Tip: This study explored the paradoxical role of Candida albicans (C. albicans) in colorectal cancer (CRC), focusing on its tumor-modulating effects through metabolic interactions. A novel prognostic model incorporating five mRNA markers, EHD4, LIME1, GADD45B, TIMP1, and FDFT1, was developed, providing a framework for CRC risk assessment. The C. albicans metabolite mixture demonstrated a significant inhibitory effect on CRC initiation by reducing cell viability, altering gene expression, impairing migratory and invasive abilities, and promoting apoptosis without affecting reactive oxygen species accumulation. The study highlighted the potential of C. albicans metabolites as a therapeutic avenue in CRC management.
- Citation: Zhang HL, Zhao R, Wang D, Mohd Sapudin SN, Yahaya BH, Harun MSR, Zhang ZW, Song ZJ, Liu YT, Doblin S, Lu P. Candida albicans and colorectal cancer: A paradoxical role revealed through metabolite profiling and prognostic modeling. World J Clin Oncol 2025; 16(4): 104182
- URL: https://www.wjgnet.com/2218-4333/full/v16/i4/104182.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i4.104182
Candida albicans (C. albicans), a commensal yeast widely present on gastrointestinal mucosal surfaces, has recently gained recognition as a significant opportunistic pathogen, particularly in immunocompromised individuals[1]. Its association with increased mortality in colorectal cancer (CRC) highlights the intricate and multifactorial nature of CRC, which is shaped by genetic predispositions, delayed diagnosis, tumor localization, dietary nitrites and nitrates, tumor aggressiveness, and resistance to therapies[2]. Increasing attention has been directed toward the role of C. albicans in CRC progression, with its presence in tumor tissues becoming a central focus of study.
Extensive research has revealed the direct contributions of C. albicans to CRC pathogenesis, primarily through immune disruption and chronic inflammation. In a cohort study of 52 patients with newly diagnosed CRC and 52 matched controls, elevated levels of C. albicans were identified in the gut microbiota of patients with CRC, suggesting that fungal dysbiosis may facilitate sporadic CRC development[3]. Supporting this, findings in Dectin-3 knockout (Dectin-3 -/-) mice demonstrated accelerated CRC progression associated with an increased gut fungal burden, particularly of C. albicans. Fecal microbiota transplants from Dectin-3 -/- tumor-bearing mice further confirmed the role of C. albicans in exacerbating CRC malignancy, while antifungal treatments significantly reduced tumor burden. Both in vitro and in vivo analyses indicated that Dectin-3 deficiency impaired macrophage-mediated clearance of C. albicans, resulting in uncontrolled fungal proliferation[4].
Sequencing of the ITS1 region in fecal samples from patients with CRC revealed significantly elevated levels of C. albicans. Dectin-1, another C-type lectin receptor, recognizes β-1,3-glucan in fungal cell walls and is expressed in immune cells such as dendritic cells, macrophages, and monocytes. Research suggests that C. albicans can stimulate the proliferation of intestinal epithelial cells by activating the Wnt signaling pathway, a critical mechanism in CRC development. In murine models, C. albicans infection activated Wnt signaling, suggesting that intestinal epithelial cells sense C. albicans through Dectin-1, thereby promoting tumorigenesis[5]. These findings highlight how C. albicans overgrowth in the gut contributes to a protumorigenic microenvironment characterized by chronic inflammation, immune evasion, and epithelial disruption. However, the precise molecular mechanisms by which C. albicans contributes to CRC remain incompletely understood.
Recent findings indicated that C. albicans may engage in alternative oncogenic pathways beyond its established inflammatory and immunomodulatory roles. In rat models of CRC treated with chemotherapy, Lactobacillus plantarum, and C. albicans, significant tumor reduction was observed, accompanied by nuclear condensation, a hallmark of apoptosis. Additionally, treated groups exhibited significantly lower serum levels of interferon (IFN)-γ, interleukin (IL)-4, and transforming growth factor beta compared to control groups, suggesting that the combination of C. albicans and Lactobacillus plantarum may hold therapeutic potential in CRC treatment[6]. These findings implied that metabolic byproducts of C. albicans could influence tumor cell proliferation, invasion, and metastasis, though the precise mechanisms remain unclear.
This study aimed to develop a prognostic model for CRC based on C. albicans-regulated mRNA expression profiles. By focusing on a metabolic mixture rather than individual metabolites, this approach better replicates the complex in vivo environment where cells interact with a variety of metabolites simultaneously. Understanding the effects of such metabolic mixtures on CRC cell behavior carries significant clinical relevance. Our in vitro analyses emphasized evaluating the impact of C. albicans metabolic mixtures on CRC cell invasion and migration while also identifying key components within these mixtures. This work provided a foundation for innovative therapeutic strategies targeting fungal metabolites in CRC prevention and treatment.
Gene expression profiles influenced by C. albicans were retrieved from the Gene Expression Omnibus (GEO) dataset (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE42606. Data collection occurred on August 16, 2023, comprising 25 samples infected with C. albicans for 4 h and 34 samples infected for 24 h. Additionally, mRNA expression profiles and clinical data were obtained from The Cancer Genome Atlas CRC Dataset (TCGA-CRC; https://portal.gdc.cancer.gov/), including 44 samples of normal and tumor tissues from a total of 571 specimens. The dataset was randomly divided into a training set (70%) and an internal validation set (30%) to ensure robust model construction and validation. For external validation, dataset GSE41258, containing 390 CRC mRNA profile samples and associated clinical data, was retrieved from the GEO database on the same date. Samples with incomplete clinical information or a survival duration of less than 10 days were excluded to improve the reliability and accuracy of the analysis.
Differential expression analysis of mRNA matrices between 4-h and 24-h C. albicans infection samples was performed using the limma package. Significantly differentially expressed mRNAs were identified based on criteria of |log2 fold change| > 1 and a false discovery rate < 0.05.
Data analysis in this study was conducted using R software version 4.1.0. The glmnet (v2.0.18) and survival (v2.44.1.1) packages were employed for univariate and multivariate Cox proportional hazards model regression as well as least absolute shrinkage and selection operator (LASSO) regression. Univariate Cox regression was initially performed to evaluate the relationship between mRNA expression levels and overall survival (OS), with P values below 0.05 considered statistically significant. mRNAs meeting this criterion were subjected to LASSO regression for feature selection. Subsequently, multivariate Cox regression was applied to assess the prognostic significance and hazard ratios of the predictive model, with 95% confidence intervals calculated. The prognostic risk score was determined using the formula Σ (expmRNAn × βmRNA), incorporating mRNA expression values and their corresponding coefficients. This facilitated the classification of samples into high-risk and low-risk groups. Kaplan-Meier survival analysis and log-rank tests, performed using the survminer package with a significance level of P < 0.05, were used to evaluate the prognostic significance of risk scores across cohorts. Additionally, the time receiver operating characteristic (ROC) package generated time-dependent ROC curves to calculate area under the curve (AUC) values, providing a metric for model accuracy. These analyses demonstrated the validated efficacy of the developed mRNA prognostic models in predicting survival outcomes and assessing risk in patients with CRC, with all metrics achieving significance at P < 0.05.
Gene enrichment analyses, including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, were conducted using microbial informatics tools (https://www.bioinformatics.com.cn/, accessed August 20, 2023). A significance threshold of P < 0.05 was applied to identify enriched Gene Ontology terms and KEGG pathways. The R package GSVA was utilized to compute enrichment scores (ES) based on Z-scores for metabolic gene sets obtained from the KEGG database. Samples were divided into two groups based on expression levels. The top 30% were designated as the high expression group, while the bottom 30% were classified as the low expression group. Differential GSVA scores between these cohorts were analyzed using the limma package, enabling a robust comparison of metabolic gene set activity across expression-defined subgroups.
To ensure data quality and consistency, immune infiltration profiles for all TCGA samples were obtained from the publicly accessible TIMER2.0 database. Spearman correlation coefficients were calculated and visualized through heatmaps, offering a comprehensive overview of relationships between distinct immune cell types and gene expression across multiple algorithms. Samples within the top 30% of gene expression levels were categorized as the high expression group, while those in the bottom 30% were classified as the low expression group. Differential analysis was conducted using the limma package to compute log2 fold changes for each gene, which were subsequently ranked by value. Gene Set Enrichment Analysis (GSEA) was performed using the clusterProfiler package, focusing on hallmark gene sets and pathways associated with KEGG metabolism. ES were calculated for each gene set, followed by significance testing and multiple hypothesis adjustments applied to the ES values to ensure robust statistical interpretation.
Kaplan-Meier survival analysis was performed with the survival package, and optimal cutoffs for high and low ex
The influence of continuous variables on risk may follow nonlinear patterns, and neglecting such nonlinearity can introduce bias into analytical results. To address this, restricted cubic spline was employed to examine the nonlinear relationship between MKI67 expression and the risk of OS, disease-specific survival (DSS), progression-free interval (PFI), and disease-free interval (DFI). In the GSE146773 dataset, 1152 U2OS FUCCI cells were sorted using fluorescence-activated cell sorting, followed by RNA-seq profiling using SMART-seq2 technology. Single-cell RNA sequencing of the U2OS FUCCI cell line allowed precise measurement of RNA expression levels and cell cycle phases at the single-cell level. Standardized RNA expression levels were plotted against pseudo-time progression derived from fluorescence intensities of cell cycle markers. Gene expression values were normalized to Z-scores using the scale function, with outliers exceeding ± 3 standard deviations excluded from the analysis. Statistical differences in gene expression among groups were assessed using the Kruskal-Wallis rank sum test.
The single-nucleotide polymorphism results were obtained using the mr_singlesnp function, and forest plots were generated with mr_forest_plot for visualization. Funnel plots were created using the mr_funnel_plot function, while sensitivity analysis was conducted using a leave-one-out approach. Scatter plots illustrating individual SNP associations were produced with the mr_scatter_plot function. Bayesian colocalization analysis was performed to assess the probability of shared causal variation between two traits using the coloc package with default settings (https://github.com/chr1swallace/coloc). Colocalization data were obtained using the ieugwasr_to_coloc function, and genetic colocalization of phenotypes was analyzed using coloc.abf. The analysis determined whether phenotypes share a causal variant within genomic regions associated with the corresponding expression quantitative trait locus genes. A colocalization probability threshold of PP.H4.abf > 80% was applied. Results were visualized using the locuscompare function from the locuscompare package. The genome-wide association study datasets utilized included ebi-a-GCST90018808 and ieu-b-4965.
The uniform manifold approximation and projection (UMAP) technique was employed to project high-dimensional data into two-dimensional space, visualized as heat maps. UMAP, as a dimensionality reduction method, retains the topological structure of the data, enabling the identification of clusters and patterns within the dataset. This approach was used to visualize gene expression data from the model, providing a clear representation of spatial organization. To characterize cellular composition at each locus on the 10x Visium slide, a deconvolution analysis integrating spatial transcriptomic and single-cell transcriptomic data was conducted, focusing specifically on the cancer type under investigation. Initially, RNA-seq data were collected from various samples of the same cancer type, followed by the con
The experiment employed the C. albicans strain BNCC263676, sourced from Beina Chuanglian Biotechnology Co., Ltd. C. albicans was cultured on Yield Monitoring (YM) agar medium at 30 °C. To induce the filamentous phase, colonies grown on YM agar at 30 °C for 24 h were transferred to YM liquid medium and incubated at 37 °C on a shaker at 250 rpm for 14-16 h. The C. albicans culture was then transferred to fresh YM liquid medium and incubated under the same conditions for an additional 3-8 h to ensure stable results. Absorbance readings of the C. albicans liquid culture were taken at 600 nm using an ELISA reader. Samples were collected from the YM liquid culture at optical density (OD) values of 0.2, 0.3, and 0.4. To prepare a metabolite mixture, the C. albicans cells were inactivated by heating at 95 °C in a water bath for 30 min.
NCM460, HT29, and HCT116 cells were harvested to create a suspension. A 100 μL volume of this suspension, containing a cell density of 1 × 104-1 ×105 cells per well, was added to each well of a 96-well plate. To minimize edge effects, sterile PBS buffer was introduced into the outer wells. The plates were incubated under standard conditions (37 °C, 5% CO2) to allow cell adherence. Interventions using C. albicans metabolite mixtures with OD values of 0.2, 0.3, and 0.4 were conducted, and cells were exposed for 12, 24, 48, and 72 h. After the medium was discarded, 10 μL of cell counting kit (CCK)-8 solution was added to each well, and the plates were incubated for 2 h. Absorbance at 450 nm was measured using an enzyme-linked spectrophotometer. This protocol established criteria for distinguishing metabolite concentrations based on OD values, which were used to identify the optimal concentration and duration for the intervention.
The Matrigel matrix was thawed overnight at 4 °C, and the microinjection apparatus, including blue and yellow tips, was precooled at -20 °C. Matrix preparation involved mixing 2.5 mL of Matrigel (Corning Incorporated, REF 354248) with 97.5 mL of serum-free medium prechilled to 4 °C. This mixture was gently blended and dispensed into the upper chambers of a 24-well Transwell plate, with 100 µL added per well. The plates were incubated at 37 °C for 30-60 min to allow the matrix to polymerize. Once the matrix had solidified, cells from the control and C. albicans metabolite mixture intervention groups, prerinsed three times with PBS, were resuspended and added to the upper chambers at 100 μL per chamber. The lower chambers were filled with 600 μL of medium containing 10% FBS. After a 48-h incubation, the chamber contents were removed, and the wells were rinsed three times with PBS. The chambers were then fixed with 4% paraformaldehyde (Lanzhou Kebao Biotechnology Co., Ltd., 35942) for 30 min. Following fixation, the chambers were washed with PBS, stained with crystal violet (Lanzhou Kebao Biotechnology Co., Ltd., 3422) for 15 min, and rinsed again with PBS. Microscopic images were captured to assess cell migration and invasion.
Total RNA from each experimental group was extracted using the Trizol method, followed by reverse transcription to synthesize cDNA in accordance with the manufacturer’s protocol. Using cDNA as the template, mRNA expression levels were quantified, with GAPDH serving as the internal reference control. Primers for mRNA analysis were obtained from Lanzhou Kebao Biotechnology Co., Ltd. (catalog no. 1174624162), with specific sequences provided in Table 1. Gene expression was analyzed using real-time quantitative PCR (RT-qPCR), which quantified mRNA levels across the sample groups to determine variations in gene expression.
| Gene (Rabbit) | Prime | Product length | Login ID |
| GAPDH | F: GCAAAGTGGATGTTGTCGCC | 132 | NM_001082253.1 |
| R: TGATGACCAGCTTCCCGTTC | |||
| FDFT1 | F: AGATTCGGAAAGGGCAAGCA | 223 | NM_001287742.2 |
| R: AACGACAGGTAGATGGGGGA | |||
| TIMP1 | F: TCAACCAGACCACCTTATACC | 296 | NM_003254.3 |
| R: GCATTCCTCACAGCCAACAG | |||
| GADD45B | F: GCCCTGCAAATCCACTTCAC | 165 | NM_015675.4 |
| R: GTTCGTGACCAGGAGACAAT | |||
| LIME1 | F: GGAAGCGCAAGTCGGACAC | 236 | NM_001305654.2 |
| R: CACGTTGGAATAGGTGGCCT | |||
| EHD4 | F: CTTCGAGAACAAGCCCATGA | 161 | NM_139265.4 |
| R: TGCCCTCAGTCTCTCCATACA |
Extracellular ATP (eATP) levels were quantitatively measured in this study. The culture medium was first removed, and approximately 0.1 mL of the medium was collected and mixed with 1 mL of the extraction solution. The mixture was thoroughly homogenized and centrifuged at 10000 g for 10 min at 4 °C. The resulting supernatant was carefully transferred to a new tube and mixed with 500 μL of chloroform. After thorough mixing, a second centrifugation at 10000 g for 3 min at 4 °C was performed. The supernatant was then kept on ice for subsequent measurement. The quantification process was as follows: (1) The UV spectrophotometer or ELISA reader was preheated for a minimum of 30 min. The wavelength was set to 340 nm, and the instrument was zeroed using distilled water; (2) A 10 μmol/mL ATP standard solution was diluted 16 times with distilled water to achieve a final concentration of 0.625 μmol/mL; and (3) A working solution was prepared using reagents in the following proportions: Reagent 2 (mL): Reagent 3 (mL): Reagent 4 (mL): Reagent 5 (mL): Reagent 6 (mL) = 1:1:0.1:0.4:0.1. The prepared samples were added to a 96-well UV plate according to the procedure outlined in Table 2.
| Reagent name (μL) | Measuring tube | Standard tube |
| Sample | 20 | |
| Standard liquid | 20 | |
| Reagent I | 128 | 128 |
| Operating fluid | 52 | 52 |
After thorough mixing, the initial absorbance (A1) was promptly recorded at 340 nm within 10 s. The reaction mixture in the colorimetric plate was then transferred to a water bath maintained at 37 °C for mammalian samples or 25 °C for other species. The mixture was incubated for precisely 3 min. At 3 min and 10 s, the second absorbance (A2) was measured. The 96-well plate was subsequently placed in an incubator set to 37 °C or 25 °C, depending on the sample type. If the plate reader featured temperature control, the corresponding settings were applied. The test sample was determined by subtracting A1 from A2 for the test tube, while the standard was calculated by subtracting A1 from A2 for the standard tube. The ATP concentration (μmol/mL) was calculated using the following formula:
ATP content = Determination ÷ (standard ÷ C standard) × [Vextraction + Vserum (pulp)]/V serum (pulp) = 6.875 × determination ÷ standard
The experiment included a control group (untreated with C. albicans metabolites) and a group treated with C. albicans metabolites. The concentration of C. albicans metabolites for screening was established based on previously optimized treatment times. Cells were washed, digested, and collected before lipid peroxidation (LPO) detection, which was carried out using a specific kit (Solarbio Science and Technology, Beijing, Lot: No. 2309002). Blank tubes, standard tubes, and test tubes were prepared according to the manufacturer’s instructions, and samples were added as directed. The samples were mixed and incubated at 45 °C for 1 h. Following incubation, the supernatant was collected through centrifugation, and the OD was measured at a wavelength of 586 nm. The LPO content was calculated using the formula:
LPO content = (test-blank)/(standard - blank) × standard substance concentration ÷ sample protein concentration
NCM460, HCT-116, and HT-29 cells were cultured in confocal dishes at a density of 1 × 104 cells per dish. Following treatment as described in Culture and metabolite acquisition of C. albicans, the cells were washed once with PBS. Subsequently, 1 mL of cell culture medium was added, followed by the addition of 1 mL of JC-1 staining solution (Solarbio Science and Technology, Beijing, Lot: No. 2311005). The cells were incubated with the staining solution for 20 min and then rinsed using the specific buffer provided in the kit. Finally, 2 mL of culture medium was added to the dishes, and the cells were analyzed using a laser confocal microscope.
NCM460, HCT-116, and HT-29 cells were cultured under optimal conditions and seeded into 6-well plates at a density of approximately 1 × 105 cells per well. The cells were organized into groups based on PCR was used to detect gene expression in the model. After the intervention, the supernatant was discarded, and the cells were digested with 0.5 mL of trypsin without EDTA. Digestion was halted by adding 1 mL of complete medium supplemented with 10% FBS. The cells were incubated for 5 min, pelleted by centrifugation, and resuspended in 1 mL of cell staining buffer. To the suspension, 5 µL of Hoechst staining solution and 5 µL of propidium iodide solution were added. The mixture was thoroughly mixed and incubated at 4 °C for 20-30 min. Apoptosis levels were then analyzed by flow cytometry (Biosharp, BL114A).
Following the grouping and interventions described in Culture and metabolite acquisition of C. albicans, reactive oxygen species levels in NCM460, HCT-116, and HT-29 cells were measured using an ROS detection kit (Beyotime, Cat NO. S0033S). DCFH-DA was diluted in serum-free medium to a final concentration of 10 mmol/L using a 1:1000 dilution ratio. After the cell culture medium was removed, at least 1 mL of the diluted DCFH-DA solution was added to each well. The cells were incubated at 37 °C for 20 min. Following incubation, the cells were rinsed three times with serum-free medium. Rosup was added to the positive control well to serve as a positive control.
All samples were collected, and 500 μL of an 80% methanol-water extraction solvent was added to each sample, followed by swirling for 3 min to ensure complete suspension. The centrifuge tubes were then rapidly frozen in liquid nitrogen for 5 min, thawed on dry ice for 5 min, and further thawed on ice for an additional 5 min. Care was taken to prevent temperature differences that could cause tube lids to burst or samples to aerosolize. This freeze-thaw process, coupled with swirling for 2 min, was repeated three times. The samples were centrifuged at 12000 rpm for 10 min at 4 °C. Subsequently, 300 μL of the supernatant was transferred to a labeled centrifuge tube and incubated at -20 °C for 30 min. The samples then underwent a second centrifugation at 12000 rpm for 3 min at 4 °C. From this, 200 μL of the supernatant was placed into the inner liner of each sample vial for instrumental analysis. Chromatographic separation was carried out using a Waters ACQUITY Premier HSS T3 column (1.8 µm, 2.1 mm × 100 mm). The mobile phases consisted of: Phase A: 0.1% formic acid in water; and Phase B: 0.1% formic acid in acetonitrile. The column was maintained at 40 °C, with a flow rate of 0.4 mL/min. A sample injection volume of 4 μL was used for analysis. Raw mass spectrometry data were converted into mzXML format using ProteoWizard software. Peak extraction, alignment, and retention time correction were conducted using XCMS software. Peaks with a missing rate exceeding 50% in any sample group were excluded, and missing values were estimated using the k-nearest neighbors method. Peak areas were corrected using the support vector regression method. The metabolites corresponding to the corrected and filtered peaks were identified by cross-referencing a laboratory-developed database and publicly available databases such as HMDB (https://hmdb.ca/) and KEGG (https://www.kegg.jp/). Substances with an identification score greater than 0.5 and a coefficient of variation (CV) below 0.3 for QC samples were retained. Data from both positive and negative modes were combined, ensuring that only metabolites with the highest quality scores and lowest CV values were included. This culminated in the creation of the all_sample_data.xlsx file, which contained the final processed data.
Statistical analyses were performed using Perl software (version 5.32.0.1-64-bit; https://strawberryperl.com/) and R software (version 4.0.3; https://www.r-project.org). A P value less than 0.05 was considered statistically significant, values below 0.01 were deemed more statistically significant, and values under 0.001 were categorized as highly statistically significant.
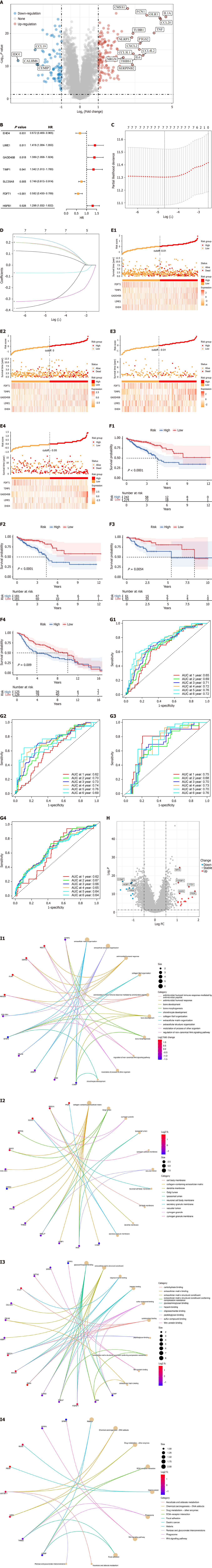
Using thresholds of false discovery rate < 0.01 and |log2 fold changes| > 1, a total of 213 differentially expressed genes were identified, comprising 135 upregulated and 78 downregulated genes (Figure 1A). In the training cohort, univariate Cox proportional hazards regression analysis was applied to 214 mRNAs, revealing that 7 mRNAs were significantly associated with prognosis (P < 0.05) (Figure 1B). These results were further refined through LASSO-Cox regression analysis, which selected five mRNAs for inclusion in the prognostic model (Figure 1C and D). A novel risk scoring formula was developed based on these selected mRNAs:
Risk score = 0.245a1 - 0.378a2 + 0.318a3 - 0.357a4 + 0.3184a5
Tag: a1:LIME1; a2:EHD4; a3:GADD45B; a4:FDFT1; a5:TIMP1
The overall risk score for each patient was calculated using this formula (Figure 1E1). Both training and testing cohorts were used to validate the reliability of the model (Figure 1E2 and 1E3). The external validation set is present in Figure 1E4. Kaplan-Meier survival analysis, along with a two-sided log-rank test, revealed that high-risk patients had significantly shorter OS compared to low-risk patients (P < 0.001) (Figure 1F1). Validation confirmed a similar OS difference between groups (P < 0.05) (Figure 1F2 and 1F3). Time-dependent ROC analysis indicated an AUC of 0.76 for the overall population (Figure 1G1). For 5-year OS prediction, the AUC was 0.76 in the training cohort and 0.70 in the testing cohort (Figures 1G2 and 1G3). Risk scores were also validated in the GEO dataset, confirming significant OS differences between high-risk and low-risk groups consistent with TCGA results (P < 0.001) (Figure 1F4). Furthermore, time-specific AUC values in the GEO dataset were 0.62 at 1 year, 0.67 at 3 years, and 0.66 at 5 years (Figure 1G4), demonstrating the robustness and successful establishment of the model.
As shown in Figure 1H, a comparative analysis between the high-risk and low-risk groups identified 17 upregulated and 12 downregulated genes (adjusted P < 0.05, |log2 fold change| > 1). Enrichment analysis of biological processes indicated significant involvement in pathways related to extracellular matrix organization, extracellular structure arrangement, and the humoral response to antimicrobial factors, among others (Figure 1I1). Molecular function enrichment analysis revealed that these genes were primarily associated with activities such as glycosaminoglycan binding, structural components of the extracellular matrix, and oligosaccharide binding (Figure 1I2). Cellular component enrichment analysis highlighted notable associations with the collagen-containing extracellular matrix, Golgi lumen, and zymogen granules (Figure 1I3). Additionally, KEGG pathway enrichment analysis demonstrated gene participation in pathways related to ascorbate and aldarate metabolism as well as DNA adduct formation linked to chemical carcinogenesis (Figure 1I4).
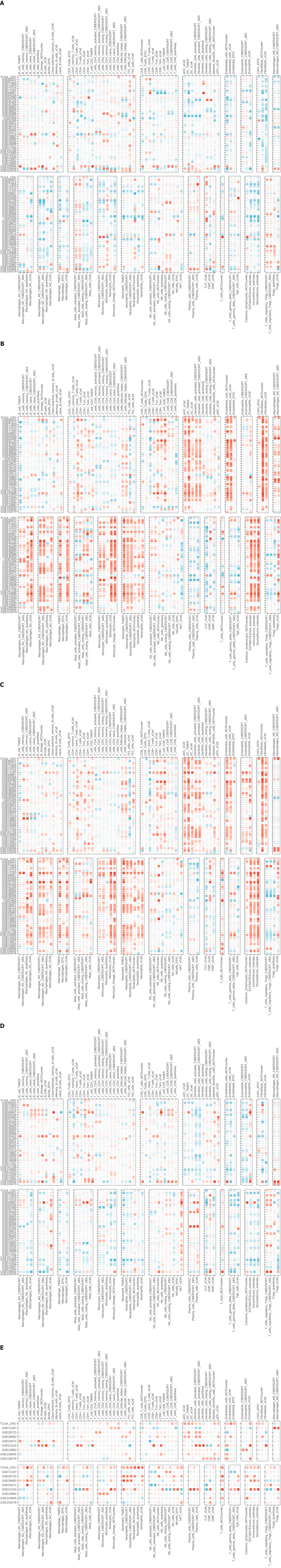
FDFT1 showed a positive correlation with B cells and CD4+ T cells, while it negatively correlated with endothelial cells, dendritic cells, fibroblasts, eosinophils, macrophages, and natural killer cells (Figure 2A). TIMP1 exhibited positive correlations with macrophages, monocytes, mast cells, neutrophils, CD8+ T cells, fibroblasts, eosinophils, and macrophages (Figure 2B). GADD45B demonstrated positive correlations with CD8+ T cells, CD4+ T cells, dendritic cells, fibroblasts, eosinophils, endothelial cells, and macrophages (M1, M2) as well as monocytes and neutrophils (Figure 2C). LIME1 was positively associated with CD8+ T cells, dendritic cells, fibroblasts, eosinophils, endothelial cells, macrophages (M0, M1, M2), neutrophils, and T cells while negatively correlated with endothelial cells and B cells (Figure 2D). EHD4 showed positive correlations with CD8+ T cells, dendritic cells, eosinophils, endothelial cells, macrophages (M0, M1, M2), and neutrophils (Figure 2E).
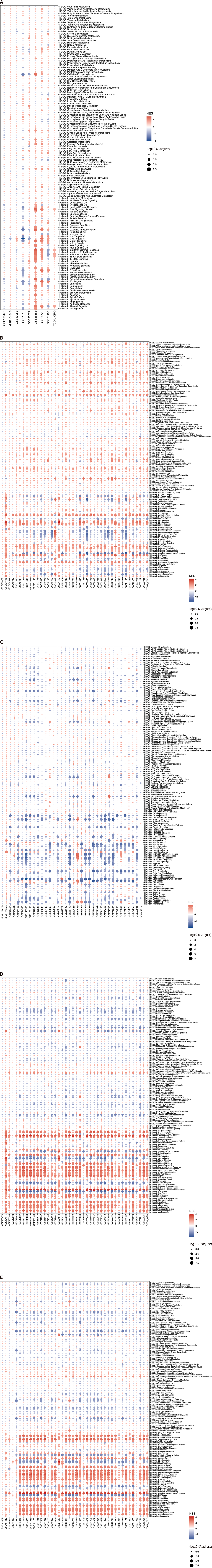
The high EHD4 expression group demonstrated enrichment in pathways related to tumor necrosis factor (TNF) signaling via nuclear factor (NF)-κB, the IFN-γ response, inflammatory response, E2F targets, and allograft rejection (Figure 3A).
Conversely, the TIMP1 high expression group was significantly enriched in pathways such as cholesterol homeostasis, porphyrin and chlorophyll metabolism, retinol metabolism, oxidative phosphorylation, drug metabolism by various enzymes, ascorbate and aldarate metabolism, Myc targets V1, mTORC1 signaling, and E2F targets. The low TIMP1 expression group exhibited enrichment in pathways including epithelial-mesenchymal transition (EMT), IFN-γ response, allograft rejection, IL-6-JAK-STAT3 signaling, and IFN-α response (Figure 3B).
The GADD45B high expression group displayed notable enrichment in pathways such as the upregulated UV response, TNF-α signaling via NF-κB, enhanced KRAS signaling, IFN-γ response, inflammatory response, IL-2-STAT5 signaling, hypoxia, apoptosis, apical junction formation, angiogenesis, and allograft rejection. Meanwhile, the low GADD45B expression group was enriched in pathways like E2F targets, G2M checkpoint, Myc targets V1, oxidative phosphorylation, ascorbate and aldarate metabolism, cytochrome P450-mediated drug metabolism, and pentose and glucuronate interconversions (Figure 3C).
The LIME1 low expression group showed significant enrichment in pathways such as pentose and glucuronate interconversions, downregulated UV response, TNF-α signaling via NF-κB, Myc targets V1, mTORC1 signaling, G2M checkpoint, E2F targets, and DNA repair (Figure 3D).
In the TIMP1 high expression group, pathways related to downregulated UV response, TNF-α signaling via NF-κB, upregulated KRAS signaling, IFN-γ response, inflammatory response, IL-6-JAK-STAT3 signaling, IL-2-STAT5 signaling, hypoxia, and EMT were notably enriched. In contrast, the low TIMP1 expression group showed enrichment in pathways involving E2F targets, G2M checkpoint, Myc targets V1, oxidative phosphorylation, and ascorbate and aldarate metabolism (Figure 3E).
Patients with CRC with high levels of FDFT1 expression demonstrated better survival outcomes compared to those with lower expression levels. Furthermore, the 25% of patients with the lowest FDFT1 expression exhibited worse survival rates compared to the 25% with the highest expression levels (Figure 4A). Patients with low TIMP1 expression showed improved survival outcomes compared to those with high TIMP1 expression. Among the top 25% of samples with the highest TIMP1 expression, survival rates were significantly lower than those observed in the bottom 25% with the lowest TIMP1 expression (Figure 4B).
Similarly, patients in the low GADD45B expression group had better survival outcomes compared to those with high GADD45B expression. The top 25% of samples with the highest GADD45B expression had poorer survival rates than the bottom 25% with the lowest expression levels (Figure 4C). The group characterized by low LIME1 expression also demonstrated improved survival compared to the high expression group. The top 25% of samples with the highest LIME1 expression exhibited worse survival rates than the bottom 25% with the lowest expression levels (Figure 4D).
Patients with high EHD4 expression had better survival outcomes compared to those with low EHD4 expression. The 25% of samples with the lowest EHD4 expression experienced worse survival rates than the 25% with the highest expression levels (Figure 4E). Notably, the highest number of deaths was recorded in the 25% of patients with the lowest EHD4 expression (Figure 4F).
In the GSE103479 dataset, LIME1 showed the highest number of fatalities among the 25% of patients with the lowest expression levels (Figure 4F). Conversely, the highest number of deaths occurred in the top 25% of patients with the highest TIMP1 expression levels (Figure 4F, G, H and I). In the GSE14333 dataset, the largest number of deaths was observed in the 25% of patients with the lowest expression levels. However, for TIMP1, the greatest number of fatalities occurred in the 25% of patients with the highest expression levels (Figure 4J, K and L). For FDFT1, the highest number of deaths was recorded among the 25% of patients with both the lowest and highest expression levels (Figure 4M1, M2 and N).
In the FDFT1 high expression group, pathways such as terpenoid backbone biosynthesis, caffeine metabolism, and other metabolic pathways were activated, while glycosaminoglycan biosynthesis, specifically heparan sulfate and heparin metabolic pathways, were suppressed (Figure 5A). In the TIMP1 high expression group, metabolic pathways related to the biosynthesis of glycosaminoglycans, including chondroitin sulfate and dermatan sulfate, and glycosphingolipid biosynthesis (globo series) were activated. Conversely, pathways such as vitamin B6 metabolism were inhibited.
For the GADD45B high expression group, glycosaminoglycan biosynthesis pathways (chondroitin sulfate and dermatan sulfate) were activated, whereas oxidative phosphorylation and several other metabolic pathways were suppressed. In the LIME1 high expression group, pathways such as taurine and hypotaurine metabolism were activated, while pathways like biotin metabolism were inhibited. For EHD4, galactose metabolism pathways were activated in the high expression group, whereas pathways such as biotin metabolism were suppressed (Figure 5A).
The effect of EHD4 on CRC survival risk was linear (Figure 5B). The effects of FDFT1 on DSS, PFI, and DFI survival risks in CRC were also linear (Figure 5C). The impact of GADD45B on DSS, PFI, and DFI survival risks was linear (Figure 5D). The influence of LIME1 on DSS and PFI survival risks was linear (Figure 5E). The effect of TIMP1 on DSS and PFI survival risks was linear (Figure 5F). The expressions of EHD4, GADD45B, LIME1, and TIMP1 were influenced by cell cycle progression, while FDFT1 expression remained unaffected (Figure 5G-K).
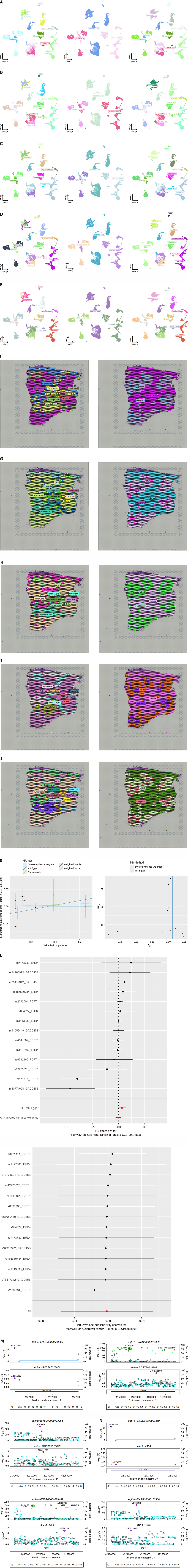
UMAP analysis revealed distinct clustering of cell populations based on their expression profiles. These groups corresponded to different cell types or states, including mast cells, M1 macrophages, B cells, myofibroblasts, epithelial cells, enteric glial cells, CD4Tn cells, CD8Tex cells, and fibroblasts (Figure 6A-E). The spatial expression patterns of EHD4, FDFT1, GADD45B, LIME1, and TIMP1 indicated similar localization to tumor cells. In CRC, these genes appear to be predominantly expressed by tumor cells (Figure 6F-J).
Forest plot depicting causal effect estimates and their confidence intervals for EHD4, GADD45B, FDFT1, and individual SNPs is presented in Figure 6L. The scatter points in the funnel plot are relatively symmetrical, indicating low heterogeneity (Figure 6K). Genetic analysis showed that EHD4, GADD45B, FDFT1, and CRC share genetic causal variations (Figure 6M and N).
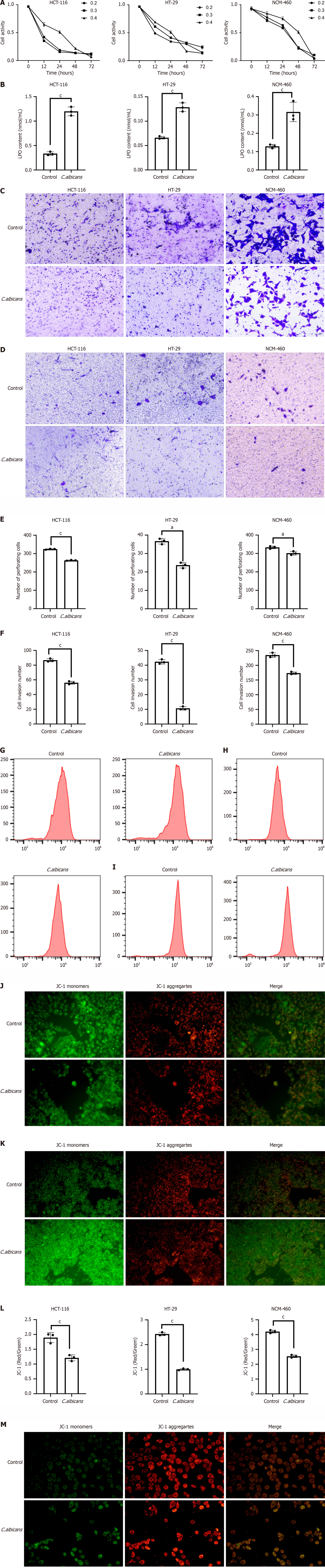
The cytotoxic effects of the C. albicans metabolite mixture on cell lines were evaluated using the CCK-8 assay to measure cell viability. The results demonstrated a dose-dependent and time-dependent reduction in cell viability. For HT29 cells treated with the metabolite mixture at an OD value of 0.4 for 24 h, cell viability decreased to approximately 53%, confirming significant cytotoxicity. These conditions (OD = 0.4, 24 h) were used for subsequent experiments. Similarly, HCT116 cells exposed to an OD value of 0.4 for 24 h exhibited a viability rate of approximately 54%, further validating these parameters for follow-up assays. For NCM460 cells, a slightly lower OD value of 0.3 with 24-h exposure resulted in a viability rate of approximately 57%, supporting the use of this concentration and duration for additional experiments (Figure 7A).
Compared to the control group, the group treated with the C. albicans metabolite mixture, as well as the LPO-treated group, showed a significant increase in intracellular LPO levels (P < 0.05), as depicted in Figure 7B.
Scratch assay results indicated that the healing rate in the C. albicans metabolite mixture-treated group was significantly lower than that of the control group (P < 0.01). These findings suggest that the C. albicans metabolite mixture effectively inhibits the migration of CRC cells (Figure 7C-F).
No significant changes in ROS levels were observed in CRC cells treated with the C. albicans metabolite mixture compared to the control group, as shown in Figure 7G-I.
Immunofluorescence staining results revealed a notable presence of the C. albicans metabolite mixture in HCT-116, HT-29, and NCM-460 cells. Prominent green fluorescence, accompanied by minimal red fluorescence, indicated a decline in mitochondrial membrane potential during the early stages of apoptosis. Quantitative analysis showed that the mito
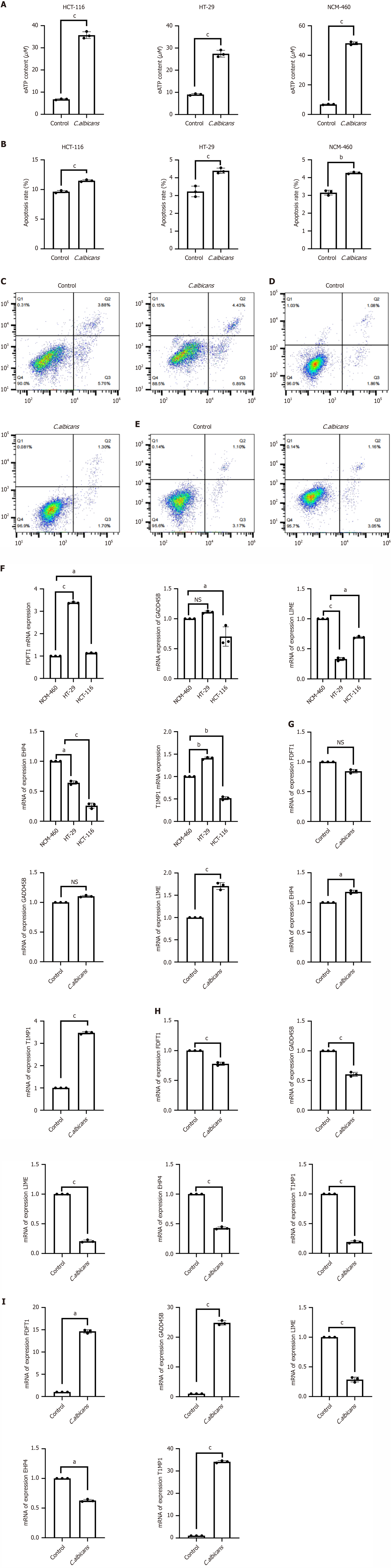
After intervention, eATP levels significantly increased in the experimental group compared to the control group in HCT116, HT29, and NCM460 cells (P < 0.001) (Figure 8A).
Flow cytometry analysis (Figure 8B-E) demonstrated a significantly higher apoptosis rate in the group treated with the C. albicans metabolite mixture compared to the control group (P < 0.001).
RT-qPCR analysis revealed significantly elevated levels of FDFT1 (P < 0.001) in the CRC cell lines HT29 and HCT116 compared to normal colorectal epithelial cells. TIMP1 expression increased significantly in HT29 cells (P < 0.05), while it decreased in HCT116 cells (P < 0.05). GADD45B levels were significantly reduced in HCT116 cells (P < 0.05), as were EHD4 and LIME1 expression levels (P < 0.05) (Figure 8F). Following treatment with the C. albicans metabolite mixture, FDFT1 expression in HCT116 cells showed no significant change (P > 0.05). However, levels of TIMP1 (P < 0.05), GADD45B (P > 0.05), LIME1 (P < 0.01), and EHD4 (P < 0.05) significantly increased (Figure 8G). In HT29 cells, the intervention significantly decreased expression levels of FDFT1, TIMP1, GADD45B, LIME1, and EHD4 (P < 0.001) (Figure 8H). In NCM460 cells, post-intervention analysis revealed significant increases in FDFT1 (P < 0.05), TIMP1 (P < 0.01), and GADD45B (P < 0.01). In contrast, LIME1 (P < 0.01) and EHD4 (P < 0.01) expression levels significantly decreased (Figure 8I).
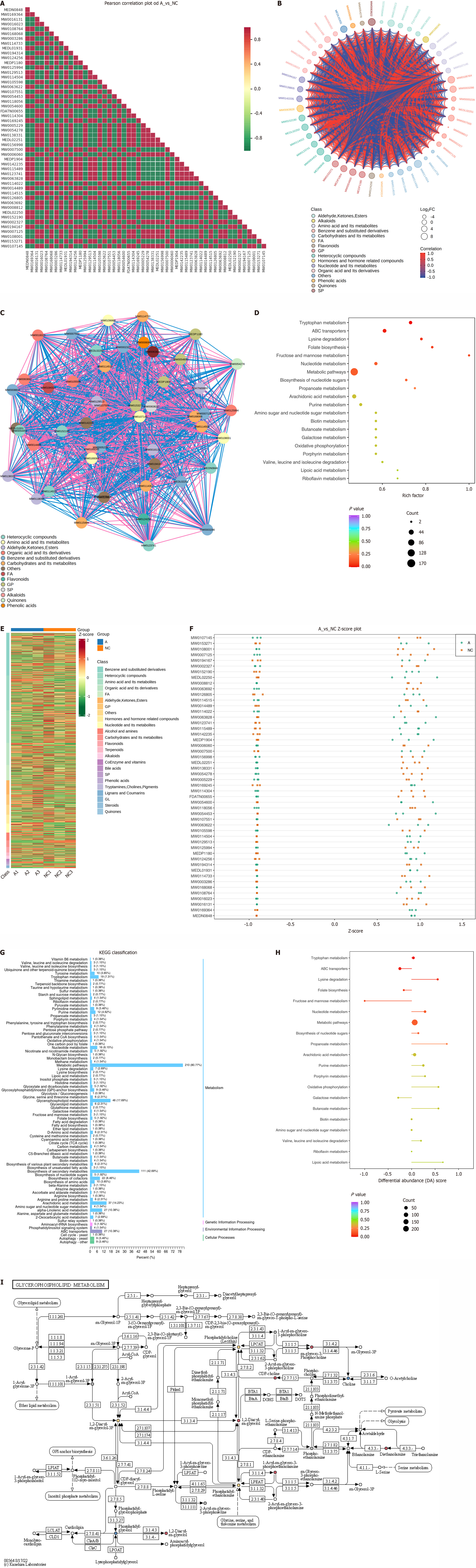
The findings indicated a high degree of overlap in the total ion flow curves, indicating consistent retention times and peak intensities across samples. This consistency demonstrated excellent signal stability when the same sample was analyzed at different intervals (Figure 9A). In the extracted ion chromatogram, all signals were identified as noise peaks, with no discernible signal peaks corresponding to internal standard substances at the expected times (Figure 9B). Pearson correlation analysis of the QC samples showed a strong correlation (|r| approaching 1), reflecting improved stability in the detection process and improved data quality (Figure 9C).
The composition of metabolites varied across samples, with heterocyclic compounds, amino acids and their metabolites, and benzene derivatives being the most abundant classes (Figure 9D). In QC samples, more than 85% of substances exhibited a CV below 0.5, further confirming the stability of the experimental data (Figure 9E). Principal component analysis demonstrated a clear tendency for metabolome separation among groups, with no significant intragroup differences detected within the sample metabolomes (Figure 9F). Cluster heat map analysis was performed for all samples, and the results were visualized using an R script (Figure 9G). Model evaluation using permutation analysis showed that the Q2 value exceeded 0.9, signifying an excellent model. A total of 109 random grouping models were generated, with the optimal model achieving a superior interpretation rate of the Y matrix compared to the OPLS-DA model (P < 0.05) (Figure 9J).
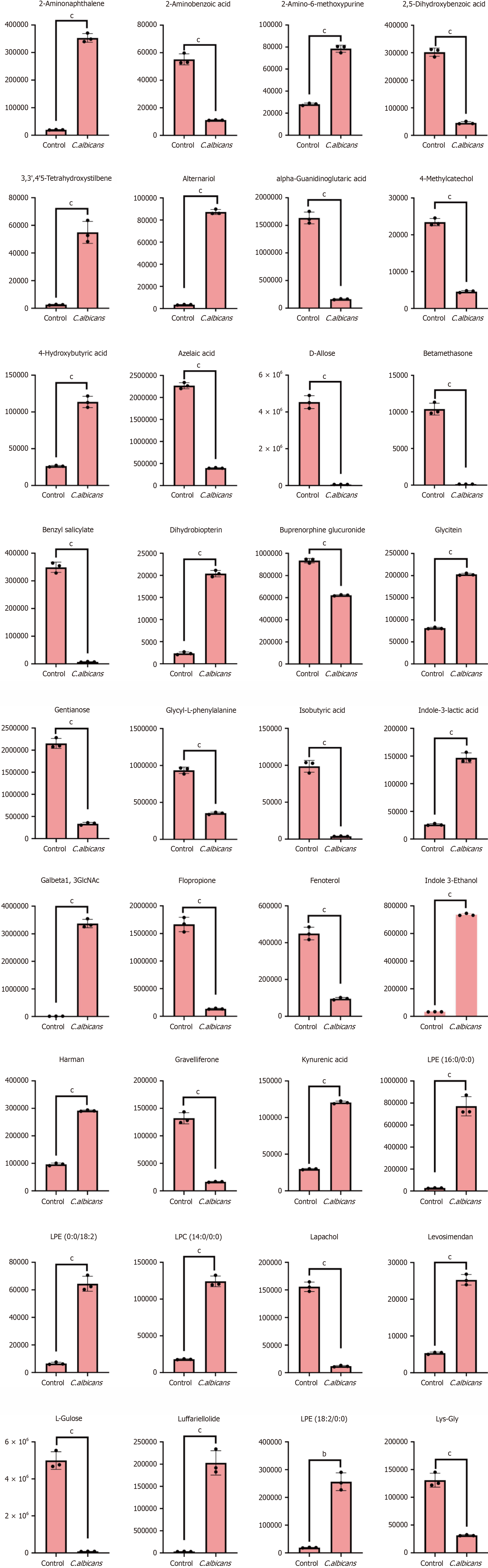
The S-plot for OPLS-DA highlighted the relationship between metabolites and principal components (Figure 9H). The horizontal axis represents the covariance between principal components and metabolites, while the vertical axis indicates the correlation coefficient between these components and the metabolites. Metabolites closer to the upper right and lower left corners exhibit greater significance in their differences. Green dots signify metabolites with a VIP value ≤ 1. To enhance clarity and provide an intuitive representation of metabolic differences, metabolites in the comparison group were evaluated based on their fold change values. The dynamic distribution of metabolite differences was plotted in ascending order of fold change values, with the top 10 upregulated and downregulated metabolites labeled for emphasis (Figure 9I). The volcano plot depicts the distribution of differential metabolites, showing 516 upregulated and 531 downregulated metabolites. A detailed table of metabolites is provided in Annex 1 (Figure 9K). The scatter diagram visualizes the relative differences in content across various categories of metabolites between the two groups (Figure 9L). Differential metabolites for each comparison group are clearly presented. Hierarchical cluster analysis was performed on samples from different comparison groups to generate a cluster tree, illustrating the similarity between samples. Samples within the same cluster exhibit greater similarity (Figure 9M).
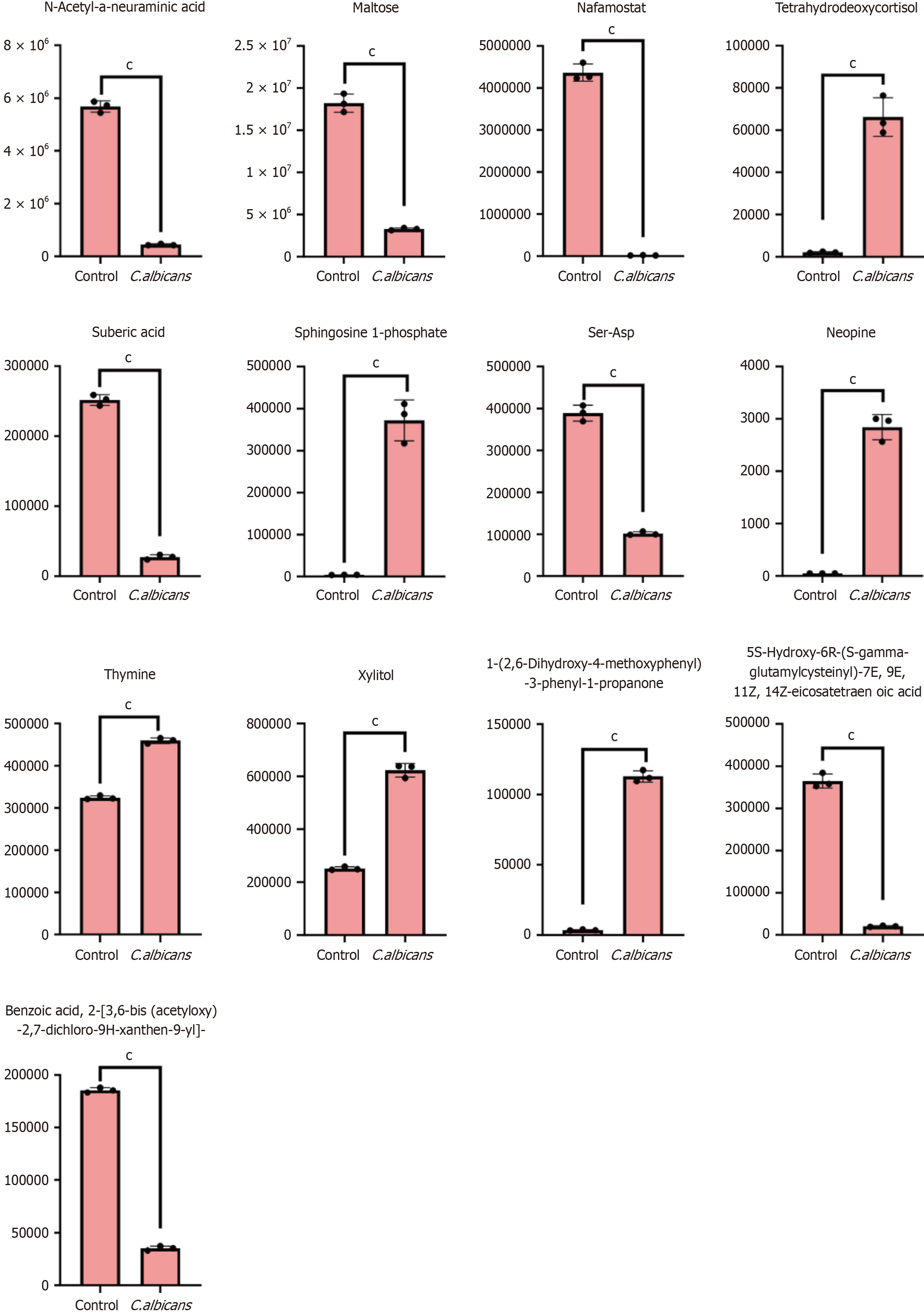
The Pearson correlation method was applied to analyze the relationships between differential metabolites identified based on the screening criteria. The results are visualized in Figure 10A. Figure 10B presents the correlation network of the differential metabolites, where red lines represent positive correlations and blue lines indicate negative correlations. The thickness of the lines corresponds to the absolute value of the Pearson correlation coefficient (|r|), with thicker lines signifying higher |r| values. By default, all differential metabolites are mapped. When the number of differential metabolites exceeds 50, the top 50 metabolites with the highest variable importance in projection values are displayed (Figure 10C). The core metabolites identified were glycyl-L-phenylalanine, lysophosphatidylethanolamine (0:0/18:2), and sphingosine 1-phosphate.
KEGG analysis revealed that the main pathways enriched involved metabolic processes (Figure 10D). Differential metabolite clustering heat maps indicated that Glycerol(GL), steroids, quinones, lignans, and coumarins were dominant categories (Figure 10E). The Z-value chart provided a visual representation of the distribution of differential metabolites across different groups (Figure 10F). Pathway analysis of the differential metabolites aligned with KEGG results, showing significant enrichment in metabolic pathways (Figure 10G and H). Metabolite interactions within organisms were mapped to form distinct pathways, annotated using the KEGG database (Figure 10I). In this diagram, red circles re
EHD4, encoded by the Eps15 Homology Domain 4 gene, represents one of the four EHD proteins identified in mammals (EHD1 to EHD4), which share an amino acid sequence identity ranging from 70% to 86%[7,8]. These findings suggest that EHD4 may serve as an independent prognostic marker for CRC[9]. Additionally, the roles of miR-4701-3p and miR-4793-3p in inducing apoptosis in CRC cells were explored. Using MirTarBase, 62 shared targets of these microRNAs were identified, including SMARCA5, MBD3, VPS53, and EHD4[10]. High expression of GADD45B has been identified as an independent prognostic marker, correlating with reduced OS and progression-free survival (PFS) in patients with stage II CRC. This highlights the potential benefit of adjuvant chemotherapy in this subgroup[11].
In a cohort with a pretest probability of 20%, TIMP1 findings yielded post-test probabilities of 56% for positive cases and 9% for negative cases, indicating moderate to high diagnostic accuracy for TIMP1 as a biomarker in CRC screening[12]. A supplementary study involving 43 patients with CRC and 24 healthy participants confirmed significantly elevated serum TIMP1 levels in patients with CRC. This elevation was associated with factors such as female sex, tumor localization in the colon, dedifferentiation, and higher platelet counts[13]. Moreover, increased TIMP1 expression was observed in CRC tumor tissues and lymph node metastases, suggesting its involvement in CRC progression[14].
The downregulation of FDFT1 is strongly linked to advanced malignancy and poor outcomes in CRC, establishing it as a critical tumor suppressor. Mechanistically, FDFT1 exerts tumor-suppressive effects by interfering with the AKT/mTOR/HIF1α signaling pathway[15]. Additionally, widespread downregulation of FDFT1 in CRC tissues has been documented across multiple studies[16,17]. Conversely, the role of LIME1 in CRC remains largely unexplored.
A prognostic model incorporating four key genes, CXCL8, IL13RA2, MELK, and POP1, has been developed to improve survival predictions in patients with CRC. This model offers potential insights into CRC therapies related to pyroptosis[18]. Another prognostic model, based on five genes (RPX, CXCL13, MMP-10, FABP4, CLDN23), provides an alternative strategy for CRC prognostication[19]. β-glucan, derived from the C. albicans cell membrane, demonstrated dose-dependent reductions in mesenchymal stem cell viability within 48 h, inducing apoptosis in cancer cells[20]. In oral squamous cell carcinoma, C. albicans was shown to promote the proliferation of WSU-HN4, WSU-HN6, and CAL27 cell lines via the TLR2/MyD88 signaling pathway, as confirmed by CCK-8 assays. Furthermore, oral squamous cell carcinoma cells exposed to zymosan exhibited significantly higher C. albicans densities compared to the control group. Additionally, C. albicans β-glucan was found to physically block the antifungal activity of Sanguisorba officinalis derivatives, such as sodium sanguinarine. Exposure to β-glucan significantly enhanced the phagocytic capacity of C. albicans while simultaneously inhibiting its growth[21].
KEGG pathway enrichment analysis revealed that the identified genes are primarily associated with ascorbate and aldarate metabolism, the formation of DNA adducts linked to chemical carcinogenesis, and various other signaling pathways. Chemical carcinogenesis is a critical mechanism of cancer development, wherein exposure to specific chemicals in vivo or in vitro leads to the formation of DNA adducts. These adducts interfere with DNA replication and repair, resulting in accumulated DNA damage and erroneous repair processes. This disruption promotes genetic mutations that impair growth regulation, apoptosis, and cellular repair mechanisms, ultimately driving cancer initiation and progression[22].
In human tissues, a positive correlation exists between higher levels of DNA adducts in blood cells or target organs and an increased cancer risk. This risk elevation remains consistent regardless of whether DNA adducts are measured in blood cell DNA or in target tissue DNA. Typically, individuals with the highest DNA adduct levels exhibit a cancer risk increase of less than tenfold. However, the presence of an additional significant risk factor can substantially amplify this risk[23]. Further studies indicate that DNA adduct levels in CRC tissues are significantly higher than in control samples. This increase is accompanied by upregulated pathways involved in ascorbic acid and uronate metabolism and fatty acid degradation, which may contribute to enhanced regulatory T cell-mediated immunosuppression[24]. The metabolism of alpha-linolenic, linoleic, and arachidonic acids also appears to suppress the proinflammatory functions of CD4+ TCM and CD8+ TEM cells in conditions such as psoriasis and psoriatic arthritis. This suppression facilitates regulatory T cell-mediated immunosuppression in psoriatic arthritis[25]. These findings align with the immunoinfiltration patterns observed in the present study.
C. albicans, a commonly encountered fungal pathogen in humans, typically colonizes the skin and mucosal surfaces of most healthy individuals[26]. Previous studies have demonstrated that the upregulation of FDFT1 inhibited the proliferative, migratory, and invasive capacities of 786-O cells, while its inhibition reduced cellular movement[27,28]. In HT29 and HCT116 CRC cell lines, FDFT1 expression was significantly elevated, which may enhance its invasive and migratory abilities. However, following metabolite intervention, FDFT1 levels markedly decreased in HT29 and HCT116 cells while significantly increasing in NCM460 cells. This change is likely associated with a reduction in invasive and migratory potential, as supported by related studies[29].
In HT29 cells, TIMP1 expression increased significantly, whereas in HCT116 cells, its expression was notably lower compared to NCM460 cells. TIMP1 is typically associated with the inhibition of tissue metalloproteinases, and its upregulation is generally linked to reduced cell invasion and migration. Conversely, TIMP1 downregulation may promote these processes[17]. However, earlier research has shown that increased TIMP1 expression promotes the in vivo growth of HCT116 and HT29 human CRC cells[30]. Moreover, reduced TIMP1 expression has been demonstrated to impede the proliferation, migration, and invasion of colon cancer cells. Despite these findings, further investigation is necessary to clarify the relationship between TIMP1 overexpression and the migratory and invasive characteristics of HCT116 and HT29 cells.
In HCT116 cells, GADD45B expression was significantly lower, while HT29 cells showed a slight increase in its levels. GADD45B plays a critical role in the DNA damage response and cell cycle regulation. In HT29 cells, reduced GADD45B expression may suppress invasion and migration, whereas its upregulation in HCT116 cells appears to promote these processes. Following intervention with C. albicans, GADD45B expression decreased in HT29 cells, suggesting a potential inhibitory effect on invasion and migration, while little change was observed in HCT116 cells. Overexpression of GADD45B serves as an independent prognostic marker, correlating with poorer survival outcomes, including reduced PFS and OS in patients with epithelial ovarian cancer. Higher levels of GADD45B are associated with venous infiltration, lymphatic invasion, and peritoneal metastasis. Conversely, lower GADD45B expression reduces motility in endometrial stromal sarcoma and SKOV3 cell lines. KEGG and GSEA analyses suggested that EMT is a potential downstream pathway influenced by GADD45B. Downregulation of GADD45B has been linked to increased E-cadherin levels and reduced expression of N-cadherin and vimentin, indicating a shift away from EMT-associated characteristics[14,31].
Regarding EHD4 and LIME1, expression levels were significantly reduced in both HCT116 and HT29 cells. However, following intervention, EHD4 and LIME1 expression notably increased in HCT116 cells while remaining low in HT29 cells. Elevated expression of EHD4 and LIME1 may inhibit invasion and migration in HCT116 cells, whereas reduced levels may limit these activities in HT29 cells. While these alterations in gene expression influence the invasiveness and motility of CRC cells, further mechanistic investigations and experimental validations are needed to clarify the precise underlying pathways.
Following the intervention, the experimental group displayed significantly higher levels of eATP compared to the control group. Elevated eATP concentrations can affect cancer cells through various direct and indirect mechanisms. Notably, purinergic receptors are often overexpressed in tumor cells. In CRC cells, purinergic receptors such as A2B, P2X4, P2Y1, P2Y2, and P2Y11 are predominantly expressed, with substantial upregulation of P2Y1 and P2Y2 observed across all CRC cell lines compared to normal colon cells (HCEC-1CT). Furthermore, eATP has been shown to induce cell death in CRC more effectively than adenosine[32]. These findings suggest that metabolites produced by C. albicans may enhance CRC cell death and suppress proliferation by promoting the release of eATP.
Previous studies indicated that elevated ATP concentrations (1–10 mmol/L) and its analogs, such as AMP-PNP and ATPγS, exert a PKC inhibitory effect in Caco-2 cells similar to that of GF109203X, thereby hindering cell proliferation during the S phase[33]. Further research highlights the role of STC2 in modulating PKC activity, influencing claudin-1 gene expression, and enhancing EMT-associated factors, including ZEB1, ZO-1, Slug, Twist, and MMP-9. A reduction in STC2 levels decreases cell motility; however, treatment with the PKC inhibitor Go 6983 restores motility in cells with silenced STC2. Conversely, 231 HM cells demonstrated limited migration and invasion capabilities[34].
In addition, ATP has been shown to reduce the expression of P2Y11 purinergic receptors and CXCR4, thereby inhibiting breast cancer cell migration and the development of bone metastases[35]. By inducing CRC cell death and arresting proliferation during the S phase, elevated eATP levels may effectively limit CRC cell invasion and migration. This effect could be mediated by metabolites derived from C. albicans. Recent evidence has linked ATP-induced cell death to a distinct form of cellular demise increasingly associated with cancer progression[36]. We propose that C. albicans metabolites may regulate CRC invasion, migration, and progression through ATP-induced cell death, warranting further investigation into this mechanism.
The reference strains C. albicans, C. parapsilosis, C. glabrata (Cp2), and C. krusei were observed to cleave TIMP1 into fragments, reducing its ability to inhibit MMP-9 collagenase. This activity suggests that these fungal strains could influence tissue inflammation by altering the balance of host MMP-9 and its inhibitors[37]. In a fungal keratitis model, corneas inoculated with C. albicans exhibited infections within the first day, with an average clinical score of 8.2 ± 0.8. Microarray analyses revealed that MMP-8, -9, -10, -12, -13, -19, and TIMP1 were upregulated between 5- to 375-fold on day one, while RT-PCR showed increases ranging from 3- to 78-fold. This upregulation, observed in both the corneal epithelium and stroma, coincided with an acute influx of inflammatory cells. Interestingly, mechanical injury alone did not affect the expression of MMP-8 and MMP-13, which increased by more than 100-fold within 1 week following the onset of fungal keratitis[38].
FDFT1 functions as a critical tumor suppressor in CRC by downregulating the AKT/mTOR/HIF1α signaling pathway, thereby exerting tumor-suppressive effects[15]. The combination of mTOR inhibitors with fasting has demonstrated a synergistic effect, significantly inhibiting CRC cell proliferation. Additionally, FDFT1 exhibits anti-tumor properties in CRC, potentially through the regulation of ISCU expression to promote iron efflux[16]. The administration of 3β-hydroxy-12-oleanene-27-acid has been shown to significantly reduce the growth of xenograft tumors in nude mice. This treatment decreases FDFT1 expression in tumor tissues and impacts biomarkers associated with autophagy, cell cycle regulation, apoptosis, and iron efflux[39].
In patients with CRC, serum TIMP1 levels were significantly higher than in healthy controls. Elevated TIMP1 concentrations were associated with female gender, tumor localization in the colon, low tumor differentiation, and increased platelet counts in whole blood[13]. Furthermore, plasma levels of both TIMP1 and MMP-9 were significantly higher in patients with CRC compared to control groups. Among patients with metastatic disease, plasma TIMP1 levels were elevated in both systemic and portal circulation compared to those with localized disease[40].
Adjuvant chemotherapy is associated with extended PFS compared to patients who do not receive this treatment. Elevated expression of GADD45B serves as an independent prognostic indicator for decreased OS and PFS in stage II CRC. This suggests that patients with stage II CRC with high GADD45B expression may particularly benefit from adjuvant chemotherapy[41]. Levels of GADD45B mRNA and protein are significantly higher in CRC tissues compared to adjacent non-cancerous tissues, with this upregulation correlating with recurrence and mortality among patients with CRC. Kaplan-Meier survival analysis demonstrated that GADD45B overexpression is linked to significantly reduced disease-free survival.
Additionally, Cox multivariate analysis identified GADD45B overexpression and TNM stage as critical factors influencing patient survival. Despite its association with poor prognosis, GADD45B also functions as a tumor suppressor gene; in normal colorectal tissue, it can induce apoptosis in CRC cell lines, potentially via the p53-mediated apoptotic pathway[11].
EHD2 exhibits a strong correlation with various clinicopathological parameters and is associated with improved OS. Both univariate and multivariate analyses indicated that EHD2 acts as an independent prognostic factor[9]. Further investigation into EHD4 is needed to understand its mechanisms, functions, and potential clinical applications.
FDFT1 plays a key role in regulating cholesterol composition in cell membranes and coordinating immune cell functions. One of its interacting proteins may act as a mediator in pathways related to immune evasion[29]. Aberrant FDFT1 expression has been observed in tumor-infiltrating lymphocytes[16].
TIMP1, a secretory protein that inhibits MMPs, is heavily involved in inflammatory processes and is linked to increased immune cell infiltration[41]. In anorectal cancer, activation of the IL-6-JAK2-STAT3-TIMP1 signaling pathway aligns with adaptive immune responses mediated by Th1 and Th17 cells. IL-6 promotes TIMP1 production in M1 macrophages via phosphorylation of JAK2 and STAT3, subsequently increasing the expression of Th1 and Th17 markers, such as IL-17 and IFN-γ[42,43].
The suppression of Gadd45a and Gadd45b is linked to impaired macrophage chemotactic responses to lipopolysaccharide. Mice lacking Gadd45b (Gadd45b-/-) exhibit weakened tumor immune surveillance, characterized by reduced levels of IFN-γ, granzyme B, and CCR5 in tumor-infiltrating CD8+ T cells. When stimulated with TCR or IL-12/IL-18, these cells show decreased activation of p38 MAP kinase, though ERK and JNK activation remain unaffected, resulting in reduced IFN-γ production. Additionally, lower mRNA levels of T-BET and Eomes in Gadd45b-/- CD8+ T cells highlight the critical role of Gadd45b in Th1 lineage differentiation[44,45].
In knockout models for EHD1, EHD3, and EHD4, CD4+ T cells proliferate in response to antigens but exhibit decreased secretion of IL-2. These knockout mice also display milder symptoms of experimental autoimmune encephalomyelitis, a condition associated with reduced recycling of the TCR-CD3 complex. This leads to increased lysosomal targeting and lower surface expression of CD4+ T cells[46]. Collectively, these findings suggest that metabolite mixtures derived from C. albicans may influence the onset and progression of CRC by modulating immune cell infiltration dynamics.
Moderate ROS levels contribute to cellular damage, DNA mutations, and inflammation, fostering cancer initiation and progression. Conversely, excessive ROS levels induce cancer cell death, exerting anti-cancer effects. In CRC cells, ROS levels are generally elevated compared to normal cells, creating a therapeutic opportunity to selectively induce cancer cell death while sparing healthy tissue[47]. The ferroptosis inducer RSL3 triggers CRC cell death by increasing ROS levels in HCT116, LoVo, and HT29 cells within 24 h. Treatment with RSL3 significantly raises ROS levels and transferrin ex
Both STAT5a and STAT5b play key roles in CRC cell growth by regulating cell cycle progression and apoptosis through downstream STAT signaling pathways. STAT5b, in particular, has a more pronounced effect on CRC apoptosis compared to STAT5a (P < 0.05), as it reduces mitochondrial membrane potential and increases ROS production[49]. Additionally, withaferin A has been shown to induce apoptosis in human CRC cells through ROS-dependent mito
Kynurenic acid, a metabolite derived from tryptophan, has been shown to inhibit the proliferation of various cancer cell lines, including CRC, renal carcinoma, and glioblastoma. This inhibition is mediated by a reduction in the phosphorylation of AKT, ERK1/2, and p38 kinases, particularly in HT-29 cells[51]. Betamethasone, traditionally utilized as an antipsychotic, has recently gained attention for its efficacy in alleviating refractory nausea and vomiting in patients undergoing chemotherapy[52]. Similarly, D-allose, a rare sugar and C-3 epimer of D-glucose, promotes the expression of the tumor suppressor gene TXNIP, leading to G1 cell cycle arrest and inhibiting cancer cell growth[53]. Glycitein also exhibits therapeutic potential against colon cancer[54].
Sphingosine-1-phosphate (S1P), a bioactive lipid derived from mammalian sphingolipids, plays a key role in sustained STAT3 activation in inflammation-associated CRC epithelial and tumor cells. Elevated IL-6 levels stimulate STAT3-mediated activation of sphingosine kinases (SphK1 and SphK2), resulting in increased S1P production. S1P, in turn, activates STAT3 through a positive autocrine signaling loop. The crosstalk among IL-6, STAT3, and the sphingolipid pathway is a critical component of inflammation-driven CRC tumorigenesis and progression[55].
Levosimendan, initially approved for heart failure treatment as a calcium sensitizer, has demonstrated potential in oncology due to its pharmacological effects, including phosphodiesterase-3 inhibition, nitric oxide production, and ROS reduction. Studies suggest that levosimendan can inhibit cancer cell migration and improve radiosensitivity in hypoxic cells[56].
Xylitol, widely recognized for its anti-caries properties, has shown dose-dependent anti-proliferative effects on cancer cell lines such as A549, Caki, NCI-H23, HCT-15, HL-60, K562, and SK MEL-2. Notably, the IC50 value of xylitol in human gingival fibroblasts is higher than that in cancer cells, indicating selective toxicity toward cancer cells. Additionally, xylitol-induced autophagy in A549 cells can be blocked by the autophagy inhibitor 3-methyladenine[57].
Thymine DNA glycosylase (TDG) enhances Wnt signaling through its interaction with TCF4 and the coactivator CREB-binding protein/p300. Both in vitro and in vivo studies demonstrate that TDG knockdown via shRNA effectively reduces CRC cell proliferation. Tumor tissues from patients with CRC exhibit significantly higher TDG levels compared to adjacent normal tissues[58]. Collectively, these findings highlight the therapeutic potential of these metabolites and agents as biomarkers and treatment targets in CRC.
This study successfully established and validated a prognostic model linking C. albicans with CRC using CRC datasets. This model provided a foundation for further in vitro and in vivo studies to explore these genes as potential therapeutic targets for CRC. In vitro experiments using the CCK8 assay demonstrated that C. albicans metabolite mixtures significantly reduced CRC cell viability in a dose-dependent and time-dependent manner. Furthermore, these metabolites effectively inhibited CRC cell invasion and migration. Treatment with the C. albicans metabolite mixture led to a notable elevation in eATP levels in CRC cells compared to the control group. Interestingly, while the metabolic mixture did not alter ROS accumulation in CRC cells, it caused a reduction in mitochondrial membrane potential, an increase in intracellular LPO content, and enhanced apoptosis rates.
Metabolomic analysis revealed the upregulation of 516 metabolites and the downregulation of 531 metabolites in response to the C. albicans metabolic mixture. This study highlighted the dual role of C. albicans in CRC progression: While it promotes CRC development, its metabolic mixture exhibits inhibitory effects on tumorigenesis and holds therapeutic potential. Moreover, the findings offer fresh insights into the mechanisms by which C. albicans contributes to CRC onset, highlighting the intricate relationship between microbes and cancer. The detailed metabolite profiling opens new avenues for personalized treatment strategies in CRC.
| 1. | Lopes JP, Lionakis MS. Pathogenesis and virulence of Candida albicans. Virulence. 2022;13:89-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 222] [Article Influence: 74.0] [Reference Citation Analysis (0)] |
| 2. | Tepus M, Yau TO. Non-Invasive Colorectal Cancer Screening: An Overview. Gastrointest Tumors. 2020;7:62-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 3. | Starý L, Mezerová K, Vysloužil K, Zbořil P, Skalický P, Stašek M, Raclavský V. Candida albicans culture from a rectal swab can be associated with newly diagnosed colorectal cancer. Folia Microbiol (Praha). 2020;65:989-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 4. | Zhu Y, Shi T, Lu X, Xu Z, Qu J, Zhang Z, Shi G, Shen S, Hou Y, Chen Y, Wang T. Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J. 2021;40:e105320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 94] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 5. | Wang Y, Ren Y, Huang Y, Yu X, Yang Y, Wang D, Shi L, Tao K, Wang G, Wu K. Fungal dysbiosis of the gut microbiota is associated with colorectal cancer in Chinese patients. Am J Transl Res. 2021;13:11287-11301. [PubMed] |
| 6. | Shams K, Larypoor M, Salimian J. The immunomodulatory effects of Candida albicans isolated from the normal gastrointestinal microbiome of the elderly on colorectal cancer. Med Oncol. 2021;38:140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Jones T, Naslavsky N, Caplan S. Eps15 Homology Domain Protein 4 (EHD4) is required for Eps15 Homology Domain Protein 1 (EHD1)-mediated endosomal recruitment and fission. PLoS One. 2020;15:e0239657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Lin YS, Huang KY, Yu HC, Lu MC, Fan CJ, Huang Tseng HY, Jhuang BY, Liu SQ, Lai NS, Lin TH, Huang HB. Identification of phostensin in association with Eps 15 homology domain-containing protein 1 (EHD1) and EHD4. Biochem Biophys Res Commun. 2020;531:236-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 9. | Guan C, Lu C, Xiao M, Chen W. EHD2 Overexpression Suppresses the Proliferation, Migration, and Invasion in Human Colon Cancer. Cancer Invest. 2021;39:297-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 10. | Wang H, Ma Y, Lin Y, Chen R, Xu B, Deng J. SHU00238 Promotes Colorectal Cancer Cell Apoptosis Through miR-4701-3p and miR-4793-3p. Front Genet. 2019;10:1320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Wang L, Xiao X, Li D, Chi Y, Wei P, Wang Y, Ni S, Tan C, Zhou X, Du X. Abnormal expression of GADD45B in human colorectal carcinoma. J Transl Med. 2012;10:215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Meng C, Yin X, Liu J, Tang K, Tang H, Liao J. TIMP-1 is a novel serum biomarker for the diagnosis of colorectal cancer: A meta-analysis. PLoS One. 2018;13:e0207039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 13. | Niewiarowska K, Pryczynicz A, Dymicka-Piekarska V, Gryko M, Cepowicz D, Famulski W, Kemona A, Guzińska-Ustymowicz K. Diagnostic significance of TIMP-1 level in serum and its immunohistochemical expression in colorectal cancer patients. Pol J Pathol. 2014;65:296-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, Wu L, Zhang T, Sun X, Zhong L, Zhou C, Wang Z, Peng Z, Chen J, Wang X. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 212] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 15. | Weng ML, Chen WK, Chen XY, Lu H, Sun ZR, Yu Q, Sun PF, Xu YJ, Zhu MM, Jiang N, Zhang J, Zhang JP, Song YL, Ma D, Zhang XP, Miao CH. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11:1869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 174] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 16. | Ma YS, Wu ZJ, Zhang HW, Cai B, Huang T, Long HD, Xu H, Zhao YZ, Yin YZ, Xue SB, Li L, Liu CL, Xie RT, Tian LL, Liu JB, Wu XM, Fu D. Dual Regulatory Mechanisms of Expression and Mutation Involving Metabolism-Related Genes FDFT1 and UQCR5 during CLM. Mol Ther Oncolytics. 2019;14:172-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Jiang H, Tang E, Chen Y, Liu H, Zhao Y, Lin M, He L. Squalene synthase predicts poor prognosis in stage I-III colon adenocarcinoma and synergizes squalene epoxidase to promote tumor progression. Cancer Sci. 2022;113:971-985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Li R, Zhang S, Liu G. Identification and validation of a pyroptosis-related prognostic model for colorectal cancer. Funct Integr Genomics. 2022;23:21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 19. | Hong J, Lin X, Hu X, Wu X, Fang W. A Five-gene Signature for Predicting the Prognosis of Colorectal Cancer. Curr Gene Ther. 2021;21:280-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Peymaeei F, Sadeghi F, Safari E, Khorrami S, Falahati M, Roudbar Mohammadi S, Roudbary M. Candida albicans Beta-Glucan Induce Anti- Cancer Activity of Mesenchymal Stem Cells against Lung Cancer Cell Line: An In-Vitro Experimental Study. Asian Pac J Cancer Prev. 2020;21:837-843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Chen X, Luo Q, Ding J, Yang M, Zhang R, Chen F. Zymosan promotes proliferation, Candida albicans adhesion and IL-1β production of oral squamous cell carcinoma in vitro. Infect Agent Cancer. 2020;15:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Garner RC. The role of DNA adducts in chemical carcinogenesis. Mutat Res. 1998;402:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 74] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 23. | Poirier MC. Linking DNA adduct formation and human cancer risk in chemical carcinogenesis. Environ Mol Mutagen. 2016;57:499-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 24. | Al-Saleh I, Arif J, El-Doush I, Al-Sanea N, Jabbar AA, Billedo G, Shinwari N, Mashhour A, Mohamed G. Carcinogen DNA adducts and the risk of colon cancer: case-control study. Biomarkers. 2008;13:201-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Peng L, Chen L, Wan J, Liu W, Lou S, Shen Z. Single-cell transcriptomic landscape of immunometabolism reveals intervention candidates of ascorbate and aldarate metabolism, fatty-acid degradation and PUFA metabolism of T-cell subsets in healthy controls, psoriasis and psoriatic arthritis. Front Immunol. 2023;14:1179877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 26. | Wang Q, Wang Z, Xu C, Wu D, Wang T, Wang C, Shao J. Physical impediment to sodium houttuyfonate conversely reinforces β-glucan exposure stimulated innate immune response to Candida albicans. Med Mycol. 2024;62. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 27. | Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2011;10:112-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 553] [Cited by in RCA: 641] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 28. | Huang R, Zhang C, Wang X, Zou X, Xiang Z, Wang Z, Gui B, Lin T, Hu H. Identification of FDFT1 as a potential biomarker associated with ferroptosis in ccRCC. Cancer Med. 2022;11:3993-4004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 29. | Ha NT, Lee CH. Roles of Farnesyl-Diphosphate Farnesyltransferase 1 in Tumour and Tumour Microenvironments. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 30. | Gong Y, Scott E, Lu R, Xu Y, Oh WK, Yu Q. TIMP-1 promotes accumulation of cancer associated fibroblasts and cancer progression. PLoS One. 2013;8:e77366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 100] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 31. | Gong L, Cai L, Li G, Cai J, Yi X. GADD45B Facilitates Metastasis of Ovarian Cancer Through Epithelial-Mesenchymal Transition. Onco Targets Ther. 2021;14:255-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Dillard C, Borde C, Mohammad A, Puchois V, Jourdren L, Larsen AK, Sabbah M, Maréchal V, Escargueil AE, Pramil E. Expression Pattern of Purinergic Signaling Components in Colorectal Cancer Cells and Differential Cellular Outcomes Induced by Extracellular ATP and Adenosine. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Yaguchi T, Saito M, Yasuda Y, Kanno T, Nakano T, Nishizaki T. Higher concentrations of extracellular ATP suppress proliferation of Caco-2 human colonic cancer cells via an unknown receptor involving PKC inhibition. Cell Physiol Biochem. 2010;26:125-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Hou J, Wang Z, Xu H, Yang L, Yu X, Yang Z, Deng Y, Meng J, Feng Y, Guo X, Yang G. Stanniocalicin 2 suppresses breast cancer cell migration and invasion via the PKC/claudin-1-mediated signaling. PLoS One. 2015;10:e0122179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 35. | Liu X, Riquelme MA, Tian Y, Zhao D, Acosta FM, Gu S, Jiang JX. ATP Inhibits Breast Cancer Migration and Bone Metastasis through Down-Regulation of CXCR4 and Purinergic Receptor P2Y11. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Zhang HL, Sandai D, Zhang ZW, Song ZJ, Babu D, Tabana Y, Dahham SS, Adam Ahmed Adam M, Wang Y, Wang W, Zhang HL, Zhao R, Barakat K, Harun MSR, Shapudin SNM, Lok B. Adenosine triphosphate induced cell death: Mechanisms and implications in cancer biology and therapy. World J Clin Oncol. 2023;14:549-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (1)] |
| 37. | Pärnänen P, Meurman JH, Sorsa T. The effects of Candida proteinases on human proMMP-9, TIMP-1 and TIMP-2. Mycoses. 2011;54:325-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 38. | Yuan X, Mitchell BM, Wilhelmus KR. Expression of matrix metalloproteinases during experimental Candida albicans keratitis. Invest Ophthalmol Vis Sci. 2009;50:737-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 39. | Tu J, Meng X, Wang J, Han Z, Yu Z, Sun H. 3β-Hydroxy-12-oleanen-27-oic Acid Exerts an Antiproliferative Effect on Human Colon Carcinoma HCT116 Cells via Targeting FDFT1. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 40. | Jensen SA, Vainer B, Bartels A, Brünner N, Sørensen JB. Expression of matrix metalloproteinase 9 (MMP-9) and tissue inhibitor of metalloproteinases 1 (TIMP-1) by colorectal cancer cells and adjacent stroma cells--associations with histopathology and patients outcome. Eur J Cancer. 2010;46:3233-3242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Zhao Z, Gao Y, Guan X, Liu Z, Jiang Z, Liu X, Lin H, Yang M, Li C, Yang R, Zou S, Wang X. GADD45B as a Prognostic and Predictive Biomarker in Stage II Colorectal Cancer. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 42. | Allen JR, Ge L, Huang Y, Brauer R, Parimon T, Cassel SL, Sutterwala FS, Chen P. TIMP-1 Promotes the Immune Response in Influenza-Induced Acute Lung Injury. Lung. 2018;196:737-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Ding X, Cao Y, Xing Y, Ge S, Lin M, Li J. TIMP-1 Mediates Inflammatory and Immune Response to IL-6 in Adult Orbital Xanthogranulomatous Disease. Ocul Immunol Inflamm. 2020;28:288-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Salerno DM, Tront JS, Hoffman B, Liebermann DA. Gadd45a and Gadd45b modulate innate immune functions of granulocytes and macrophages by differential regulation of p38 and JNK signaling. J Cell Physiol. 2012;227:3613-3620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 45. | Ju S, Zhu Y, Liu L, Dai S, Li C, Chen E, He Y, Zhang X, Lu B. Gadd45b and Gadd45g are important for anti-tumor immune responses. Eur J Immunol. 2009;39:3010-3018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 46. | Iseka FM, Goetz BT, Mushtaq I, An W, Cypher LR, Bielecki TA, Tom EC, Arya P, Bhattacharyya S, Storck MD, Semerad CL, Talmadge JE, Mosley RL, Band V, Band H. Role of the EHD Family of Endocytic Recycling Regulators for TCR Recycling and T Cell Function. J Immunol. 2018;200:483-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Lin S, Li Y, Zamyatnin AA Jr, Werner J, Bazhin AV. Reactive oxygen species and colorectal cancer. J Cell Physiol. 2018;233:5119-5132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 48. | Sui X, Zhang R, Liu S, Duan T, Zhai L, Zhang M, Han X, Xiang Y, Huang X, Lin H, Xie T. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front Pharmacol. 2018;9:1371. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 490] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 49. | Du W, Wang YC, Hong J, Su WY, Lin YW, Lu R, Xiong H, Fang JY. STAT5 isoforms regulate colorectal cancer cell apoptosis via reduction of mitochondrial membrane potential and generation of reactive oxygen species. J Cell Physiol. 2012;227:2421-2429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 50. | Xia S, Miao Y, Liu S. Withaferin A induces apoptosis by ROS-dependent mitochondrial dysfunction in human colorectal cancer cells. Biochem Biophys Res Commun. 2018;503:2363-2369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 51. | Walczak K, Dąbrowski W, Langner E, Zgrajka W, Piłat J, Kocki T, Rzeski W, Turski WA. Kynurenic acid synthesis and kynurenine aminotransferases expression in colon derived normal and cancer cells. Scand J Gastroenterol. 2011;46:903-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 52. | Suzuki M, Komuro K, Ohara K. Olanzapine and Betamethasone Are Effective for the Treatment of Nausea and Vomiting due to Metastatic Brain Tumors of Rectal Cancer. Case Rep Gastroenterol. 2014;8:13-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 53. | Noguchi C, Kamitori K, Hossain A, Hoshikawa H, Katagi A, Dong Y, Sui L, Tokuda M, Yamaguchi F. D-Allose Inhibits Cancer Cell Growth by Reducing GLUT1 Expression. Tohoku J Exp Med. 2016;238:131-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 54. | Xiang T, Jin W. Mechanism of Glycitein in the Treatment of Colon Cancer Based on Network Pharmacology and Molecular Docking. Lifestyle Genom. 2023;16:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 55. | Nguyen AV, Wu YY, Lin EY. STAT3 and sphingosine-1-phosphate in inflammation-associated colorectal cancer. World J Gastroenterol. 2014;20:10279-10287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 56. | Ribeiro E, Vale N. Understanding the Clinical Use of Levosimendan and Perspectives on its Future in Oncology. Biomolecules. 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 57. | Park E, Park MH, Na HS, Chung J. Xylitol induces cell death in lung cancer A549 cells by autophagy. Biotechnol Lett. 2015;37:983-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 58. | Xu X, Yu T, Shi J, Chen X, Zhang W, Lin T, Liu Z, Wang Y, Zeng Z, Wang C, Li M, Liu C. Thymine DNA glycosylase is a positive regulator of Wnt signaling in colorectal cancer. J Biol Chem. 2014;289:8881-8890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |