Published online Feb 24, 2024. doi: 10.5306/wjco.v15.i2.195
Peer-review started: December 15, 2023
First decision: December 22, 2023
Revised: January 5, 2024
Accepted: January 30, 2024
Article in press: January 30, 2024
Published online: February 24, 2024
Processing time: 66 Days and 18.4 Hours
Interferon-gamma (IFN-γ) plays a dual role in cancer; it is both a pro- and an antitumorigenic cytokine, depending on the type of cancer. The deregulation of the IFN-γ canonic pathway is associated with several disorders, including vulnerability to viral infections, inflammation, and cancer progression. In particular, the interplay between lung adenocarcinoma (LUAD) and viral infections appears to exist in association with the deregulation of IFN-γ signaling. In this mini-review, we investigated the status of the IFN-γ signaling pathway and the expression level of its components in LUAD. Interestingly, a reduction in IFNGR1 expression seems to be associated with LUAD progression, affecting defenses against viruses such as severe acute respiratory syndrome coronavirus 2. In addition, alterations in the expression of IFNGR1 may inhibit the antiproliferative action of IFN-γ signaling in LUAD.
Core Tip: IFNGR1 is a transmembrane receptor required for interferon-gamma (IFN-γ) signaling. IFNGR1 expression is deregulated in lung adenocarcinoma, affecting IFN-γ signaling to promote cancer progression and reduce antiviral responses. Thus, the status of IFNGR1 expression may be critical in the detection of this cancer, and the restoration of its homeostasis may help control tumor progression and improve defense against viral infections.
- Citation: Tecalco-Cruz AC, Medina-Abreu KH, Oropeza-Martínez E, Zepeda-Cervantes J, Vázquez-Macías A, Macías-Silva M. Deregulation of interferon-gamma receptor 1 expression and its implications for lung adenocarcinoma progression. World J Clin Oncol 2024; 15(2): 195-207
- URL: https://www.wjgnet.com/2218-4333/full/v15/i2/195.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i2.195
According to data provided by the Global Cancer Observatory and recently published analyses, in 2020, there were more than 9.9 million deaths by cancer worldwide. Lung cancer was the leading cause of death (18%) in both males and females, and lung adenocarcinoma (LUAD) was the histological type with the highest incidence in both men (39%) and women (57%)[1,2]. Changes in lung cancer incidence patterns that reflect the increase in LUAD may be attributed to several risk factors, including cigarette smoking, exposure to environmental pollution, cooking oil fumes, indoor charcoal burning, and nonsmoker exposure[3,4]. Studies of mechanistic insights at the molecular level in LUAD have shown the presence of alterations in the signaling pathways that drive its initiation and progression. The intracellular signaling disruptions collectively contribute to the aggressive phenotype, invasive nature, and metastatic propensity of LUAD[3,4].
The interferon-gamma (IFN-γ) signaling pathway is among the most deregulated signaling pathways in LUAD. IFN-γ is a cytokine that plays a pivotal role in immune responses, the orchestration of leukocyte trafficking, antiviral and antibacterial defense, and the modulation of cellular proliferation and apoptosis[5-8]. IFN-γ signaling is thought to trigger antitumoral activities and has protumorigenic effects depending on the cancer context. Hence, IFN-γ can induce apoptosis in some cellular contexts, whereas in others, IFN-γ can induce the expression of programmed death ligand 1 (PD-L1), favoring its binding to its receptor PD-1 on activated T cells and suppressing their cytotoxic effect[9]. IFN-γ-induced actions occur when it binds to its receptor complex (IFNGRs), promoting the activation of its canonical signaling pathway with the phosphorylation of signal transducer and activator of transcription 1 (STAT1), which acts as a transcription factor to mediate IFN-γ-dependent gene expression; therefore, IFNGR homeostasis is crucial for the signaling of this interferon[5,10,11]. In this mini-review, we focus on describing and analyzing the relevance of IFN-γ receptor 1 (IFNGR1) in LUAD and its implications for IFN-γ signaling and the progression and complication of this cancer type.
IFN-γ signal is transduced through a heterotetrameric receptor complex comprising two IFNGR1 and two IFNGR2. This receptor complex induces antiviral, proapoptotic, and antiproliferation activities via the JAK/STAT1 pathway[12]. Janus kinase (JAK) 1 and JAK2 bind to the intracellular regions of IFNGR1 and IFNGR2, respectively. After IFN-γ is recognized by its receptors, JAKs are activated via transphosphorylation[13-16]. JAKs phosphorylate IFNGR1, generating a docking site for STAT1 proteins. These are also phosphorylated by JAKs, forming P-STAT1 dimers that are translocated to the nucleus to regulate tissue-specific gene expression[17-20].
IFNGR1 and IFNGR2 are central to the signaling of IFN-γ. IFNGR1 recognizes and binds IFN-γ, whereas IFNGR2 interacts mainly with IFNGR1 and promotes intracellular signaling. The interaction between IFN-γ and IFNGR1:IFNGR2 induces JAK2 autophosphorylation, followed by JAK1 transphosphorylation by JAK2[21]. IFNGR1 is phosphorylated in Y440 by activated JAK kinases, generating a docking site for the interaction of STAT1 via its Src-homology 2 domain[18]. STAT1 is phosphorylated on Y701 by JAK2, promoting its homodimerization, and association with the gamma-activated site (GAS) element on the regulatory regions of IFN-γ-regulated genes to modulate their expression[21]. Interferon regulatory factor 1 (IRF-1) is a primary IFN-γ target gene that encodes for a transcription factor that recognizes interferon-sensitive response element elements, modulating the expression of a second cascade of IFN-γ target genes (Figure 1)[13].
IFNGR1 may exhibit moderate expression, while IFNGR2 has lower expression levels and depends on external stimuli for regulation[22]. IFNGRs play a pivotal role in the immune response against viral diseases[23-27]. For instance,
In the context of cancer, tumor cells exhibit variations in the levels of IFNGR1 and IFNGR2. For example, deficiencies in IFNGR1 and IFNGR2 expression can occur in acute myeloid leukemia[36]. The deficiencies, overexpression, and polymorphisms of IFNGR1/2 may collectively impact IFN-γ signaling, affecting immune responses to infectious diseases and the predisposition to cancer[37-39]. In particular, research has indicated that variations in IFNGR1 expression affect the response to IFN-γ in different temporal and spatial contexts[40].
The altered expression and abundance of IFNGRs have been reported in diverse cancer contexts. Interestingly, the University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) dataset indicates significant statistical changes in the expression of IFNGR1 and/or IFNGR2 in the majority of cancer types (20/35; Table 1). The expression of IFNGR1 displays an increase (8 cancers), a decrease (9 cancers), or no change (3 cancers) with respect to normal tissue. Of these, the expression of both IFNGR1/2 receptors is upregulated (8 cancers) or downregulated (2 cancers) compared to normal tissue, but IFNGR1 is downregulated and IFNGR2 is upregulated in 4 cancers. Whereas IFNGR1 can be up- or downregulated, IFNGR2 is mainly upregulated in cancers compared to healthy tissue (15/20 upregulated, 2/20 downregulated, and 3/20 no change). These data suggest that the deregulation of IFNGR1 can be differential to IFNGR2 in a manner dependent on cancer type[41,42].
| Cancer type | IFNGR1 expression | IFNGR2 expression |
| Bladder urothelial carcinoma | No change | Upregulated |
| Breast invasive carcinoma | Downregulated | Upregulated |
| Cholangiocarcinoma | Upregulated | Upregulated |
| Colon adenocarcinoma | Downregulated | Downregulated |
| Esophageal carcinoma | Upregulated | Upregulated |
| Glioblastoma multiforme | Upregulated | Upregulated |
| Head and neck squamous cell carcinoma | Upregulated | Upregulated |
| Kidney chromophobe | Downregulated | Downregulated |
| Kidney renal clear cell carcinoma | Upregulated | Upregulated |
| Kidney renal papillary cell carcinoma | Upregulated | Upregulated |
| Liver hepatocellular carcinoma | Upregulated | Upregulated |
| Lung adenocarcinoma | Downregulated | Upregulated |
| Lung squamous cell carcinoma | Downregulated | Upregulated |
| Pancreatic adenocarcinoma | Downregulated | No change |
| Prostate adenocarcinoma | No change | Upregulated |
| Rectum adenocarcinoma | Downregulated | No change |
| Skin cutaneous melanoma | Downregulated | No change |
| Stomach adenocarcinoma | Upregulated | Upregulated |
| Thyroid carcinoma | No change | Upregulated |
| Uterine corpus endometrial carcinoma | Downregulated | Upregulated |
In addition, some studies have demonstrated the relevance of changes in the expression of IFNGRs in cancer. For example, IFNGR2 upregulation by RUNX1 transcription factor is associated with growth, migration, and invasion, along with a poor prognosis of low-grade glioma[43]. However, principally, the deregulation of IFNGR1 has been reported in some cancer types.
It has been reported that patients with breast cancer exhibiting elevated levels of IFN-γ and/or IFNGR1 may undergo tumor rejection, whereas those with intermediate levels may experience tumor recurrence[44]. Reduced IFNGR1 expression has also been observed in patients with mammary tumors, suggesting a decrease in IFN-γ signaling[45]. In the context of ovarian cancer, patients whose tumors express high levels of IFNGR1 have a significantly better survival rate than those whose tumors have low levels[46]. The loss of this IFN-γ receptor results in a poor prognosis for patients whose cancer is more aggressive, and the benefit of treatment with IFN-γ is reduced or nonexistent[33]. Thus, changes in IFNGR1 expression appear to be particularly strongly related to the progression of specific cancer types.
Interestingly, polymorphisms in the IFNGR1 promoter are correlated with susceptibility to diseases such as leishmaniasis, tuberculosis, leprosy, and hepatitis[47]. Polymorphisms of this receptor are also associated with some cancer types; for example, polymorphisms in IFNGR1 have been associated with gastric cancer[48] and rectal cancer[49]. For example, IFNGR1 rs3799488 polymorphism is associated with a risk of developing rectal cancer[50], whereas the presence of rs2234711 in the IFNGR1 promoter is associated with an increased risk of developing colorectal cancer[51]. Moreover, the risk of developing colon and rectal cancer is associated with polymorphisms in IFN-γ and its receptors, the influence of other genes related to inflammation, the use of nonsteroidal anti-inflammatory drugs, and smoking[50]. Nevertheless, a longer overall survival has been observed in patients with variations in the IFNGR1 promoter region (rs2234711, rs9376267) diagnosed with metastatic colorectal cancer under treatment with bevacizumab-based chemo
Furthermore, the polymorphisms on IFNGR1 have been associated with early stage breast cancer with depression[53], chronic lymphocytic leukemia[54], classic infantile Kaposi’s sarcoma[55,56] and hepatocellular carcinoma[57].
In addition, IFNGR1 protein can be a target of posttranslational modifications[58]. For example, IFNGR1 can be ubiquitinated by STUB1, an E3 ubiquitin-protein ligase that negatively regulates IFNGR1, thereby reducing IFN-γ sensing. STUB1 depletion increases IFNGR1 abundance and enhances IFN-γ response, promoting an IFN-γ–STAT1–IRF1 axis to induce the activation of genes associated with antigen processing and presentation[59]. The abundance of IFNGR1 can also be regulated by palmitoylation, leading to accelerated lysosomal degradation of IFNGR1. The palmitoylated cysteine in IFNGR1 acts as a signal for its interaction with AP3D1, targeting it to lysosomes. Hence, when AP3D1 is downregulated, IFNGR1 levels are increased, and when IFNGR1 palmitoylation is pharmacologically inhibited, IFNGR1 is stabilized. Moreover, high optineurin protein levels have been positively associated with the survival of melanoma patients, but the loss of optineurin facilitates IFNGR1 binding to AP3D1 and increases AP3D1-mediated IFNGR1 lysosomal sorting and degradation[60]. Additionally, MUC1-C is a protein with transmembrane domains that protects epithelial niches; however, its prolonged activation can promote oncogenesis and the epithelial-mesenchymal transition (EMT) of castration-resistant prostate cancer cells. Interestingly, MUC1 associates with expression of IFNGR1, STAT1 and IRF1. Moreover, it has been reported that the downregulation of MUC1 leads to the activation of FBXW—an E3 ubiquitin-protein ligase—for IFNGR1 degradation via the ubiquitin–proteasome system[61]. Phosphorylation is another posttranslational modification that can alter the stability of IFNGR1. For example, glycogen synthase kinase 3 beta (GSK3β) phosphorylates IFNGR1, which protects this receptor from proteasomal degradation by ubiquitin, increasing its stability and promoting IFN-γ signaling and IFN-γ-induced inflammation; therefore, GSK3β has been proposed as an anticancer target[58,62]. Specifically, high levels of phosphorylated GSK3β are detected in LUAD; accordingly, the inactivation of GSK3β may be useful as a pharmacological treatment for this cancer type[63]. Glycosylation is another posttranslational modification of IFNGR1, which has been proposed as a signal necessary for IFN-γ signaling, but also as a signal associated with the stability of the receptor[64].
The deregulation of IFNGR1 can affect IFN-γ-induced molecular mechanisms and cellular responses. Hence, IFN-γ has been reported to induce downregulation of IFNGR1 in myeloid cells, reducing their response to IFN-γ inflammatory stimuli and promoting anti-inflammatory effects. In addition, reduced IFNGR1 expression decreases the antiproliferative effects induced by IFN-γ, suggesting that the downregulation of IFNGR1 may diminish sensitivity to IFN-γ in myeloid and nonlymphoid cells[49]. Moreover, cancer cells can express IFN-γ-induced PD-L1 to bind to its receptor PD-1 on T cells, promoting resistance to the host immune system. Consequently, the downregulation of IFNGR1 may result in reduced PD-L1/CD274 gene expression; this is associated with resistance to treatment against PDL1/PD1 in melanoma and colorectal cancer[9].
IFNGR1 expression is commonly downregulated in human colorectal cancers and mouse intestinal adenoma models. Particularly, colorectal cancer patients have a more prolonged median survival when they express higher IFNGR1 levels, suggesting that IFN-γ signaling is critical for maintaining a tumor-inhibitory microenvironment in the context of colon cancer[60,65]. Genes involved in inflammation, such as Hif1a, and genes encoding matrix metalloproteinases, such as MMP3, MMP7, and MMP9, are inhibited by IFN-γ under normal conditions. The expression of these genes is enhanced by the absence of IFNGR1 in murine models of human familial adenomatous polyposis, which is regarded as a premalignant lesion for colon cancer. In contrast, tumor suppressor genes, such as Cdx2, Cdhr2, and Cdhr5, are negatively regulated by IFNGR1 downregulation, leading to the M1 phenotype in tumor-associated M2 macrophages[66]. Therefore, altered IFNGR1 expression is associated with dysregulated IFN-γ signaling, which may enhance tumor progression. IFN-γ inhibits β-catenin activity and induces apoptosis in colon cancer cells. However, IFNGR1 deficiency affects IFN-γ signaling, increasing the invasiveness of intestinal tumors with the development of anemia in mice, considering that IFN-γ regulates hematopoiesis[49].
Aside from IFNGR1’s function as a transmembrane receptor, it has been discovered that the IFNGR1 receptor can translocate to the nucleus in uterine cancer cells treated with IFN-γ, whereas the IFNGR2 subunit is not endocytosed or transported to the cell nucleus. IFNGR1 does not have a DNA-binding domain, so its association with STAT1 allows it to bind to GAS elements located in the promoter region of IFN-γ-activated genes, such as IRF-1 and indolamine dioxygenase (IDO), increasing their expression[67,68] .Thus, the activity of IFNGR1 as a transmembrane receptor and as a nuclear protein may be affected by its deregulation.
Despite the dysregulation of IFNGR1 in several cancer types, particularly in LUAD, the alterations of this receptor have been related to cancer progression and complications such as viral infections. This suggests the multifunctional implications of IFN-γ signaling in the context of lung cancer, as discussed below.
IFN-γ signaling is central to the gene expression that participates in the remodeling of LUAD’s tumor microenvironment. For instance, in one study, seven IFN-γ response genes (CD74, CSF2RB, PTPN6, MT2A, NMI, LATS2, and PFKP) were identified and proposed as valuable prognostic predictors of LUAD[69]. Nevertheless, it has been demonstrated that IFN-γ can activate distinct signaling pathways dependent on its levels. Hence, low levels of IFN-γ activate the intercellular adhesion molecule-1 (ICAM1)–PI3K–AKT–Notch1 axis, promoting cancer stem-like properties. By contrast, the JAK1–STAT1–caspase pathway is activated by high levels of IFN-γ, inducing apoptosis in lung cancer cells[70]. Furthermore, the activation of the JAK2–STAT1 and PI3K–AKT pathways in LUAD cells treated with IFN-γ has been shown. This is important since an antiproliferative action has been associated with the IFN-γ/JAK2–STAT1 axis, whereas PI3K-AKT signaling is associated with protumor activity. Hence, a reduction in PD-L1 expression was observed in response to PI3K inhibition, enhancing the IFN-γ-dependent antiproliferative signaling. Therefore, the antitumoral activity triggered by IFN-γ may be enhanced by blocking PI3K signaling. Thus, in response to IFN-γ in LUAD, crosstalk between the JAK2–STAT1 and PI3K–AKT pathways is generated, suggesting PI3K as a target for therapeutic treatment of LUAD[71]. Moreover, IFN-γ enhances the expression of JMJD3, a histone demethylase that reduces H3K27 trimethylation on the ZEB1 gene promoter, resulting in ZEB1 transcription. IFN-γ-induced ZEB1 expression is associated with the EMT, migration, and metastasis of LUAD cells in vivo. The reduction of ZEB1 inhibits EMT, migration, and metastasis, but does not affect the expression of STAT1 and IRF1 or the antitumor effects of the IFN-γ–STAT1–IRF1 axis. This study suggests that the negative regulation of ZEB1 may inhibit the protumoral activities of IFN-γ signaling, favoring its antitumoral activities[72].
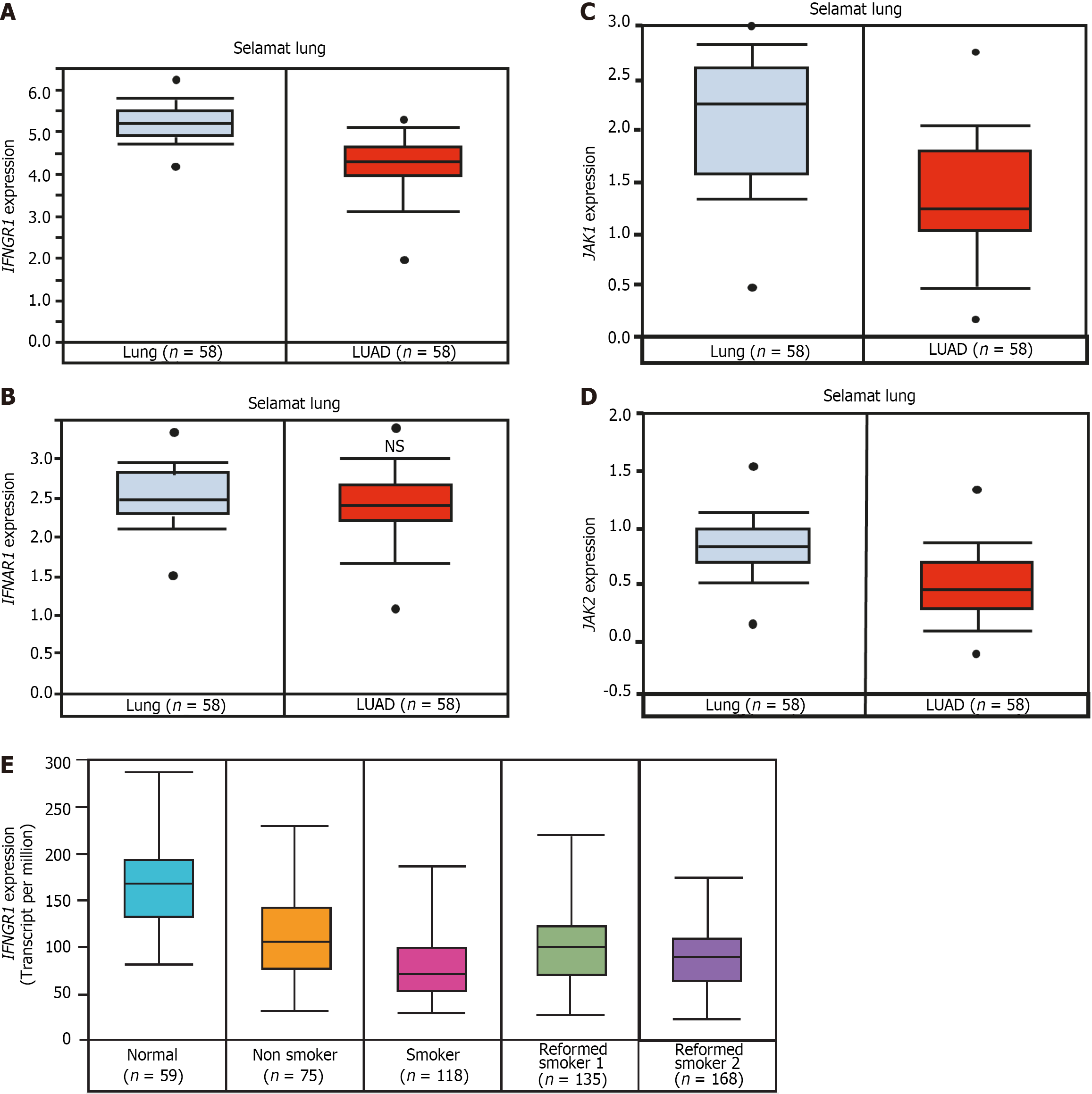
The Selamat dataset from Oncomine, and the Cancer Genome Atlas from UALCAN, indicate that IFNGR1 gene expression is significantly reduced in the tumors of patients with LUAD compared to healthy lung tissue[42] (Figure 2A). By contrast, IFNAR1 (receptor 1 for IFN-α) expression shows no significant changes in LUAD patients compared to healthy lung tissue (Figure 2B). The gene expression of JAK1 and JAK2 is also downregulated in the tumors of patients with LUAD compared to normal lung tissues (Figure 2C and D). These data suggest that the IFN-γ signaling pathway is desensitized in LUAD because of the downregulation of its components—IFNGR1, JAK1, and JAK2—whereas IFN-α/β signaling may remain active.
Interestingly, IFNGR1 expression is significantly lower in smokers’ LUAD tumors than in those of nonsmokers and reformed smokers (Figure 2E), but the expression of JAK1 and JAK2 is similar in the tumors of patients with LUAD who have different smoking habits. These data indicate that the reduction in IFNGR1 in LUAD patients with smoking habits affects IFN-γ signaling.
Emerging evidence supports the idea that the tumor microenvironment affects LUAD progression and clinical outcome[73]. IFN-γ within the tumor microenvironment can positively regulate immune checkpoint molecules, such as IDO1[74]. Moreover, inactivation of IFNGR1 affects a small subset of lung cancer and prevents response to IFNg[75]. All data suggest that IFNGR1 expression may be a useful biomarker for LUAD; nevertheless, additional studies are required to define its prognosis value and its possible cooperative correlation with JAK1/2 expression as a biomarker.
Because IFN-γ is central to immune responses, a higher susceptibility to viral infections and an elevated risk of cancer have been associated with IFNGR1 deficiency[76,77]. IFN-γ signaling deregulation via IFNGR1 downregulation is associated with lung cancer progression and seems to have an influence on susceptibility to viral infections (e.g., infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)], as discussed in the following sections.
The role that IFN-γ signaling plays in viral respiratory infection and lung cancer may depend on the expression status of IFNGR1. For example, a relationship between lung cancer and infections with SARS-CoV-2 seems to exist. SARS-CoV-2 is responsible for coronavirus disease 2019 (COVID-19), which has caused millions of deaths worldwide[78]. COVID-19 symptoms range from those similar to the common cold to more severe manifestations, such as lung injury, damaged alveoli, acute respiratory failure, acute respiratory distress syndrome, and injury to other organs[79]. The SARS-CoV-2 spike (S) glycoprotein can bind to angiotensin-converting enzyme 2 (ACE-2), promoting the virus’s entry into cells. Higher ACE-2 expression translates to higher receptors for SARS-CoV-2, favoring infection. Moreover, the same SARS-CoV-2 virus leads to the upregulation of ACE-2[79]. Some comorbidities, such as chronic obstructive lung disease, diabetes, and hypertension, have also been associated with an increase in the expression of ACE-2[78]. Similarly, patients with lung cancer display high levels of ACE-2[80]; therefore, lung cancer poses a higher risk of SARS-CoV-2 infection and its complications[79]. Researchers have thus proposed that the SARS-CoV-2 receptor ACE-2 is upregulated in lung cancer cells, increasing the risk of COVID-19[81].
ACE-2 is an interferon-stimulated gene[82] with several binding sites for transcription factor STAT in its promoter region[80]. Histone modifiers, such as HAT1, HDAC2, and KDM5B, may regulate ACE-2 expression[78]. Moreover, the deregulation of ACE-2 expression may be associated with a reduction in DNA methylation[81]. IFNs can induce a novel truncated ACE-2 isoform[83]. The mechanism for ACE-2 regulation is critical since it is upregulated in lung cancer patients who have a lower survival rate[84]. Furthermore, LUAD patients are reportedly more susceptible to SARS-CoV-2 infection than lung squamous cell carcinoma patients[85].
In addition, the lung tumor microenvironment may promote the invasion of viruses, thereby increasing the severity of COVID-19[86]. Smokers are at a higher risk of severe complications and have higher mortality rates associated with COVID-19 than nonsmokers[87]. Interestingly, Selamat lung datasets from Oncomine indicate a reduction in IFNGR1 and JAK1/2 but an increase in ACE-2 expression in LUAD tumors compared to healthy counterparts (Figure 3A). IFNGR2 and DNAJA3 (a modulator of IFN-γ signaling) are upregulated, whereas the expression of IFN-γ, which encodes IFN-γ, shows no significant change in either condition. Thus, the reduction of IFNGR1 expression in LUAD affects IFN-γ signaling, whereas the expression of the SARS-CoV-2 receptor ACE-2 increases, which may also exacerbate susceptibility to COVID-19. Nevertheless, the downregulation of IFNGR1 may also facilitate several viral infections in cancer contexts.
Viral infections can also promote a higher incidence of lung cancer. For example, infection with immunosuppressive pathogens, including human immunodeficiency virus, reduces the number of CD4+T cells, which is associated with a higher incidence of lung cancer[88]. Moreover, an association with LUAD has been proposed for patients who have recovered from SARS-CoV-2 infection. Upon infection with SARS-CoV-2, the cellular landscape is primarily inflammatory. Viral RNA may be detected by pattern recognition receptors found in endosomes, such as TLR3 and TLR7[89]. Furthermore, the recognition of SARS-CoV-2 mRNA by the cytosolic receptors, retinoic acid-inducible gene I and melanoma differentiation-associated gene 5, leads to the activation of NF-kB, which, in turn, can translocate to the cell nucleus and trigger the transcription of mRNAs encoding for proinflammatory cytokines, such as IL-1β, IL6, and TNF-α[90,91]. IFN-γ is another essential cytokine with antiviral properties that plays a crucial role during viral infections[92]. However, the inflammatory response is sometimes prolonged, resulting in tissue damage[93]. The production of anti-inflammatory molecules [IL10 and growth factors, e.g., vascular endothelial growth factor (VEGF)] is induced as a means of controlling inflammation and limiting tissue damage; however, these anti-inflammatory factors can promote a tumoral microenvironment[94]. In addition, hypoxia occurs during SARS-CoV-2 infection owing to interstitial and alveolar edema caused by increased permeability in the lung capillaries[95]. Under hypoxic conditions, hypoxia-inducible factor (HIF-1) is activated. HIF-1 comprises two subunits—HIF-1α and HIF-1β. To compensate for these hypoxic conditions, HIF-1a can activate the synthesis of other genes, including hematopoietic hormone, VEGF, enzymes involved in glycolysis, and glucose carrier proteins[96]. Nonetheless, HIF-1α also promotes the vascularization of solid tumors, including lung tumors[97]. The expression status of IFNGR1 in recovered SARS-CoV-2 patients remains to be clarified, which may help elucidate the relationship among IFNGR1, LUAD, and viral infections.
IFN-γ is a cytokine that fulfills a dual function in cancer. In some cancer types, IFN-γ prevents tumor development. However, IFN-γ is also known to promote metastasis, thereby evading the immune system. The mechanisms responsible for this duality remain elusive; however, some molecular mechanisms implicated in LUAD have been proposed, including changes in IFN-γ concentration and the simultaneous crosstalk between pro- and antitumoral pathways activated by IFN-γ. Interestingly, the information contained in public databases indicates that the expression of type I IFN-α/β receptor (IFNRA1) is not significantly affected compared to the expression of IFNGR1, which is lower in the tumors of LUAD patients than in healthy tissues. Furthermore, the expression of JAK1/2 is affected in LUAD. Interestingly, LUAD patients with a smoking habit have lower IFNGR1 expression, indicating that IFN-γ signaling is affected, altering its antiviral, antiproliferative, and proapoptotic actions. These data suggest that the IFNGR1 signaling is altered in lung cancer patients and more affected in LUAD patients who have a smoking habit.
Moreover, IFN-γ is known for its antiviral properties. The entry of SARS-CoV-2 into the cells is favored by the upregulation of ACE-2, whereas IFN-γ signaling pathways and its antiviral activities are reduced in LUAD. This may explain the greater susceptibility to SARS-CoV-2 among lung cancer patients, particularly LUAD patients. The reduction of IFN-γ signaling also implies a decrease in IFN-γ-dependent antitumoral actions, promoting lung cancer progression. Thus, IFN-γ administration or high levels of endogenous IFN-γ may have no effect on lung cancer cells due to a lack of IFNGR1, and downstream pathway components (JAK1/2), limiting its antitumoral and antiviral functions in these cells. These IFN-γ signaling elements may be restored to enhance protection against SARS-CoV-2 and regulate cancer progression.
The reduction of IFNGR1 in LUAD may also influence pathways that increase the risk of SARS-CoV-2 infection or other infections that may be more aggressive in LUAD patients than in healthy individuals (Figure 3B). Interestingly, patients who have contracted COVID-19 may be at higher risk for lung cancer, considering the inflammation conditions during infection. Importantly, IFN-γ, HIF-1, and the simple fact of a cigarette smoking habit increase ACE-2 levels. This is undesirable in the context of SARS-CoV-2 infection, as the upregulation of ACE-2 has been associated with lung cancer[82,84,98]. Further studies on the molecular mechanisms that control the expression and function of IFNGR1 in LUAD and other cancer types are warranted.
In conclusion, the expression of IFNGR1 and JAK1/2 is affected in LUAD. IFNGR1 is the first component in IFN-γ signaling. Consequently, a decrease in IFNGR1 inhibits the antitumoral and antiviral actions of IFN-γ. Therefore, patients with LUAD display lower IFNGR1 levels that promote cancer progression; this seems to be associated with several complications, including a greater risk for infections (such as COVID-19) and, ultimately, poor outcomes. Novel therapies restoring IFNGR1 expression could be used as new approaches for LUAD in personalized medicine.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu TF, China S-Editor: Liu JH L-Editor: A P-Editor: Zhang XD
| 1. | Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021;134:783-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1624] [Cited by in RCA: 1799] [Article Influence: 449.8] [Reference Citation Analysis (1)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64681] [Article Influence: 16170.3] [Reference Citation Analysis (177)] |
| 3. | Zhang Y, Vaccarella S, Morgan E, Li M, Etxeberria J, Chokunonga E, Manraj SS, Kamate B, Omonisi A, Bray F. Global variations in lung cancer incidence by histological subtype in 2020: a population-based study. Lancet Oncol. 2023;24:1206-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 131] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 4. | Burns DM, Anderson CM, Gray N. Do changes in cigarette design influence the rise in adenocarcinoma of the lung? Cancer Causes Control. 2011;22:13-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2208] [Cited by in RCA: 2216] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 6. | Young HA, Hardy KJ. Role of interferon-gamma in immune cell regulation. J Leukoc Biol. 1995;58:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 275] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 7. | Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2690] [Cited by in RCA: 3031] [Article Influence: 137.8] [Reference Citation Analysis (0)] |
| 8. | Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, Thiyagarajan S, Chatterjee A. Comprehensive network map of interferon gamma signaling. J Cell Commun Signal. 2018;12:745-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 9. | Lv C, Yuan D, Cao Y. Downregulation of Interferon-γ Receptor Expression Endows Resistance to Anti-Programmed Death Protein 1 Therapy in Colorectal Cancer. J Pharmacol Exp Ther. 2021;376:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Nishida Y, Maeda Y, Hara A, Arima T, Kimura E, Yamashita S, Uyama E, Mita S, Uchino M. Adenovirus-mediated murine interferon-gamma receptor transfer enhances the efficacy of IFNγ in vivo. Biochem Biophys Res Commun. 2002;290:1042-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res. 2011;17:6118-6124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 494] [Cited by in RCA: 460] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 12. | Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-γ in tumor progression and regression: a review. Biomark Res. 2020;8:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 815] [Cited by in RCA: 767] [Article Influence: 153.4] [Reference Citation Analysis (0)] |
| 13. | Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front Immunol. 2018;9:847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 762] [Cited by in RCA: 876] [Article Influence: 125.1] [Reference Citation Analysis (0)] |
| 14. | Gibbs VC, Williams SR, Gray PW, Schreiber RD, Pennica D, Rice G, Goeddel DV. The extracellular domain of the human interferon gamma receptor interacts with a species-specific signal transducer. Mol Cell Biol. 1991;11:5860-5866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Hemmi S, Merlin G, Aguet M. Functional characterization of a hybrid human-mouse interferon gamma receptor: evidence for species-specific interaction of the extracellular receptor domain with a putative signal transducer. Proc Natl Acad Sci U S A. 1992;89:2737-2741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Hibino Y, Kumar CS, Mariano TM, Lai DH, Pestka S. Chimeric interferon-gamma receptors demonstrate that an accessory factor required for activity interacts with the extracellular domain. J Biol Chem. 1992;267:3741-3749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Kalina U, Ozmen L, Di Padova K, Gentz R, Garotta G. The human gamma interferon receptor accessory factor encoded by chromosome 21 transduces the signal for the induction of 2',5'-oligoadenylate-synthetase, resistance to virus cytopathic effect, and major histocompatibility complex class I antigens. J Virol. 1993;67:1702-1706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Kotenko SV, Izotova LS, Pollack BP, Mariano TM, Donnelly RJ, Muthukumaran G, Cook JR, Garotta G, Silvennoinen O, Ihle JN. Interaction between the components of the interferon gamma receptor complex. J Biol Chem. 1995;270:20915-20921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 129] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Muthukumaran G, Donnelly RJ, Ebensperger C, Mariano TM, Garotta G, Dembic Z, Poast J, Baron S, Pestka S. The intracellular domain of the second chain of the interferon-gamma receptor is interchangeable between species. J Interferon Cytokine Res. 1996;16:1039-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Pestka S, Kotenko SV, Muthukumaran G, Izotova LS, Cook JR, Garotta G. The interferon gamma (IFN-gamma) receptor: a paradigm for the multichain cytokine receptor. Cytokine Growth Factor Rev. 1997;8:189-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 128] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Briscoe J, Rogers NC, Witthuhn BA, Watling D, Harpur AG, Wilks AF, Stark GR, Ihle JN, Kerr IM. Kinase-negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J. 1996;15:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 797] [Cited by in RCA: 798] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 23. | Döffinger R, Jouanguy E, Dupuis S, Fondanèche MC, Stephan JL, Emile JF, Lamhamedi-Cherradi S, Altare F, Pallier A, Barcenas-Morales G, Meinl E, Krause C, Pestka S, Schreiber RD, Novelli F, Casanova JL. Partial interferon-gamma receptor signaling chain deficiency in a patient with bacille Calmette-Guérin and Mycobacterium abscessus infection. J Infect Dis. 2000;181:379-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 131] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 24. | Dorman SE, Holland SM. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J Clin Invest. 1998;101:2364-2369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 313] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 25. | Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova JL. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guérin infection. N Engl J Med. 1996;335:1956-1961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 629] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 26. | Jouanguy E, Lamhamedi-Cherradi S, Altare F, Fondanèche MC, Tuerlinckx D, Blanche S, Emile JF, Gaillard JL, Schreiber R, Levin M, Fischer A, Hivroz C, Casanova JL. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guérin infection and a sibling with clinical tuberculosis. J Clin Invest. 1997;100:2658-2664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 279] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 27. | Pierre-Audigier C, Jouanguy E, Lamhamedi S, Altare F, Rauzier J, Vincent V, Canioni D, Emile JF, Fischer A, Blanche S, Gaillard JL, Casanova JL. Fatal disseminated Mycobacterium smegmatis infection in a child with inherited interferon gamma receptor deficiency. Clin Infect Dis. 1997;24:982-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 28. | Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1238] [Cited by in RCA: 1257] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 29. | Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, Levin M. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941-1949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 859] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 30. | Jouanguy E, Altare F, Lamhamedi-Cherradi S, Casanova JL. Infections in IFNGR-1-deficient children. J Interferon Cytokine Res. 1997;17:583-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 31. | Roesler J, Kofink B, Wendisch J, Heyden S, Paul D, Friedrich W, Casanova JL, Leupold W, Gahr M, Rösen-Wolff A. Listeria monocytogenes and recurrent mycobacterial infections in a child with complete interferon-gamma-receptor (IFNgammaR1) deficiency: mutational analysis and evaluation of therapeutic options. Exp Hematol. 1999;27:1368-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 109] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 32. | Suzuki Y, Orellana MA, Schreiber RD, Remington JS. Interferon-gamma: the major mediator of resistance against Toxoplasma gondii. Science. 1988;240:516-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 886] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 33. | Dighe AS, Richards E, Old LJ, Schreiber RD. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity. 1994;1:447-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 459] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 34. | Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. 1998;95:7556-7561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 1067] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 35. | Tannenbaum CS, Hamilton TA. Immune-inflammatory mechanisms in IFNgamma-mediated anti-tumor activity. Semin Cancer Biol. 2000;10:113-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Khaznadar Z, Boissel N, Agaugué S, Henry G, Cheok M, Vignon M, Geromin D, Cayuela JM, Castaigne S, Pautas C, Raffoux E, Lachuer J, Sigaux F, Preudhomme C, Dombret H, Dulphy N, Toubert A. Defective NK Cells in Acute Myeloid Leukemia Patients at Diagnosis Are Associated with Blast Transcriptional Signatures of Immune Evasion. J Immunol. 2015;195:2580-2590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Jüliger S, Bongartz M, Luty AJ, Kremsner PG, Kun JF. Functional analysis of a promoter variant of the gene encoding the interferon-gamma receptor chain I. Immunogenetics. 2003;54:675-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | Zhou J, Chen DQ, Poon VK, Zeng Y, Ng F, Lu L, Huang JD, Yuen KY, Zheng BJ. A regulatory polymorphism in interferon-gamma receptor 1 promoter is associated with the susceptibility to chronic hepatitis B virus infection. Immunogenetics. 2009;61:423-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Canedo P, Corso G, Pereira F, Lunet N, Suriano G, Figueiredo C, Pedrazzani C, Moreira H, Barros H, Carneiro F, Seruca R, Roviello F, Machado JC. The interferon gamma receptor 1 (IFNGR1) -56C/T gene polymorphism is associated with increased risk of early gastric carcinoma. Gut. 2008;57:1504-1508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 40. | Ucer U, Bartsch H, Scheurich P, Pfizenmaier K. Biological effects of gamma-interferon on human tumor cells: quantity and affinity of cell membrane receptors for gamma-IFN in relation to growth inhibition and induction of HLA-DR expression. Int J Cancer. 1985;36:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 41. | Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 1325] [Article Influence: 441.7] [Reference Citation Analysis (0)] |
| 42. | Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 4228] [Article Influence: 528.5] [Reference Citation Analysis (0)] |
| 43. | Zhang X, Chu H, Cheng Y, Ren J, Wang W, Liu X, Yan X. Identification of RUNX1 and IFNGR2 as prognostic-related biomarkers correlated with immune infiltration and subtype differentiation of low-grade glioma. Biomol Biomed. 2023;23:405-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 44. | Payne KK, Manjili MH. Adaptive immune responses associated with breast cancer relapse. Arch Immunol Ther Exp (Warsz). 2012;60:345-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Tecalco-Cruz AC, Macías-Silva M, Ramírez-Jarquín JO, Méndez-Ambrosio B. Identification of genes modulated by interferon gamma in breast cancer cells. Biochem Biophys Rep. 2021;27:101053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 46. | Duncan TJ, Rolland P, Deen S, Scott IV, Liu DT, Spendlove I, Durrant LG. Loss of IFN gamma receptor is an independent prognostic factor in ovarian cancer. Clin Cancer Res. 2007;13:4139-4145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 47. | Kearney SJ, Delgado C, Eshleman EM, Hill KK, O'Connor BP, Lenz LL. Type I IFNs downregulate myeloid cell IFN-γ receptor by inducing recruitment of an early growth response 3/NGFI-A binding protein 1 complex that silences ifngr1 transcription. J Immunol. 2013;191:3384-3392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 48. | Marcos-Pinto R, Dinis-Ribeiro M, Carneiro F, Wen X, Lopes C, Figueiredo C, Machado JC, Ferreira RM, Reis CA, Canedo P, Durães C, Ferreira J, Pedroto I, Areias J. First-degree relatives of early-onset gastric cancer patients show a high risk for gastric cancer: phenotype and genotype profile. Virchows Arch. 2013;463:391-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Zhang C, Hou D, Wei H, Zhao M, Yang L, Liu Q, Zhang X, Gong Y, Shao C. Lack of interferon-γ receptor results in a microenvironment favorable for intestinal tumorigenesis. Oncotarget. 2016;7:42099-42109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 50. | Slattery ML, Lundgreen A, Bondurant KL, Wolff RK. Interferon-signaling pathway: associations with colon and rectal cancer risk and subsequent survival. Carcinogenesis. 2011;32:1660-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 51. | Lu S, Pardini B, Cheng B, Naccarati A, Huhn S, Vymetalkova V, Vodickova L, Buchler T, Hemminki K, Vodicka P, Försti A. Single nucleotide polymorphisms within interferon signaling pathway genes are associated with colorectal cancer susceptibility and survival. PLoS One. 2014;9:e111061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | Qin W, Zhao B, Wang D, Liu J, Zhou Y, Zhu W, Huang Y, Qiu H, Yuan X. A Genetic Variant in CD274 Is Associated With Prognosis in Metastatic Colorectal Cancer Patients Treated With Bevacizumab-Based Chemotherapy. Front Oncol. 2022;12:922342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 53. | Lan B, Lv D, Sun X, Yang M, Zhang L, Ma F. Genetic Variations in IFNGR1, BDNF and IL-10 May Predict the Susceptibility to Depression and Anxiety in Chinese Women With Breast Cancer. Clin Breast Cancer. 2022;22:674-680. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 54. | Sava GP, Speedy HE, Houlston RS. Candidate gene association studies and risk of chronic lymphocytic leukemia: a systematic review and meta-analysis. Leuk Lymphoma. 2014;55:160-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 55. | Ding H, Wang G, Yu Z, Sun H, Wang L. Role of interferon-gamma (IFN-γ) and IFN-γ receptor 1/2 (IFNγR1/2) in regulation of immunity, infection, and cancer development: IFN-γ-dependent or independent pathway. Biomed Pharmacother. 2022;155:113683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 71] [Reference Citation Analysis (0)] |
| 56. | Jackson CC, Dickson MA, Sadjadi M, Gessain A, Abel L, Jouanguy E, Casanova JL. Kaposi Sarcoma of Childhood: Inborn or Acquired Immunodeficiency to Oncogenic HHV-8. Pediatr Blood Cancer. 2016;63:392-397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 57. | Aref S, Zaki A, El Mahdi EM, Adel E, Bahgat M, Gouda E. Predictive Value of Interferon γ Receptor Gene Polymorphisms for Hepatocellular Carcinoma Susceptibility. Asian Pac J Cancer Prev. 2021;22:1821-1826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 58. | Londino JD, Gulick DL, Lear TB, Suber TL, Weathington NM, Masa LS, Chen BB, Mallampalli RK. Post-translational modification of the interferon-gamma receptor alters its stability and signaling. Biochem J. 2017;474:3543-3557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 59. | Ng S, Lim S, Sim ACN, Mangadu R, Lau A, Zhang C, Martinez SB, Chandramohan A, Lim UM, Ho SSW, Chang SC, Gopal P, Hong LZ, Schwaid A, Fernandis AZ, Loboda A, Li C, Phan U, Henry B, Partridge AW. STUB1 is an intracellular checkpoint for interferon gamma sensing. Sci Rep. 2022;12:14087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Du W, Hua F, Li X, Zhang J, Li S, Wang W, Zhou J, Liao P, Yan Y, Li G, Wei S, Grove S, Vatan L, Zgodziński W, Majewski M, Wallner G, Chen H, Kryczek I, Fang JY, Zou W. Loss of Optineurin Drives Cancer Immune Evasion via Palmitoylation-Dependent IFNGR1 Lysosomal Sorting and Degradation. Cancer Discov. 2021;11:1826-1843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 61. | Hagiwara M, Fushimi A, Bhattacharya A, Yamashita N, Morimoto Y, Oya M, Withers HG, Hu Q, Liu T, Liu S, Wong KK, Long MD, Kufe D. MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer. Oncoimmunology. 2022;11:2029298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 62. | Tsai CC, Kai JI, Huang WC, Wang CY, Wang Y, Chen CL, Fang YT, Lin YS, Anderson R, Chen SH, Tsao CW, Lin CF. Glycogen synthase kinase-3beta facilitates IFN-gamma-induced STAT1 activation by regulating Src homology-2 domain-containing phosphatase 2. J Immunol. 2009;183:856-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 63. | Zheng H, Saito H, Masuda S, Yang X, Takano Y. Phosphorylated GSK3beta-ser9 and EGFR are good prognostic factors for lung carcinomas. Anticancer Res. 2007;27:3561-3569. [PubMed] |
| 64. | Krug J, Rodrian G, Petter K, Yang H, Khoziainova S, Guo W, Bénard A, Merkel S, Gellert S, Maschauer S, Spermann M, Waldner M, Bailey P, Pilarsky C, Liebl A, Tripal P, Christoph J, Naschberger E, Croner R, Schellerer VS, Becker C, Hartmann A, Tüting T, Prante O, Grützmann R, Grivennikov SI, Stürzl M, Britzen-Laurent N. N-glycosylation Regulates Intrinsic IFN-γ Resistance in Colorectal Cancer: Implications for Immunotherapy. Gastroenterology. 2023;164:392-406.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 39] [Reference Citation Analysis (0)] |
| 65. | Lawson KA, Sousa CM, Zhang X, Kim E, Akthar R, Caumanns JJ, Yao Y, Mikolajewicz N, Ross C, Brown KR, Zid AA, Fan ZP, Hui S, Krall JA, Simons DM, Slater CJ, De Jesus V, Tang L, Singh R, Goldford JE, Martin S, Huang Q, Francis EA, Habsid A, Climie R, Tieu D, Wei J, Li R, Tong AHY, Aregger M, Chan KS, Han H, Wang X, Mero P, Brumell JH, Finelli A, Ailles L, Bader G, Smolen GA, Kingsbury GA, Hart T, Kung C, Moffat J. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586:120-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 302] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 66. | Nakanishi Y, Nakatsuji M, Seno H, Ishizu S, Akitake-Kawano R, Kanda K, Ueo T, Komekado H, Kawada M, Minami M, Chiba T. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in ApcMin/+ mouse polyps. Carcinogenesis. 2011;32:1333-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 67. | Ahmed CM, Johnson HM. IFN-gamma and its receptor subunit IFNGR1 are recruited to the IFN-gamma-activated sequence element at the promoter site of IFN-gamma-activated genes: evidence of transactivational activity in IFNGR1. J Immunol. 2006;177:315-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 68. | Subramaniam PS, Larkin J 3rd, Mujtaba MG, Walter MR, Johnson HM. The COOH-terminal nuclear localization sequence of interferon gamma regulates STAT1 alpha nuclear translocation at an intracellular site. J Cell Sci. 2000;113 ( Pt 15):2771-2781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 69. | Liu Y, Liang L, Ji L, Zhang F, Chen D, Duan S, Shen H, Liang Y, Chen Y. Potentiated lung adenocarcinoma (LUAD) cell growth, migration and invasion by lncRNA DARS-AS1 via miR-188-5p/ KLF12 axis. Aging (Albany NY). 2021;13:23376-23392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 70. | Song M, Ping Y, Zhang K, Yang L, Li F, Zhang C, Cheng S, Yue D, Maimela NR, Qu J, Liu S, Sun T, Li Z, Xia J, Zhang B, Wang L, Zhang Y. Low-Dose IFNγ Induces Tumor Cell Stemness in Tumor Microenvironment of Non-Small Cell Lung Cancer. Cancer Res. 2019;79:3737-3748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 117] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 71. | Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 72. | Yang J, Wang X, Huang B, Liu R, Xiong H, Ye F, Zeng C, Fu X, Li L. An IFNγ/STAT1/JMJD3 Axis Induces ZEB1 Expression and Promotes Aggressiveness in Lung Adenocarcinoma. Mol Cancer Res. 2021;19:1234-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 73. | Zheng S, Luo X, Dong C, Zheng D, Xie J, Zhuge L, Sun Y, Chen H. A B7-CD28 family based signature demonstrates significantly different prognoses and tumor immune landscapes in lung adenocarcinoma. Int J Cancer. 2018;143:2592-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 74. | Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1409] [Article Influence: 128.1] [Reference Citation Analysis (0)] |
| 75. | Alburquerque-Bejar JJ, Navajas-Chocarro P, Saigi M, Ferrero-Andres A, Morillas JM, Vilarrubi A, Gomez A, Mate JL, Munoz-Marmol AM, Romero OA, Blecua P, Davalos V, Esteller M, Pros E, Llabata P, Torres-Diz M, Esteve-Codina A, Sanchez-Cespedes M. MYC activation impairs cell-intrinsic IFNγ signaling and confers resistance to anti-PD1/PD-L1 therapy in lung cancer. Cell Rep Med. 2023;4:101006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 76. | de Vor IC, van der Meulen PM, Bekker V, Verhard EM, Breuning MH, Harnisch E, van Tol MJ, Wieringa JW, van de Vosse E, Bredius RG. Deletion of the entire interferon-γ receptor 1 gene causing complete deficiency in three related patients. J Clin Immunol. 2016;36:195-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 77. | Lee EY, Schultz KL, Griffin DE. Mice deficient in interferon-gamma or interferon-gamma receptor 1 have distinct inflammatory responses to acute viral encephalomyelitis. PLoS One. 2013;8:e76412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 78. | Pinto BGG, Oliveira AER, Singh Y, Jimenez L, Gonçalves ANA, Ogava RLT, Creighton R, Schatzmann Peron JP, Nakaya HI. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J Infect Dis. 2020;222:556-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 269] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 79. | Gupta I, Rizeq B, Elkord E, Vranic S, Al Moustafa AE. SARS-CoV-2 Infection and Lung Cancer: Potential Therapeutic Modalities. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 80. | Gottschalk G, Knox K, Roy A. ACE2: At the crossroad of COVID-19 and lung cancer. Gene Rep. 2021;23:101077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 81. | Zhang H, Quek K, Chen R, Chen J, Chen B. Expression of the SAR2-Cov-2 receptor ACE2 reveals the susceptibility of COVID-19 in non-small cell lung cancer. J Cancer. 2020;11:5289-5292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 82. | Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J; HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016-1035.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1890] [Cited by in RCA: 1795] [Article Influence: 359.0] [Reference Citation Analysis (0)] |
| 83. | Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM, Ring TJ, Kee C, Doldan P, Tyrrell DL, Mendoza JL, Boulant S, Prokunina-Olsson L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet. 2020;52:1283-1293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 84. | Samad A, Jafar T, Rafi JH. Identification of angiotensin-converting enzyme 2 (ACE2) protein as the potential biomarker in SARS-CoV-2 infection-related lung cancer using computational analyses. Genomics. 2020;112:4912-4923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 85. | Kong Q, Xiang Z, Wu Y, Gu Y, Guo J, Geng F. Analysis of the susceptibility of lung cancer patients to SARS-CoV-2 infection. Mol Cancer. 2020;19:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 86. | Malkani N, Rashid MU. SARS-COV-2 infection and lung tumor microenvironment. Mol Biol Rep. 2021;48:1925-1934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 87. | Jacobs M, Van Eeckhoutte HP, Wijnant SRA, Janssens W, Joos GF, Brusselle GG, Bracke KR. Increased expression of ACE2, the SARS-CoV-2 entry receptor, in alveolar and bronchial epithelium of smokers and COPD subjects. Eur Respir J. 2020;56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 88. | de Groot PM, Wu CC, Carter BW, Munden RF. The epidemiology of lung cancer. Transl Lung Cancer Res. 2018;7:220-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 448] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 89. | Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Strazzabosco G, Fernandez M, Caccuri F, Caruso A, Rizzo R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 131] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 90. | Faraj SS, Jalal PJ. IL1β, IL-6, and TNF-α cytokines cooperate to modulate a complicated medical condition among COVID-19 patients: case-control study. Ann Med Surg (Lond). 2023;85:2291-2297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 91. | Thorne LG, Reuschl AK, Zuliani-Alvarez L, Whelan MVX, Turner J, Noursadeghi M, Jolly C, Towers GJ. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021;40:e107826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 153] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 92. | Todorović-Raković N, Whitfield JR. Between immunomodulation and immunotolerance: The role of IFNγ in SARS-CoV-2 disease. Cytokine. 2021;146:155637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 93. | Liu QQ, Cheng A, Wang Y, Li H, Hu L, Zhao X, Wang T, He F. Cytokines and their relationship with the severity and prognosis of coronavirus disease 2019 (COVID-19): a retrospective cohort study. BMJ Open. 2020;10:e041471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 94. | Alfadda AA, Rafiullah M, Alkhowaiter M, Alotaibi N, Alzahrani M, Binkhamis K, Siddiqui K, Youssef A, Altalhi H, Almaghlouth I, Alarifi M, Albanyan S, Alosaimi MF, Isnani A, Nawaz SS, Alayed K. Clinical and biochemical characteristics of people experiencing post-coronavirus disease 2019-related symptoms: A prospective follow-up investigation. Front Med (Lausanne). 2022;9:1067082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 95. | Jahani M, Dokaneheifard S, Mansouri K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J Inflamm (Lond). 2020;17:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 96. | Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1963] [Cited by in RCA: 1948] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 97. | Zhao Y, Xing C, Deng Y, Ye C, Peng H. HIF-1α signaling: Essential roles in tumorigenesis and implications in targeted therapies. Genes Dis. 2024;11:234-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 73] [Reference Citation Analysis (0)] |
| 98. | Benowitz NL, Goniewicz ML, Halpern-Felsher B, Krishnan-Sarin S, Ling PM, O'Connor RJ, Pentz MA, Robertson RM, Bhatnagar A. Tobacco product use and the risks of SARS-CoV-2 infection and COVID-19: current understanding and recommendations for future research. Lancet Respir Med. 2022;10:900-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |