Published online Jul 24, 2020. doi: 10.5306/wjco.v11.i7.428
Peer-review started: March 2, 2020
First decision: April 2, 2020
Revised: May 8, 2020
Accepted: May 27, 2020
Article in press: May 27, 2020
Published online: July 24, 2020
Processing time: 142 Days and 9 Hours
MUTYH is a base excision repair enzyme, it plays a crucial role in the correction of DNA errors from guanine oxidation and may be considered a cell protective factor. In humans it is an adenine DNA glycosylase that removes adenine misincorporated in 7,8-dihydro-8-oxoguanine (8-oxoG) pairs, inducing G:C to T:A transversions. MUTYH functionally cooperates with OGG1 that eliminates 8-oxodG derived from excessive reactive oxygen species production. MUTYH mutations have been linked to MUTYH associated polyposis syndrome (MAP), an autosomal recessive disorder characterized by multiple colorectal adenomas. MAP patients show a greatly increased lifetime risk for gastrointestinal cancers. The cancer risk in mono-allelic carriers associated with one MUTYH mutant allele is controversial and it remains to be clarified whether the altered functions of this protein may have a pathophysiological involvement in other diseases besides familial gastrointestinal diseases. This review evaluates the role of MUTYH, focusing on current studies of human neoplastic and non-neoplastic diseases different to colon polyposis and colorectal cancer. This will provide novel insights into the understanding of the molecular basis underlying MUTYH-related pathogenesis. Furthermore, we describe the association between MUTYH single nucleotide polymorphisms (SNPs) and different cancer and non-cancer diseases. We address the utility to increase our knowledge regarding MUTYH in the light of recent advances in the literature with the aim of a better understanding of the potential for identifying new therapeutic targets. Considering the multiple functions and interactions of MUTYH protein, its involvement in pathologies based on oxidative stress damage could be hypothesized. Although the development of extraintestinal cancer in MUTYH heterozygotes is not completely defined, the risk for malignancies of the duodenum, ovary, and bladder is also increased as well as the onset of benign and malignant endocrine tumors. The presence of MUTYH pathogenic variants is an independent predictor of poor prognosis in sporadic gastric cancer and in salivary gland secretory carcinoma, while its inhibition has been shown to reduce the survival of pancreatic ductal adenocarcinoma cells. Furthermore, some MUTYH SNPs have been associated with lung, hepatocellular and cervical cancer risk. An additional role of MUTYH seems to contribute to the prevention of numerous other disorders with an inflammatory/degenerative basis, including neurological and ocular diseases. Finally, it is interesting to note that MUTYH could be a new therapeutic target and future studies will shed light on its specific functions in the prevention of diseases and in the improvement of the chemo-sensitivity of cancer cells.
Core tip: This review focuses on the role of MUTYH, linked to MUTYH associated polyposis syndrome, on human neoplastic and non-neoplastic diseases, different to polyposis and colorectal cancer, considering its involvement in disorders based on oxidative stress damage. MUTYH pathogenic variants are independent predictors of poor prognosis in sporadic gastric, salivary gland secretory, pancreatic ductal, lung and cervical cancers, whereas some variants are involved in degenerative disorders, including neurological and ocular diseases. This provides novel insights for identifying the molecular basis of MUTYH pathogenesis in the light of recent advances, to better understand its potential in disease prevention and identify new therapeutic targets.
- Citation: Curia MC, Catalano T, Aceto GM. MUTYH: Not just polyposis. World J Clin Oncol 2020; 11(7): 428-449
- URL: https://www.wjgnet.com/2218-4333/full/v11/i7/428.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i7.428
Biallelic germline MUTYH (MutY homolog Escherichia coli, homolog of MYH, hMYH) gene variants were historically identified in a fraction of Adenomatous Polyposis Coli (APC) mutation-negative cases with a phenotype overlapping with attenuated familial adenomatous polyposis (AFAP) or classical familial adenomatous polyposis (FAP) (MIM# 604933)[1,2]. In humans, MUTYH protein is considered a cell protective factor able to counteract oxidative damage. It is a DNA glycosylase involved in the restoration of post-replicative mispairs in double-stranded DNA, where MUTYH recognizes and specifically removes adenine or 2-hydroxyadenines misincorporated in 7,8-dihydro-8-oxoguanine (8-oxoG) pairs with adenine or cytosine inducing G:C to T:A transversions as a consequence of DNA replication errors or DNA recombination[3-5].
In 1996 E. coli mutY gene (mutY) was cloned and characterized for the first time[6]. It encodes a protein involved in the bacterial repair system together with mutM, homologous of human 8-Oxoguanine glycosylase 1 (OGG1), and mutT that hydrolyzes 8-oxo-dGTP[7-9]. In E. coli MutY, MutM and MutT compose the GO (formally known as Guanine Oxidation) system responsible for removing and correcting mutations due to adducts[7]. Human MUTYH and OGG1 act like their E. coli homologs MutT and MutM to avoid the mutagenic consequences induced by 8-oxoG. Moreover, MUTYH shows 41% of homology to the E. coli corresponding protein[6]. In 1998, the sequence of SpMYH gene, the mutY homolog of the yeast Schizosaccharomyces pombe, was identified[10]. The SpMYH protein, that removes misincorporated G from 8-oxoG, shows 28% and 31% identity to E. coli MutY and human MUTYH, respectively[10,11]. In 2002, the role of the MUTYH gene in inherited predisposition to colorectal cancer (CRC) was identified in a British family, suggesting that an inefficient MUTYH protein and a defective base excision repair (BER) increase the incidence of somatic G > T transversions in APC gene[1]. Subsequent studies described germline biallelic mutations involved in MUTYH associated polyposis (MAP), a recessive heritable colorectal polyposis with an increased risk of CRC[12,13]. Further research explored MUTYH modifications associated with a hypermutable phenotype involving different organs and the development of various diseases, including cancers, through the accumulation of unrepaired 8-oxoG under conditions of oxidative stress.
In this review we evaluate biochemical and functional aspects as well as the role of MUTYH in BER. Moreover, we focus on current and updated studies on human diseases induced by alteration of MUTYH through research that associate it with neoplastic and non-neoplastic pathologies different to colon polyposis and CRC. This will produce novel insights into the understanding of the molecular basis underlying MUTYH-related pathogenesis. Furthermore, we describe the association between MUTYH single nucleotide polymorphisms (SNPs) and different cancer and non-cancer diseases.
We address the utility to increase our knowledge regarding MUTYH in light of recent advances in the literature with the aim of a better understanding of the potential for identifying new therapeutic targets.
In mammals, oxidized bases are continuously induced by endogenous reactive oxygen species (ROS) and more abundantly after oxidative stress. In the genome the oxidized bases are mutagenic due to their incorrect coupling property[14,15]. Moreover, to prevent the establishment of a new mutated base, the error must be repaired before DNA replication. In fact, the oxidized bases present in the model filament do not impede the replicative polymerases to prevent mutation[16]. BER is one of the most important DNA repair pathways, which improves intrinsic DNA damage due to metabolic processes[17-19] and plays a very important role in maintaining the integrity of the genomic structure. Therefore, the functions of its main components are highly conserved during evolution[20]. The repair mechanisms of DNA damage are sustained by proteins belonging to different signaling pathways. Among these pathways, the BER plays a crucial role in the correction of DNA errors due to oxidation, deamination and alkylation[5]. The BER pathway consists of a series of coordinated enzymatic events: Initially the specific DNA glycosylases recognize and exclude damaged bases or abasic sites and then, in a subsequent step, different sequential proteins correct the DNA through the template-direct insertion of one or more nucleotides, starting from the damaged site.
In mammals, MUTYH is functionally associated with OGG1 to eliminate and replace the oxidized guanine with a guanine[5,9,21,22]. The incorrect incorporation of 8-oxoG into DNA is initially recognized by the bifunctional glycosylase OGG1, that recognizes 8-oxoG coupled to the C base. This glycosylase breaks down 8-oxoG and subsequently catalyzes its elimination from which an abasic site is formed at 5’ of the nascent DNA[23]. If the excision due to the OGG1 action does not occur correctly, the persistence of 8-oxoG in the nascent DNA leads to the formation of damaged pairs of 8oxoG: A, which is fairly stable and can easily escape the correction activity by DNA polymerase. At this point the role of MUTYH is fundamental because the elimination of the correct base A induces the formation of an 8-oxoG: C base pair, offering OGG1 another opportunity to eliminate the 8-oxoG lesion before the mutation becomes permanent in the nascent DNA[23]. Finally, Nudix hydrolase (NUDT1, alias MutT human homolog 1 or MTH1) intervenes in the GO repair pathway to hydrolyze the nucleoside triphosphates 8-oxoG (8-oxoGTP) into monophosphates (8-oxoGMP) in order to prevent their incorporation into the genome. In this way NUDT1 also prevents that the removal of base A, from the DNA strand model, becoming pro-mutagenic after the action of MUTYH[23].
MUTYH is a bifunctional A/G-specific adenine DNA glycosylase[24], and is one of five BER-initiating enzymes in mammalian cells. Its glycosylase function has been known for at least thirty years and consists, as already mentioned, of eliminating the adenines or the 2-hydroxy-adenines (2-OH-A) paired with respect to 8-oxoG[25]. 8-oxoG is an oxidized form of the guanine base derived from ROS, is a stable product on DNA and has a lower oxidation potential than guanine. Traditionally it is considered one of the main mutagenic lesions of DNA with evolutionary and adaptive meaning[26]. In fact, 8-oxoG manages to couple with both adenine and cytosine during DNA replication; in this way it has the potential ability to cause a high transversion rate from G:C to T:A and A:T to G:C[27,28].
MUTYH, together with the other enzymes that repair the 8-oxo-G mis-pairing is a phylogenetically conserved protein (as MutY). Indeed, MUTYH homologous proteins have been identified both in prokaryotes and eukaryotes confirming the importance of its function in the protective mechanism from oxidative damage to DNA[15,29].
In mammals, the connection between C and N-terminal MUTYH domains, called the interdomain connector (IDC), is significantly longer than that of its bacterial counterpart[30]. Furthermore, even among mammals the homology of the IDC sequence is variable (i.e. H. Sapiens shares 78% of MUTYH with M. musculus) and this region has probably evolved within eukaryotic homologs giving way to MUTYH to interact with other enzymes of the reparative response to DNA damage[3,28,31]. Although the role of MUTYH glycosylase has been biochemically well defined for some time[32], new functional elements have recently been discovered: In fact, in addition to the Fe-S cluster, the need for a cofactor of the Zn2+ ion for the recognition and repair of damage has been demonstrated[31].
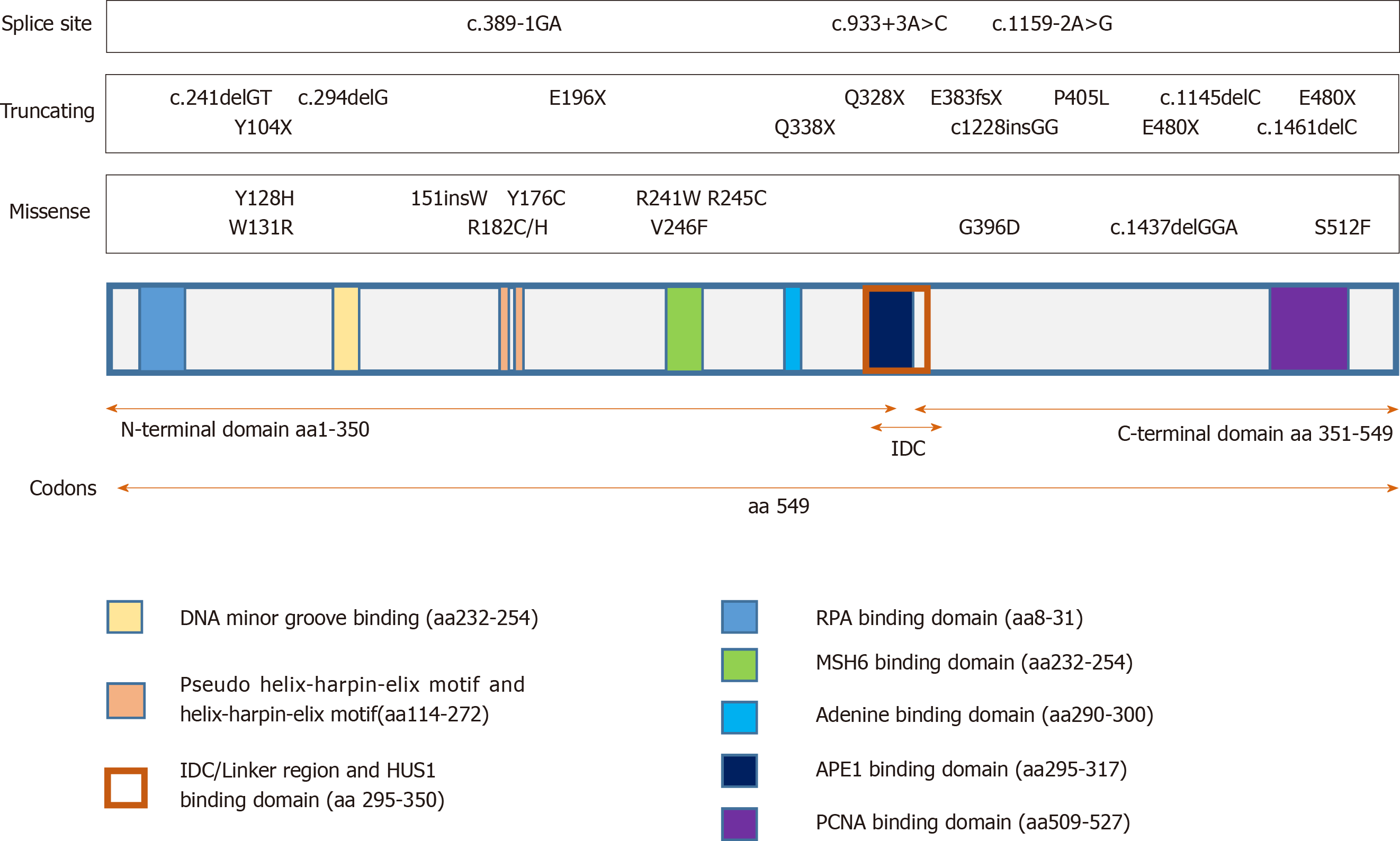
To perform its functions, MUTYH associates with various partner proteins[33]. This is necessary to ensure effective BER in coordination with other cellular processes. Figure 1 shows the known functional domains of MUTYH protein (data derived from the Ensembl genome database[34] and are representative of MUTYH well-known typical mutations). MUTYH consists of two globular domains linked by a flexible IDC, the N-terminal harbors the mitochondrial targeting signal (MTS), an adenine recognition domain, the helix-hairpin-helix-GPT (HhH-GPT) domain and a [4Fe-4S]+2 cluster cofactor coordinated to 4 Cys residues[35]; the switching between the reduced and oxidized form allows sensing of the DNA helical integrity damage. Peculiar characteristics of mammals in the IDC are 3 Cys coordinated with a Zn+2 ion; this “zinc linchpin” region could play an important role in coordinating the N-terminal and C-terminal domains of MUTYH for the correct 8oxoG: A recognition/excision and subsequent repair[30,31]. In fact, C-terminal domains are necessary for 8-oxoG recognition[36]. Evidence suggests that post-translational modifications in MUTYH may be a mechanism to modulate the BER pathway[37]. Indeed, phosphorylation of Ser524, on the PCNA binding site, modifies the affinity for the DNA substrate; recently it has been demonstrated that MUTYH protein levels may be regulated by momo-ubiquitination E3 ligase Mule (also called Huwe1, ArfBP1 and Lasu1), with target 5 Lys in the C-terminal[38]. Poly [ADP-ribose] polymerase 1 (PARP1) and p53 may regulate MUTYH expression and participate in MUTYH-mediated cell death[22,39,40]. MUTYH also displays crosstalk with the mismatch repair complex MSH2/MSH6[41,42], interacts with sirtuin (SIRT6) and may be modulated by acetylation[43]. It directly associates with PCNA and co-localizes with PCNA to nucleus replication foci during G1/S phase[44]; this improves polymerase processivity[45].
The protein interaction domains and other MUTYH motifs are shown in Figure 1; all reported information are available in the Entrez Gene NCBI database, the http//Ensembl.org database[46], and the ClinVar database [http://simple-clinvar.broadinstitute.org][47]. Rad9-Hus1-Rad1 complex (9–1–1) facilitates ATR-mediated Chk1 phosphorylation and activation, which in turn elicit cellular responses such as arresting cells in the G2/M phase[48]. The N-terminal 32-aa peptide also contains a binding site for replication protein A, indicating that some isoforms of MUTYH may participate in replication-coupled repair[49]. In the rat brain, specific mitochondrial isoforms of MUTYH protein have been observed: They are developmentally regulated and induced by respiratory hypoxia in the hippocampus[50].
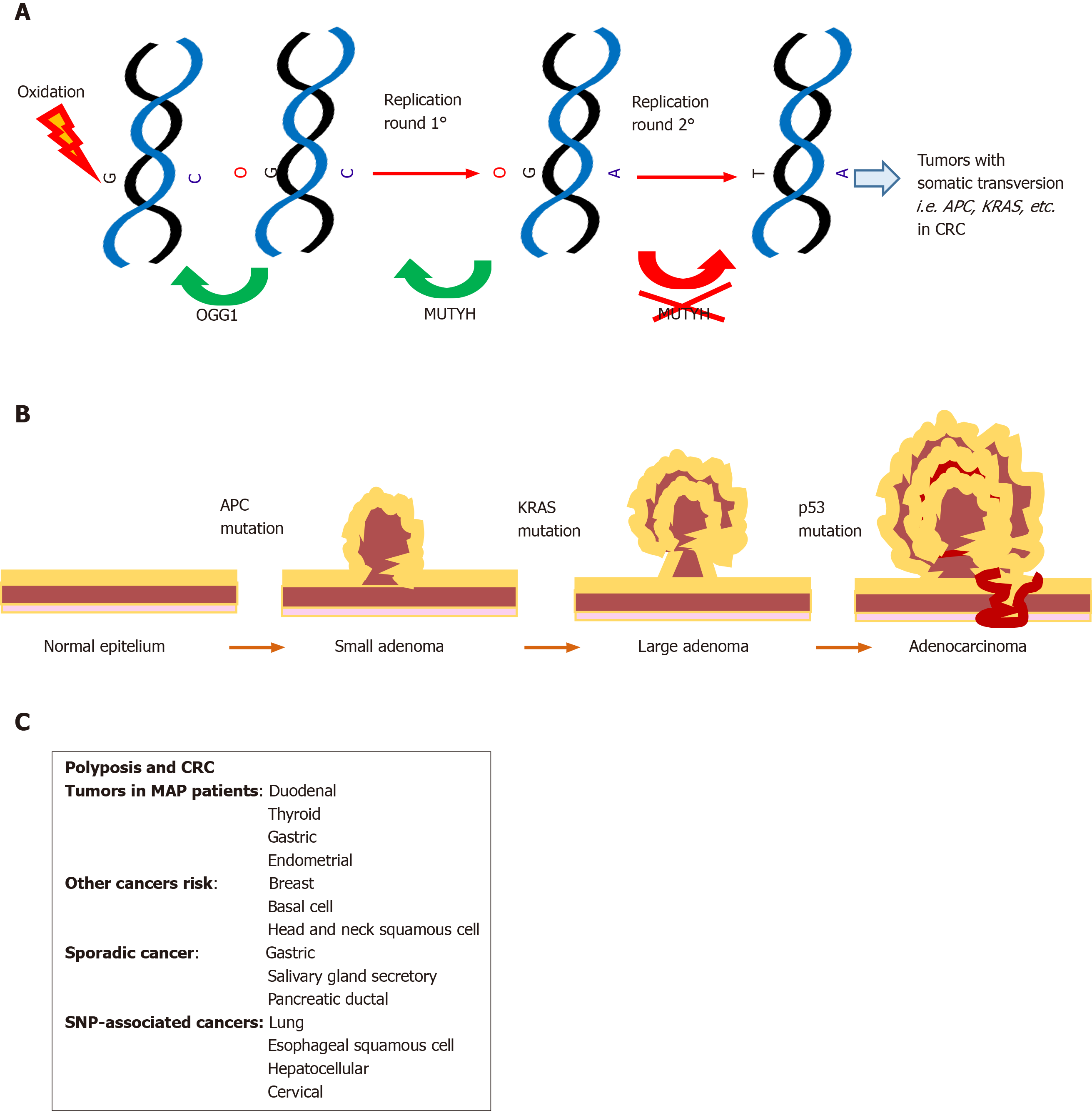
Initially, MUTYH mutations have been linked to the MAP syndrome known as MUTYH associated polyposis[1], an autosomal recessive disorder characterized by multiple colorectal adenomas. In the intestinal epithelium, progenitor cells with loss of MUTYH function can escape from programmed cell death, accumulate Kras and APC, thereby promoting carcinogenesis[28] (Figure 2); although later SNP variants that decreased the affinity and catalytic activity on 8oxoG: A[51] were also associated with other complex pathologies (see below).
MUTYH gene is located on chromosome 1 (p32.1-p34.3) (seq ref NM_001128425.1.) and consists of 16 exons[6,52]. In human cells, three primary transcripts (α, β, γ) were experimentally identified; from these multiple transcripts were generated by alternative splicing and exon skipping[53]. Transcription initiation and splicing of the MUTYH pre-mRNA produces at least thirteen mRNA isoforms and nine protein isoforms with different 5’-terminal mRNA and N-terminal protein sequences[53]. Of the two major MUTYH isoforms, MUTYH 1 (p57) is the product of the α transcript and contains the mitochondrial targeting signal (MTS) at its N-terminus to establish it in mitochondria, whereas MUTYH 2 (p60) is produced from the β or γ transcript, lacks the MTS and is localized in the nucleus[54]. Moreover, there is an overlapping gene, Target Early Growth Response 1 (EGR1) member 1 (TOE1) at the 5’ end of MUTYH, and the transcription of these two genes proceeds in opposite directions[28]. TOE1 inhibits cell growth rate and cell cycle, induces Cyclin Dependent Kinase Inhibitor 1A (CDKN1A) expression as well as Transforming Growth Factor-beta (TGF-beta) expression, mediates the inhibitory growth effect of EGR1 involved in the maturation of snRNAs and snRNA 3'-tail processing[55,56]. The immediate early gene and transcription factor EGR1 is an important mediator and regulator of synaptic plasticity and neuronal activity in both physiological and pathological conditions[57].
Considering that a total of 3081 public variants and 352 unique variants are currently listed on the public databases, it can easily be assumed that much still needs to be defined on the functions of MUTYH, especially at the level of specific tissues, even if most of the MUTYH mutations have been detected in polyposis associated with MUTYH and in other types of gastrointestinal carcinoma. It remains to be clarified whether there are hormesis effects in the functions of all MUTYH isoforms and whether particular alternative splicings have a pathophysiological involvement in other diseases besides gastrointestinal diseases.
Until 2002, mutations in a second gene, MUTYH, besides APC, were found in patients that presented with AFAP, in which no APC gene mutation was identified. MUTYH, together with APC germline defects, are responsible for the majority of clinically well-characterized patients with FAP and AFAP phenotype, and with more than 30 colorectal adenomas[58,59]. In particular mutations of MUTYH are found in 7%-12% and 40% of FAP and AFAP patients, respectively, and in 26% of patients with multiple colorectal adenomas (MRCA)[59-61]. In MRCA, MUTYH mutations differed according to the number of colorectal adenomas, being more frequent in patients with 30-99 adenomas than in those with 10-29 adenomas[59].
Single polyps undergo carcinogenesis but the exact route to carcinoma seems to differ between conditions. Patients with MAP can present with serrated adenomas, hyperplastic/sessile serrated polyps, and mixed (hyperplastic and adenomatous) polyps[62]; duodenal adenomas are common, with an increased risk of duodenal cancer[63,64]. In addition, gastric fundic gland polyps are common and although a higher risk for gastric cancer (GC) than in the general population was observed, the trend was not significant[64]. Extraintestinal reported features include thyroid nodules and benign adrenal lesions[65,66].
MAP patients are characterized by a greatly increased lifetime risk for CRC (43%-63% at age 60 years) that increases to 80%-90% in the absence of timely surveillance[67,68]. In patients with MAP, colon cancer is right-sided in 29%-69% of cases and metachronous or synchronous colon cancers occurred in 23%-27%[69,70].
At first, the two non-conservative MUTYH mutations c.536A > G, p.Tyr179Cys (Y179C) and c.1187G > A, p.Gly396Asp (G396D) were found to reduce the removal of A from G:A mismatches, suggesting that a defect in MUTYH activity leads to accumulated mutations in patients with FAP-like phenotype, referred to as MAP or FAP2; this is the unique mechanism to induce CRC with a recessive mode of inheritance[1]. The polyposis of this syndrome tends to be less pronounced than FAP. However, the penetrance is very high, showing almost 100% penetrance for cancer by the age of 55 years[71,72]. The prevalence of MAP is around 1% in all CRC cases[73]. This estimate rises to 2% when young-onset CRC patients are considered and to 5%–25% in CRC patients with somatic Kras G12C substitution[67].
In contrast to APC, no relationship has been observed between the location of the mutation and the phenotype of the disease. Mutations located throughout the entire MUTYH have been described in MAP, but only two missense mutations, p.Tyr179Cys and p.Gly396Asp, are the most prevalent in Caucasians and account for approximately 75% of the reported mutations in MAP patients[74]. Other population-specific MUTYH mutations have been found[34,52,69,75].
A large meta-analysis study defined the CRC risk associated with bi-allelic mutations in the MUTYH gene as 28-fold (95%CI: 6.95–115) following logistic regression analysis[76]. Bi-allelic carriers of the p.Gly396Asp variant and p.Tyr179Cys/ p.Gly396Asp compound heterozygotes were also significantly associated with a similar increase in CRC risk [OR = 23.1 (95%CI: 3.15–169) and 21.6 (95%CI: 2.94–159), respectively]. On the other hand, the risk associated with one MUTYH mutant allele is controversial[67,73,77,78]. Furthermore, the differences in the frequency of individual MUTYH variants in the population have made it difficult to replicate study findings. In particular, with regard to MUTYH p.Tyr179Cys, because of the rarity of the variant (allele frequency 0.002), the evidence presented raises the possibility of a mono-allelic effect for this variant[76]. Nevertheless, a two-fold increase in CRC risk was observed in mono-allelic carriers, thus providing evidence of a mono-allelic effect of the MUTYH gene, and was demonstrated in other studies[68,79-82].
For years it remained unclear whether variants in the MUTYH gene were associated with an increased risk of other cancers beyond that of the colorectal and the results were controversial.
Several studies have reported an increased risk of bladder, ovarian, skin, breast and endometrial cancer for biallelic mutation carriers[64,83-85] and a slightly increased cumulative risk in MUTYH heterozygotes for gastric, liver, endometrial and breast cancer[64,84,86,87].
Breast cancer: Previous studies on the association between MUTYH variants and the risk of breast cancer (BC) produced controversial results[83,88,89] and it is equally unclear if BC is increased in women with MAP. The risk was found to be higher, with a median age at diagnosis of 53 years (range 45-76) in the study by Vogt et al[64]. Six heterozygous MUTYH mutations, including p.Tyr179Cys, p.Gly396Asp and p.Pro405Leu (c.1214C > T, P405L), resulted in an association in families with both BC and CRC, but not polyposis[84] even if two years later an extensive case-control study did not confirm the association between MUTYH variants and BC, but did not exclude BC susceptibility[89]. An interesting result is the finding of one male with biallelic germline MUTYH pathogenic variants in a large cohort of males with BC[90]. In the same study, no biallelic MUTYH mutations been reported in women from large BC cohorts. In a case-control study, 930 Jewish women with a high prevalence of MUTYH mutations were investigated for the two variants p.Gly396Asp and p.Tyr179Cys, and patients with BC revealed a 6.7% prevalence of p.Gly396Asp[91].
Basal cell carcinoma: Is a relatively benign skin cancer caused by UV exposure and it is known that the DNA repair mechanisms in the skin are interconnected to protect against UV mutagenesis, involving, other than nucleotide excision repair (NER) also base excision and mismatch repairs[92]. In a study by Cho et al[93], saliva samples from patients affected by basal cell carcinoma (BCC) were collected, DNA was extracted and 29 genes were analyzed for germline mutations. Individuals with a high frequency of BCC (presence of metastatic prostate, primary ovarian and BC) harbored pathogenic mutations in DNA repair genes: APC, BARD1, BRCA1, BRCA2, CDH1, CHEK2, MLH1, MSH2, MSH6, MUTYH, NBN, and PALB2. Individuals with germline mutations in DNA repair genes had an increased risk of malignancy especially other skin cancers, such as melanoma and squamous cell carcinoma, possibly due to UV exposure. These results indicate that these patients might benefit from multigene cancer-susceptibility panel testing, as performed for gastrointestinal cancer[94].
Head and neck squamous cell carcinoma: The role of BER genes has also been investigated in squamous cell carcinomas of the head and neck[95]. Development of this tumor is a multifactorial process associated with a variety of environmental risk factors such as tobacco and alcohol consumption, and, in this scenario, oxidative DNA damage and BER could be involved in the development of this tumor. MUTYH, OGG1 and MTH1 germline mutations or polymorphisms have been investigated using complete genomic sequencing in patients and controls and no pathogenic germline mutations were identified. However, common and rare new variants in the coding and adjacent intronic regions have been detected possibly indicating a minor role of the three BER genes in the tumorigenesis of sporadic head and neck squamous cell carcinoma (SCCHN)[95].
In sporadic cancers, which account for a large majority of human cancer, a multitude of genetic and environmental components cooperate. In contrast to germ-line mutations in DNA repair genes, which cause a strong deficiency in DNA repair activity in all cell types, the role of missense and SNPs in sporadic cancer is unclear because deficiencies in DNA repair are much milder. Furthermore, slow progress is due to a lack of functional and well-designed studies.
Gastric cancer:MUTYH mutations can contribute to the development of sporadic gastric cancer (GC) as documented by Kim et al[96]. They found biallelic mutations, somatic missense in one allele and loss of the remaining allele, in GC patients with H. pylori-positive advanced intestinal-type GC and lymph node metastasis. It has been found that patients with GC exhibiting low MUTYH expression showed a poor outcome when compared to those expressing high levels of MUTYH. This finding may act as an independent predictor of poor survival in GC patients[97].
An increased accumulation of 8-oxoG in nuclei and reduced expression of MUTYH are also documented in the mucosa of patients with ulcerative colitis. In this mucosa mutations of Kras but not MUTYH were found[98]. Inflamed mucosa in these patients is known to be exposed to oxidative stress; therefore, accumulation of 8-oxoG is responsible for Kras mutations, thus indicating that MUTYH plays a tumor suppressor role in ulcerative colitis as well as MAP patients[28].
Salivary gland secretory carcinoma: Pathogenic variants of MUTYH were also identified in salivary gland secretory carcinoma (SC), in particular acinic cell carcinoma, an aggressive phenotype with lymph node metastasis[99]. This type of tumor also demonstrated mutations in MLH1 and Serine/Threonine Kinase 11 (STK11), genes involved in polyposis and cancer of the colon. Missense and splice site mutations in four genes were identified as pathogenic or likely pathogenic: Serine Protease 1 (PRSS1) (c.47C > T; A16V), MLH1 (c.1151T > A; V384D), MUTYH (c.934-2A > G; splice site), and STK11 (c.842C > T; P281L). The most frequent mutations were in PRSS1 gene and this is foreseeable considering that germline and somatic PRSS1 mutations are associated with hereditary, chronic pancreatitis and pancreatic adenocarcinoma, and salivary glands are exocrine glands with a similar histology to the pancreas[100]. No significant copy number alteration was shown in these genes. Secretory carcinoma displays a mutation spectrum well described in the cancer genome study by Vogelstein et al[101]; low-grade indolent tumors usually presented mutations in PRSS1 gene, while cases with aggressive clinicopathologic features such as lymph node metastasis and tumor recurrence also revealed mutations in MUTYH and MLH1 genes. These findings may improve the diagnosis and treatment of different types of secretory carcinoma, taking into account the risk stratification of patients.
Pancreatic ductal adenocarcinoma: Pancreatic ductal adenocarcinoma (PDAC) is a highly metastatic and chemo-resistant disease which is best treated with surgery[102]. It is also characterized by extensive fibrosis, which creates a hypoxic microenvironment and consequently leads to intracellular oxidative stress. Given the function of BER enzymes in protecting cells from oxidative DNA damage, they have been identified as important regulators of resistance to a variety of chemotherapeutics. The inhibition of their activity could represent a novel therapeutic approach for PDAC in reducing survival of cancerous cells[103]. In this study, silencing of MUTYH using siRNA in a cultured PDAC cell line reduced proliferation, increased apoptosis and finally increased chemo-sensitivity in vitro, suggesting that MUTYH is a novel therapeutic target for pancreatic cancer. PDAC has not or has rarely been reported in MAP patients[64,65,87].
Genetic variations in DNA repair genes may modulate DNA repair ability and are thought to be related to cancer risk, but polymorphisms of DNA repair genes are little studied in genome-wide association studies[104,105]. The majority of studies have provided inconsistent results due to very limited coverage of DNA repair-related genes, typically evaluating only a few repair genes that play a key role in the BER pathway, such as the X-Ray Repair Cross Complementing 1 (XRCC1), OGG1, and Apurinic/Apyrimidinic Endonuclease 1 (APEX1) genes[106-108]. These three genes are associated with human tumor susceptibility and radiation toxicity[109]. In a study of nasopharyngeal carcinoma (NPC) patients, some polymorphisms were reported to have a significant correlation with the curative effect at the end of radiation therapy, but no influence on radiation toxicity has been reported[110].
Lung cancer: At first it seemed that the NER pathway had a stronger influence on lung cancer (LC) than the BER pathway[111]. Tobacco smoke accounts for a huge generation of ROS species and thereby oxidative damage. Studies on the association with LC risk focused mostly on three key genes in the BER pathway OGG1, APE1/APEX1 and XRCC1[112,113]. No association was found between cancer risk and the APE1/APEX1 p.Asp148Glu (D148E, rs3136820) and XRCC1 p.Arg280His polymorphisms, while a positive association between two gene polymorphisms, OGG1 p.Ser326Cys (S326C, rs1052133) and MUTYH c.972G > C, p.Gln324His (Q324H, rs3219489) or OGG1 p.Ser326Cys alone and the risk of LC, has been reported[112,113]. This is consistent with experimental evidence that these isoforms exhibit decreased enzyme activity. The two polymorphic variants have been extensively studied for their roles in cancer susceptibility and prognosis. The combined effect of both OGG1 p.Ser326Cys and MUTYH p.Gln324His on the risk of lung adenocarcinoma has been confirmed in a recent study performed on DNA from 326 LC cases and 330 controls by genotyping making use of polymerase chain reaction-restriction length fragment polymorphism[114]. In the presence of both variants, the risk of LC was found to be independent of tobacco smoke. In particular, it was observed that heterozygotes for MUTYH p.Gln324His exhibited a 2-fold increased risk of LC (OR = 2.35, C.I. = 1.59-3.4, P < 0.0001) in smokers and similarly in non-smokers (OR = 3.37, C.I. = 1.62-7.02, P = 0.001). Despite these findings, the association between MUTYH polymorphisms and the risk of LC remains controversial and requires further verification in a larger study population to facilitate the evaluation of multigenic effects of environmental exposure. An in vitro analysis of MUTYH p.Gln324His showed that it has reduced enzyme activity similar to that of the known cancer variant p.Gly396Asp, providing evidence that this common variant may lead to increased colorectal and LC risk[115].
Esophageal squamous cell carcinoma and GC: In a pathway-based analysis, Li et al[116] identified a significant association between several DNA repair pathway genes and the risk of esophageal squamous cell carcinoma and GC. The most significant genes were CHEK2, SMUG1 (uracil-DNA glycosylase), TP53, but MUTYH (c.504+35A > G; rs3219487) was associated with the risk of cancer. To date, of the DNA repair related genes, CHEK2 remains the only one associated with cancer to be identified by genome-wide-associated studies (GWAS).
Hepatocellular carcinoma: The involvement of MUTYH in hepatocellular carcinoma (HCC) has been investigated by Sakurada et al[117]. The authors found a significant association between the intronic MUTYH SNP rs3219487 and the risk of developing HCC. Patients with A/A or G/A genotypes had reduced mRNA levels in peripheral mononuclear cells and a higher risk of developing HCC than those with the G/G genotype (OR = 9.27, 95%CI = 2.39−32.1, P = 0.0005). Reduced enzyme activity was also confirmed in MUTYH-null mice that did not develop any tumors after an antioxidant-rich diet[117].
Cervical carcinoma: To date, few reports have focused on the potential effects of the MUTYH SNP p.Gln324His on cervical cancer. Recently this common polymorphism has been studied in cervical squamous cell carcinoma (CSCC) to ascertain its relationship with the risk of this cancer in a case-control group (400 CSCC, 400 precursor lesion CIN III and 1200 control participants)[118]. The results of the study revealed that the MUTYH p.Gln324His heterozygous CAG/CAC and homozygous CAC/CAC genotypes were associated with a significantly increased risk of cervical cancer (CIN III, OR = 1.46) and CSCC (OR = 2.34). Moreover, the authors observed a positive correlation between the proportion of homozygous CAC/CAC MUTYH genotype and malignant prognostic factors of CSCC, such as cell differentiation grade and lymph node metastasis. These findings highlight associations between the MUTYH SNP p.Gln324His and susceptibility to CSCC and support the utility of this variant as an early indicator of prognosis for patients at risk of cervical carcinoma and HPV infection[118].
Some diseases not associated with polyposis but characterized by MUTYH mutations or SNPs have recently been described. These genetic events can lead to dysregulation of MUTYH glycosylase activity with a lack of its protective antioxidant role. As organ functions reflect the various biochemical reactions of the products of gene transcription, changes in MUTYH expression may be detected in distinct parts of the body and be responsible for different diseases.
Neurodegenerative diseases: Alterations in MUTYH gene or functional involvement of the MUTYH protein in severe stress conditions can predispose to different neurodegenerative disorders. The extreme variability of phenotypes and progression displayed in these pathologies is probably linked to the type and location of the cell populations in nervous tissue, and the relationships occurring among the cells involved in the pathology.
Parkinson’s disease: Is characterized by motor deficits, mainly related to an early loss of 40%-60% of dopamine-containing neurons in the substantia nigra. The occurrence of 8-oxoG is associated with elevated expression of MTH1, OGG1, and MUTYH proteins in nuclear and mitochondrial DNA or cytoplasmic RNA in nigrostriatal dopaminergic neurons that additionally show intense and diffuse MUTYH immunostaining only in the cytoplasm[119]. The major MUTYH isoform in Parkinson’s disease brain is represented by a mitochondrial 47-kDa molecule, that might be derived from the MUTYH type α4 mRNA containing the MTS[119]. This molecule could play a role in protection of the remaining neurons against possible oxidative stress factors, including excessive oxidized lipids, proteins, and DNA[119,120]. In contrast, Nakabeppu et al[119] also suggested that the 47-kDa molecule could have no glycosylase activity in the BER mechanism as it lacks the DNA minor groove-reading motif.
Alzheimer’s disease: Results in a progressive loss of brain cells with a decline in memory and cognition. Its multifactorial etiology is probably associated with a reduction of BER efficacy, or excessive neuronal or microglial production of ROS, including 8-oxoG generated in mitochondria during dysregulated insulin-glucose metabolism[121-123]. Single-strand DNA breaks (SSBs) and 8-oxoG activate the two distinct mitochondrial DNA (mtDNA) caspase-independent/calpain and nuclear DNA (nDNA) PARP-1-dependent death signaling pathways[124]. At an early stage of Alzheimer’s disease (AD), accumulation of the amyloid β (Aβ) protein is associated with ROS production[125], that induces neurodegeneration through calcium-dependent inactivation of calpain[28,122,123]. In the later stages, the Aβ and neurofibrillary deposition blocks the repair process[125] and induces apoptosis by the PARP-1-dependent pathway. Thus, damaged neurons stimulate a delayed inflammatory response, microgliosis, which involves the adjacent neurons[28,122-124]. Sheng et al[123] demonstrated that MTH1 and OGG1 can play a preventive role to avoid harmful MUTYH-mediated accumulation of 8-oxoG in a chemical AD model of neurodegeneration. Significant down-regulation of APE1, OGG1, MUTYH, PARP1 and NeiLikeDNAGlycosylase1/NeiEndonucleaseVIII-Like 1 (NEIL1) genes in AD peripheral blood lymphocytes, as compared to healthy subjects, was independent of the methylation status of gene promoters, and probably reflected other changes occurring in the AD brain[121].
Friedreich's ataxia: Is an autosomal recessive disease of the peripheral and central nervous systems and is caused by defects in the iron-sulfur cluster biogenesis consequent to a GAA trinucleotide repeat expansion within the first intron of the FXN gene or point mutations truncating the protein[126,127]. This results in decreased levels of the small mitochondrial protein frataxin, with consequent Fe-S cluster deregulation associated with iron accumulation, mitochondria dysfunction and oxidative DNA damage due to increased 8-oxo-G in microglia[126,128,129]. This induces high levels of MUTYH and PARP-1, microglia activation, neuroinflammation and neurodegeneration, elevated levels of arachidonic acid metabolites, such as prostaglandins and thromboxane B2, maintained by cyclooxygenase (COX2) overexpression induced by ROS[126,128,130].
Huntington’s disease: Is a hereditary neurodegenerative syndrome causing uncontrolled body movements, changes in behavior and a decline in cognitive abilities. It is determined by a somatic expansion of CAG repeats ranging between 35–121 trinucleotide repeats (TNRs) in the coding region of the Huntingtin (HTT) gene. Age onset and progression of Huntington’s disease depend on the length of the CAG repeat expansion in germ and brain cells[131,132]. Mutant HTT protein shows an expanded glutamine tract and is correlated with increased ROS production. Kovtun et al[133] suggested a 'toxic oxidation' model using R6/1 transgenic mice in which DNA oxidized by ROS was subjected to an oxidation-excision cycle that was iterative with age, and caused a progressive CAG repeat expansion. Jaram et al[131,132,134] observed an aberrant BER pathway in which OGG1 slowly removes 8-oxoG that accumulates in the hairpins derived from TNR region self-anneal, resulting in expanded CAG tracts. In the presence of excessive oxidative damage, MUTYH DNA glycosylase, similar to OGG1, may modulate the TNR expansion[134].
Retinitis pigmentosa: Is an inherited disease characterized by the death of photoreceptor cells induced by oxidative DNA damage in microglia[135]. In the early phases of retinal degeneration, increased levels of 8-oxoG, SSB formation, and PARP activation stimulate microglia during BER in the rd10 mouse model. Thus, under oxidative DNA damage, MUTYH mediated microglial activation inducing retina inflammation and photoreceptor degeneration[135].
Neurofibromatosis: Is an inherited dominant disease characterized by modifications in skin pigmentation and growth of tumors along nerves near the spinal cord, or in other parts of the body. It is associated with the development of different cancers. Neurofibromatosis includes neurofibromatosis type 1 (NF1) caused by heterozygous mutation of Neurofibromin 1 (NF1) gene, and neurofibromatosis type 2 (NF2), caused by mutations in the Neurofibromin 2 (NF2) gene. Li et al[136] described a clinical case of NF2 in a 12-year-old patient carrying MUTYH c.53C > T, p.Pro18Leu (P18L)/c.74G > A, p.Gly25Asp (G25D) and Ataxia Telangiectasia And Rad3-Related (ATR) mutations inherited from her father, as well as MLH1 and Checkpoint Kinase 2 (CHEK2) mutations from her mother. As a consequence of the aberrant DNA repair mechanism, a new NF2 germline mutation occurred in the young patient.
Novel MUTYH mutations were detected in an analysis of 39 candidate genes by array and confirmatory Sanger sequencing[137]. The study involved 27 cases affected by rare mitochondrial disorders and 13 controls. At cDNA position 1307 of MUTYH, Wang et al[137] identified a heterozygous deletion (A–) regarding a single base with amino acid substitution at position 364 (GAA > GAC), frameshift and premature termination (TGA) at position 393 (p.E364Dfs393X), in a region involving the DNA-glycosylase-C domain. Another sample carried both a benign MUTYH c.1535C > T, p.Ser512Phe (S512F) variant and p.Gln324His that is considered a probable damaging SNP.
Different authors have focused their attention on the genetic correlation between MUTYH SNPs and an increased individual predisposition to some non-cancer diseases. Although most of these clinical studies should be performed on a larger population to confirm the results, the association between MUTYH p.Gln324His polymorphism and susceptibility to different pathologies has been specifically suggested. The glutamine to histidine amino acid change determines a small local modification in the electron cloud, associated with a slight reduction in DNA binding activity, that might contribute to diminished efficacy of the BER, as the p.Gln324His variant was considered 34% less active than the corresponding wild-type[138].
AD: Kwiatkowski et al[138] examined SNPs in PARP1, XRCC1, and MUTYH BER-related genes as well as the distribution of their genotypes in 110 patients with AD and 120 healthy controls from Poland. This study detected an increased risk for AD in the genotype combinations MUTYH p.Gln324His and PARP1 c.2285T > C, p.Val762Ala (V762A, rs1136410), or XRCC1 c.580C > T, p.Arg194Trp (R194W, rs1799782), or XRCC1 c.1196A > G, p.Gln399Arg (Q399R, rs25487)[138]. Lillenes et al[139] investigated the presence of SNPs in MUTYH and the other BER component genes OGG1, APE1, polymerase β (PolB) and polymerase γ (PolG), as well as the TranscriptionFactorAMitochondrial (TFAM) gene in a cross-sectional case-control study with a total of 449 individuals from Norway, including AD affected and non-affected patients and healthy controls. The MUTYH c.64G > A, p.Val22Met (V22M, rs3219484) SNP was rarely detected, while MUTYH rs3219489 had the highest minor allele frequencies. To date, no MUTYH polymorphism has been strongly associated with AD.
Keratoconus: No association was found between OGG1 p.Ser326Cys, and MUTYH p.Gln324His polymorphisms or their combined genotypes and the occurrence of keratoconus (KC), a non-inflammatory disease of the cornea, in an investigation involving 205 patients with KC and 220 controls from Poland. The authors indicated that the small sample size was the main limitation in this research[140].
Primary open angle glaucoma: Another study aimed to evaluate the association between XRCC1 p.Arg399Gln, XRCC1 p.Arg194Trp, OGG1 p.Ser326Cys, MUTYH p.Gln324His, ADP-ribosyltransferase (ADPRT), also known as PARP1, p.Ala762Val and APE1 p.Asp148Glu SNPs and the risk of primary open angle glaucoma (POAG) in 412 Polish patients and 454 healthy subjects[141]. MUTYH p.Gln324His and ADPRT p.Ala762Val genotypes had a protective role in the progression of POAG, while the combined genotypes XRCC1 p.Arg399Gln and MUTYH p.Gln324His were associated with an increase in the risk of this pathology[141]. These results confirmed the data from a previous investigation by the same research team in 170 patients with POAG and 193 healthy controls, with the aim of defining the role of the XRCC1, OGG1 and MUTYH gene polymorphisms in the development of POAG[142].
Age-related macular degeneration: Synowiec et al[143] explored the relationship between the polymorphisms MUTYH p.Gln324His, OGG1 p.Ser326Cys and age-related macular degeneration (AMD), a disease involving the macula cells, with loss of photoreceptors and retinal pigment epithelium, and probably related to oxidative DNA damage associated with aging. The study was performed using DNA from the blood of 271 patients, comprising 101 subjects with wet and 170 subjects with dry AMD, and 105 healthy individuals. AMD was positively correlated with the C allele of MUTYH p.Gln324His and the wet form of the disease. Nevertheless, this association was considered weak and medically not significant[143].
Recurrent depressive disorder: Czarny et al[144] examined the link between OGG1 p.Ser326Cys, MUTYH p.Gln324His, NEIL1 c.*589G > C (rs4462560) SNPs and the occurrence of depression in 555 Polish individuals, including 257 patients and 298 healthy controls. MUTYH c.972G/G genotype was associated with late-onset depression, while the G allele of NEIL1 was positively related to early and late-onset disease[144]. In a small number of patients, the combination of MUTYH p.Gln324His and NEIL1 c.*589G > C enhanced depression susceptibility[144]. In another investigation on recurrent depressive disorder, Czarny et al[145] analyzed 12 SNPs located in NEIL1, OGG1, MUTYH, PARP1, XRCC1, Flap Endonuclease 1 (FEN1), APEX1, DNA Ligase 1 (LIG1), and DNA Ligase 3 (LIG3) genes in 43 patients and 59 healthy controls to evaluate the presence of variants in depression onset. Only APEX1 c.-468T > G (rs1760944) significantly increased the risk of depression, while NEIL1 rs4462560 reduced the risk. Moreover, some SNPs influenced the efficiency of DNA damage repair (DRE) in depressed individuals, such as MUTYH c.972G > C, while the variants MUTYH C/G and G/G did not influence DRE. This suggested that depression is associated with DNA damage caused by increased oxidative stress related to low DRE, in part attributed to definite SNP variants. Finally, the authors acknowledged some limitations in this study, such as the use of peripheral blood mononuclear cells instead of central nervous system cells in the experiments[145].
Osteoarthritis: Is a chronic disease that involves articular joints and leads to cartilage degeneration, with consequent functional impairment. The contribution of MUTYH rs3219463 in osteoarthritis (OA) onset was evaluated in association with 5 SNPs of the Calcium Release-Activated Calcium Channel Protein 1 (ORAI1) gene in a case-control study including 350 OA patients and 350 age- and gender-matched healthy controls from the Chinese Han population[146]. Carriers of MUTYH GG or GA rs3219463 genotypes and ORAI1 T allele rs7135617 showed a higher risk of developing OA. Moreover, patients with MUTYH GG or GA rs3219463 showed increased serum MUTYH levels than subjects with AA wild-type genotype. Dysfunction of the cartilage Ca2+ signaling pathway in carriers of MUTYH G allele rs3219463 and ORAl1 T allele rs7135617 was also found[146]. Although these results suggested an association between both SNPs and OA susceptibility, the authors described some limitations in the study, such as the need for in vitro analysis to determine the mechanisms influencing OA risk factors and the small sample size not representative of the population.
Rheumatoid arthritis: Kung et al[147] evaluated the association between genotypic and allelic distributions of the four MUTYH polymorphisms rs3219463, rs3219476, rs3219489, and rs3219493 in a Taiwan Chinese population by comparing 92 individuals with rheumatoid arthritis (RA), a long-term and progressive autoimmune disorder primarily affecting the articular joint, and 192 healthy controls. The study revealed significant differences in genotypic and allelic frequencies regarding MUTYH G/A and MUTYH G/G rs3219463, as well as MUTYH T/G and MUTYH T/T rs3219476 in RA and control groups. Moreover, the genotype frequency of G/− rs3219463 showed a significant increase in RA patients expressing the rheumatoid factor. The authors concluded that rs3219463 and rs3219476 polymorphisms were significantly associated with RA susceptibility, although they highlighted the relatively small number of subjects as a limitation in their study[145]. Subsequently, another investigation found a statistically significant low risk of developing RA in MUTYH rs3219463 G carriers during the analysis of 368 Chinese Han RA patients from Taiwan and 364 healthy controls[148]. In addition, individuals with RA showed 8.8% higher serum MUTYH protein levels compared to healthy controls, suggesting an association between RA and MUTYH rs3219463 polymorphism[148]. To date, the first investigation on reverse insertion in the common AluYb8MUTYH (rs10527342) insertion/deletion SNP was conducted in a selected healthy Chinese population. Homozygous status decreased from 20 to 59 years, suggesting that this condition was related to the onset of age-related or chronic diseases or death. Moreover, the homozygosity was associated with increased levels of 8-oxoG in leukocytes, probably due to impaired DNA repair, as well as a significant increase in plasma interleukin-1[149]. Subsequent studies assessed the link between AluYb8MUTYH and different pathologic conditions.
Type 2 diabetes mellitus: The role and the incidence of AluYb8MUTYH and the other SNPs of BER genes hMTH1 c.247G > A p.Val83Met (V83M, rs4866), and c.-53G > C (rs56387615), c.-23A > G (rs1801129), and c.-18G > T (rs1801126) in the 5′-UTR of OGG1 were evaluated in type 2 diabetes mellitus (T2DM) during a case–control study in a Chinese population[150]. In the analysis by Cao et al[150] AluYb8MUTYH “A/P+P/P” genotype frequency was significantly higher in diabetic patients than in healthy controls. Moreover, the genetic combinations between the AluYb8MUTYH “A/P+P/P” and the hMTH1 G/A variant or the three heterozygous OGG1 genotypes had a synergistic effect that prejudiced the repair of DNA damage induced by oxidative stress and determined an increased risk of T2DM. A previous case-control study of 565 T2DM patients and 565 healthy controls from China found a slight increase in the percentage of AluYb8MUTYH allele P (44.7% vs 40.3%) in T2DM patients compared to controls[151].
Chronic kidney disease: A recent study of a Spanish population, including 548 patients and 174 controls, aimed to identify 38 SNPs from 31 candidate genes associated with chronic kidney disease (CKD) and other related diseases, such as hypertension, diabetes and inflammation[152]. CKD shows a progressive and irreversible loss of renal function and is associated with high genomic instability and the presence of positive markers of inflammation. Of the examined variants in BER genes, the genetic change MUTYH c.1014G > C, p.Gln338His (also known as c.972G > C, p.Gln324His), rs3219489, was not associated with CKD[152]. In contrast, an association between MUTYH rs3219489 and genomic instability was revealed by a recent study on polymorphisms in genes of BER, nucleotide excision repair, phase II metabolism, and antioxidant enzymes in 415 CKD patients and 174 controls[153].
End-stage renal disease: Cai et al[154] reported a Chinese case-control study on end-stage renal disease (ESRD) in which OGG1 Ser326Cys (c.977C > G), MTH1 p.Val83Met, MUTYH p.Gln324His (c.972G > C), and AluYb8MUTYH were examined in 337 patients on hemodialysis (HD) and 404 healthy controls. The frequency of the MUTYH c.972G/G and the AluYb8MUTYH insertion (A/P or P/P) genotypes were statistically higher in HD patients than in controls. Moreover, the combination of MUTYH c.972G/G and AluYb8MUTYH A/P or P/P genotypes or OGG1 c.977G/G further increased the risk of ESRD and the leukocyte DNA 8-oxoG levels in HD patients[154]. Another study by Cai et al[154] investigated the effects of OGG1 c.977C > G, MUTYH c.972G > C, and AluYb8MUTYH polymorphisms on chronic inflammation due to oxidative stress in HD patients, and evaluated the plasma levels of the pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-6. The study included 167 patients and 66 healthy controls. HD patients showed significantly increased levels of IL-1β and IL-6. In addition, patients carrying the c.972G/G genotype had higher IL-1β levels than patients with the MUTYH c.972C/C variant and the AluYb8MUTYH genotype was strongly related to increased levels of both IL-1β and IL-6 in HD subjects. The combination of AluYb8MUTYH with OGG1 c.977C > G or MUTYH c.972G > C genotypes was significantly associated with IL-1β and IL-6 levels in HD patients[154]. Patients with the combination of MUTYH c.972 C/G or G/G genotypes and AluYb8MUTYH A/P or P/P genotypes showed significantly higher IL-1β and IL-6 levels compared to those with c.972 C/C and A/A genotypes[154].
Idiopathic pulmonary fibrosis: Is an interstitial lung disease characterized by extracellular matrix deposition and a poor prognosis. By analyzing 277 patients and 810 healthy controls, Zhou et al[54,150] found that idiopathic pulmonary fibrosis was associated with mitochondrial reduced expression of MUTYH1 protein as well as the presence of the variant AluYb8MUTYH. Of the three genotypes observed and defined by the presence (P) or absence (A) of the AluYb8 insertion, the P/P genotype showed unstable mtDNA. Moreover, this was correlated with early age-associated accumulation of ROS-induced oxidative DNA damage in lung tissues and a reduced capacity of DNA repair leading to increased 8-oxoG in dysfunctional mitochondria[54,155]. Indeed, AluYb8MUTYH decreased the rate of mtDNA turnover, reduced the expression of type 1 MUTYH protein and affected MUTYH-induced BER in mitochondria[155].
This review highlights the alterations in MUTYH gene and/or deregulated activity of MUTYH glycosylase, which causes a lack of its anti-oxidant function against the onset of non-polyposis diseases. In order to increase our understanding of the involvement of MUTYH in the pathogenesis of diseases different to colon polyposis and CRC, we analyzed and summarized the current literature concerning this topic. We evaluated the role of MUTYH on susceptibility to developing cancers and diseases such as neurological and ocular degenerative disorders. As the mechanisms of DNA repair are very important in protecting humans from cancer and avoiding disease, we also evaluated the incidence of genetic variants and SNPs in MUTYH gene able to influence the onset of neoplastic and non-neoplastic disorders.
The risk of CRC in MAP patients was reported to range from 43% to 63% at age 60 years and up to 80%-90% in the absence of timely surveillance. It is less certain in MUTYH mono-allelic carriers even though a two-fold increase in CRC risk has been reported in some studies, which is small and not clinically relevant. This may be due to several factors, such as the rarity of the allele, and possible modifying effects such as increased risk in cases with early age of onset. MUTYH alterations have also been reported in other cancers. Mutations in this gene can contribute to the development of sporadic GC and may act as an independent predictor of poor survival in patients affected by this tumor. The same cannot be said for BC in which the association with MUTYH variants has resulted in controversial findings. Pathogenic variants of MUTYH were also identified in SC, in particular acinic cell carcinoma, an aggressive phenotype with lymph node metastasis, and in individuals with a high frequency of basal cell carcinoma. Both tumors also had mutations in other genes. Increased knowledge of the mutation spectrum may improve the diagnosis and treatment of these types of carcinoma, taking into account the risk stratification of patients. Patients affected by these two cancers might benefit from multigene cancer-susceptibility panel testing, similar to that for gastrointestinal cancer. Interestingly, MUTYH may be a new therapeutic target for PDAC, as its inhibition has been shown to reduce the survival of cancer cells and increase chemo-sensitivity in vitro.
With regard to neurological degeneration in stress conditions, the MUTYH 47-kDa molecule could have a protective anti-oxidative stress function in the remaining healthy neurons in patients with Parkinson's disease. In AD, a correlation between MUTYH c.972G > C SNP and polymorphisms of other BER genes was observed. However, another study confirmed that no MUTYH polymorphism was strongly associated with AD. In Friedreich's ataxia, high levels of MUTYH were observed as consequence of decreased levels of frataxin, responsible for Fe-S cluster deregulation and oxidative DNA damage due to increased 8-oxo-G in microglia. In Huntington's disease, the alteration of MUTYH with aberrant BER may influence the trinucleotide expansion in HTT protein correlated with increased ROS production. MUTYH might also have a role in RP, since in conditions of severe oxidative DNA damage it mediates microglial activation with retina inflammation and photoreceptor degeneration. Finally, an aberrant DNA repair mechanism resulting in MUTYH mutation has been reported in NF. SNPs of DNA repair genes are little studied in genome-wide association studies due to a very limited coverage of these genes and to small patient populations. However, recently a robust study reported a 2-fold increased risk of LC in heterozygotes for MUTYH p.Gln324His, independent of tobacco smoke. Other findings reported evidence of increased colorectal and LC risk in the presence of the same variant. The involvement of MUTYH in HCC has been well studied. A significant association between the intronic MUTYH SNP (rs3219487) has been found as well as reduced enzymatic activity in MUTYH-null mice that did not develop tumors after an antioxidant-rich diet. MUTYH p.Gln324His has been associated with significantly poor prognostic factors in cervical carcinoma, such as cell differentiation grade and lymph node metastasis. Interestingly, these findings support the use of this variant as an early indicator of prognosis for patients at risk of cervical carcinoma and HPV infection. MUTYH SNP (rs3219489) and AluYb8MUTYH insertion/deletion polymorphisms have also been suggested to significantly increase susceptibility to the development of other diseases, such as T2DM, OA and AR.
Therefore, we conclude that the involvement of MUTYH mutations and/or genetic variants or alterations in the corresponding proteins could have the potential to identify new predictive and diagnostic biomarkers for diseases other than MAP. Moreover, MUTYH modifications could represent therapeutic targets to develop more efficient treatment approaches and strategies for these diseases.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ju SQ, Tarnawski AS S-Editor: Zhang H L-Editor: Webster JR E-Editor: Wang LL
| 1. | Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-- > T:A mutations in colorectal tumors. Nat Genet. 2002;30:227-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 924] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 2. | Aceto G, Curia MC, Veschi S, De Lellis L, Mammarella S, Catalano T, Stuppia L, Palka G, Valanzano R, Tonelli F, Casale V, Stigliano V, Cetta F, Battista P, Mariani-Costantini R, Cama A. Mutations of APC and MYH in unrelated Italian patients with adenomatous polyposis coli. Hum Mutat. 2005;26:394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem. 2001;276:5547-5555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 163] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 4. | Tsai-Wu JJ, Su HT, Wu YL, Hsu SM, Wu CH. Nuclear localization of the human mutY homologue hMYH. J Cell Biochem. 2000;77:666-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 676] [Cited by in RCA: 886] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 6. | Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 285] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 7. | Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol. 1992;174:6321-6325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 549] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 8. | McLennan AG. The MutT motif family of nucleotide phosphohydrolases in man and human pathogens (review). Int J Mol Med. 1999;4:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. 2013;743-744:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 10. | Lu AL, Fawcett WP. Characterization of the recombinant MutY homolog, an adenine DNA glycosylase, from yeast Schizosaccharomyces pombe. J Biol Chem. 1998;273:25098-25105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Doi T, Yonekura S, Tano K, Yasuhira S, Yonei S, Zhang QM. The Shizosaccharomyces pombe homolog (SpMYH) of the Escherichia coli MutY is required for removal of guanine from 8-oxoguanine/guanine mispairs to prevent G:C to C:G transversions. J Radiat Res. 2005;46:205-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 12. | Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-- > T:A mutations. Hum Mol Genet. 2002;11:2961-2967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 271] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 13. | Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, Thomas HJ, Tomlinson IP. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 562] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 14. | Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 465] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 15. | Møller P, Løhr M, Folkmann JK, Mikkelsen L, Loft S. Aging and oxidatively damaged nuclear DNA in animal organs. Free Radic Biol Med. 2010;48:1275-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Rangaswamy S, Pandey A, Mitra S, Hegde ML. Pre-Replicative Repair of Oxidized Bases Maintains Fidelity in Mammalian Genomes: The Cowcatcher Role of NEIL1 DNA Glycosylase. Genes (Basel). 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1040] [Cited by in RCA: 1053] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 18. | Kim YJ, Wilson DM. Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 234] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 19. | Turgeon MO, Perry NJS, Poulogiannis G. DNA Damage, Repair, and Cancer Metabolism. Front Oncol. 2018;8:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 20. | Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 21. | Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 104] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Oka S, Leon J, Tsuchimoto D, Sakumi K, Nakabeppu Y. MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death. Oncogenesis. 2014;3:e121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Banda DM, Nuñez NN, Burnside MA, Bradshaw KM, David SS. Repair of 8-oxoG:A mismatches by the MUTYH glycosylase: Mechanism, metals and medicine. Free Radic Biol Med. 2017;107:202-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 24. | D'Agostino VG, Minoprio A, Torreri P, Marinoni I, Bossa C, Petrucci TC, Albertini AM, Ranzani GN, Bignami M, Mazzei F. Functional analysis of MUTYH mutated proteins associated with familial adenomatous polyposis. DNA Repair (Amst). 2010;9:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 25. | Au KG, Cabrera M, Miller JH, Modrich P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proc Natl Acad Sci USA. 1988;85:9163-9166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 125] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 26. | Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med. 2010;49:587-596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 27. | Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 658] [Cited by in RCA: 703] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 28. | Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Jansson K, Blomberg A, Sunnerhagen P, Alm Rosenblad M. Evolutionary loss of 8-oxo-G repair components among eukaryotes. Genome Integr. 2010;1:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 30. | Engstrom LM, Brinkmeyer MK, Ha Y, Raetz AG, Hedman B, Hodgson KO, Solomon EI, David SS. A zinc linchpin motif in the MUTYH glycosylase interdomain connector is required for efficient repair of DNA damage. J Am Chem Soc. 2014;136:7829-7832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Luncsford PJ, Chang DY, Shi G, Bernstein J, Madabushi A, Patterson DN, Lu AL, Toth EA. A structural hinge in eukaryotic MutY homologues mediates catalytic activity and Rad9-Rad1-Hus1 checkpoint complex interactions. J Mol Biol. 2010;403:351-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941-950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1016] [Cited by in RCA: 933] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 33. | UniProt. UniProtKB - E5KP25 (E5KP25_HUMAN). Available from: https://www.uniprot.org/uniprot/E5KP25. |
| 34. | Ensembl. Functional domains of MUTYH protein. Available from: https://www.ensembl.org/index.html. |
| 35. | Kairupan C, Scott RJ. Base excision repair and the role of MUTYH. Hered Cancer Clin Pract. 2007;5:199-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Chmiel NH, Golinelli MP, Francis AW, David SS. Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001;29:553-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Brinkmeyer MK, David SS. Distinct functional consequences of MUTYH variants associated with colorectal cancer: Damaged DNA affinity, glycosylase activity and interaction with PCNA and Hus1. DNA Repair (Amst). 2015;34:39-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 38. | Dorn J, Ferrari E, Imhof R, Ziegler N, Hübscher U. Regulation of human MutYH DNA glycosylase by the E3 ubiquitin ligase mule. J Biol Chem. 2014;289:7049-7058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Lapucci A, Pittelli M, Rapizzi E, Felici R, Moroni F, Chiarugi A. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol Pharmacol. 2011;79:932-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 40. | Montero J, Dutta C, van Bodegom D, Weinstock D, Letai A. p53 regulates a non-apoptotic death induced by ROS. Cell Death Differ. 2013;20:1465-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 41. | Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135-11142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 151] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 42. | Niessen RC, Sijmons RH, Ou J, Olthof SG, Osinga J, Ligtenberg MJ, Hogervorst FB, Weiss MM, Tops CM, Hes FJ, de Bock GH, Buys CH, Kleibeuker JH, Hofstra RM. MUTYH and the mismatch repair system: partners in crime? Hum Genet. 2006;119:206-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Hwang BJ, Jin J, Gao Y, Shi G, Madabushi A, Yan A, Guan X, Zalzman M, Nakajima S, Lan L, Lu AL. SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol Biol. 2015;16:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Boldogh I, Milligan D, Lee MS, Bassett H, Lloyd RS, McCullough AK. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802-2809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 109] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Boehm EM, Gildenberg MS, Washington MT. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes. 2016;39:231-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 212] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 46. | Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1061] [Cited by in RCA: 1098] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 47. | Pérez-Palma E, Gramm M, Nürnberg P, May P, Lal D. Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database. Nucleic Acids Res. 2019;47:W99-W105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 48. | Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst). 2004;3:1009-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 238] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 49. | Lee HM, Hu Z, Ma H, Greeley GH, Wang C, Englander EW. Developmental changes in expression and subcellular localization of the DNA repair glycosylase, MYH, in the rat brain. J Neurochem. 2004;88:394-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 50. | Englander EW, Hu Z, Sharma A, Lee HM, Wu ZH, Greeley GH. Rat MYH, a glycosylase for repair of oxidatively damaged DNA, has brain-specific isoforms that localize to neuronal mitochondria. J Neurochem. 2002;83:1471-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Pope MA, Chmiel NH, David SS. Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair (Amst). 2005;4:315-325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 52. | GeneCards. Summaries for MUTYH Gene. Available from: https://www.genecards.org › cgi-bin › carddisp › gene = MUTYH. |
| 53. | Boldinova EO, Khairullin RF, Makarova AV, Zharkov DO. Isoforms of Base Excision Repair Enzymes Produced by Alternative Splicing. Int J Mol Sci. 2019;20:3279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 54. | Zhou W, Sun J, Guo W, Zhuang Y, Xu L, Wang Y. AluYb8 insertion polymorphism in the MUTYH gene impairs mitochondrial DNA maintenance and affects the age of onset of IPF. Aging (Albany NY). 2019;11:933-949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 55. | De Belle I, Wu JX, Sperandio S, Mercola D, Adamson ED. In vivo cloning and characterization of a new growth suppressor protein TOE1 as a direct target gene of Egr1. J Biol Chem. 2003;278:14306-14312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 56. | Lardelli RM, Schaffer AE, Eggens VR, Zaki MS, Grainger S, Sathe S, Van Nostrand EL, Schlachetzki Z, Rosti B, Akizu N, Scott E, Silhavy JL, Heckman LD, Rosti RO, Dikoglu E, Gregor A, Guemez-Gamboa A, Musaev D, Mande R, Widjaja A, Shaw TL, Markmiller S, Marin-Valencia I, Davies JH, de Meirleir L, Kayserili H, Altunoglu U, Freckmann ML, Warwick L, Chitayat D, Blaser S, Çağlayan AO, Bilguvar K, Per H, Fagerberg C, Christesen HT, Kibaek M, Aldinger KA, Manchester D, Matsumoto N, Muramatsu K, Saitsu H, Shiina M, Ogata K, Foulds N, Dobyns WB, Chi NC, Traver D, Spaccini L, Bova SM, Gabriel SB, Gunel M, Valente EM, Nassogne MC, Bennett EJ, Yeo GW, Baas F, Lykke-Andersen J, Gleeson JG. Biallelic mutations in the 3' exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat Genet. 2017;49:457-464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 57. | Duclot F, Kabbaj M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front Behav Neurosci. 2017;11:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 249] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 58. | Nielsen M, Hes FJ, Nagengast FM, Weiss MM, Mathus-Vliegen EM, Morreau H, Breuning MH, Wijnen JT, Tops CM, Vasen HF. Germline mutations in APC and MUTYH are responsible for the majority of families with attenuated familial adenomatous polyposis. Clin Genet. 2007;71:427-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 59. | Filipe B, Baltazar C, Albuquerque C, Fragoso S, Lage P, Vitoriano I, Mão de Ferro S, Claro I, Rodrigues P, Fidalgo P, Chaves P, Cravo M, Nobre Leitão C. APC or MUTYH mutations account for the majority of clinically well-characterized families with FAP and AFAP phenotype and patients with more than 30 adenomas. Clin Genet. 2009;76:242-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 60. | Pezzi A, Roncucci L, Benatti P, Sassatelli R, Varesco L, Di Gregorio C, Venesio T, Pedroni M, Maffei S, Reggiani Bonetti L, Borsi E, Ferrari M, Martella P, Rossi G, Ponz De Leon M. Relative role of APC and MUTYH mutations in the pathogenesis of familial adenomatous polyposis. Scand J Gastroenterol. 2009;44:1092-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 61. | Aceto GM, Fantini F, De Iure S, Di Nicola M, Palka G, Valanzano R, Di Gregorio P, Stigliano V, Genuardi M, Battista P, Cama A, Curia MC. Correlation between mutations and mRNA expression of APC and MUTYH genes: new insight into hereditary colorectal polyposis predisposition. J Exp Clin Cancer Res. 2015;34:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 62. | Boparai KS, Dekker E, Van Eeden S, Polak MM, Bartelsman JF, Mathus-Vliegen EM, Keller JJ, van Noesel CJ. Hyperplastic polyps and sessile serrated adenomas as a phenotypic expression of MYH-associated polyposis. Gastroenterology. 2008;135:2014-2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 129] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 63. | Nielsen M, Poley JW, Verhoef S, van Puijenbroek M, Weiss MM, Burger GT, Dommering CJ, Vasen HF, Kuipers EJ, Wagner A, Morreau H, Hes FJ. Duodenal carcinoma in MUTYH-associated polyposis. J Clin Pathol. 2006;59:1212-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 64. | Vogt S, Jones N, Christian D, Engel C, Nielsen M, Kaufmann A, Steinke V, Vasen HF, Propping P, Sampson JR, Hes FJ, Aretz S. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology. 2009;137:1976-85.e1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 223] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 65. | Sutcliffe EG, Bartenbaker Thompson A, Stettner AR, Marshall ML, Roberts ME, Susswein LR, Wang Y, Klein RT, Hruska KS, Solomon BD. Multi-gene panel testing confirms phenotypic variability in MUTYH-Associated Polyposis. Fam Cancer. 2019;18:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 66. | Kallenberg FGJ, Bastiaansen BAJ, Nio CY, Soeters MR, Boermeester MA, Aalfs CM, Bossuyt PMM, Dekker E. Adrenal Lesions in Patients With (Attenuated) Familial Adenomatous Polyposis and MUTYH-Associated Polyposis. Dis Colon Rectum. 2017;60:1057-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 67. | Lubbe SJ, Di Bernardo MC, Chandler IP, Houlston RS. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol. 2009;27:3975-3980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 148] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 68. | Win AK, Dowty JG, Cleary SP, Kim H, Buchanan DD, Young JP, Clendenning M, Rosty C, MacInnis RJ, Giles GG, Boussioutas A, Macrae FA, Parry S, Goldblatt J, Baron JA, Burnett T, Le Marchand L, Newcomb PA, Haile RW, Hopper JL, Cotterchio M, Gallinger S, Lindor NM, Tucker KM, Winship IM, Jenkins MA. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology. 2014;146:1208-11.e1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 150] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 69. | Nielsen M, Joerink-van de Beld MC, Jones N, Vogt S, Tops CM, Vasen HF, Sampson JR, Aretz S, Hes FJ. Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH-associated polyposis. Gastroenterology. 2009;136:471-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 70. | Lipton L, Halford SE, Johnson V, Novelli MR, Jones A, Cummings C, Barclay E, Sieber O, Sadat A, Bisgaard ML, Hodgson SV, Aaltonen LA, Thomas HJ, Tomlinson IP. Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 2003;63:7595-7599. [PubMed] |
| 71. | Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23:6445-6470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 227] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 72. | Balaguer F, Castellví-Bel S, Castells A, Andreu M, Muñoz J, Gisbert JP, Llor X, Jover R, de Cid R, Gonzalo V, Bessa X, Xicola RM, Pons E, Alenda C, Payá A, Piqué JM; Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Identification of MYH mutation carriers in colorectal cancer: a multicenter, case-control, population-based study. Clin Gastroenterol Hepatol. 2007;5:379-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 108] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Croitoru ME, Cleary SP, Di Nicola N, Manno M, Selander T, Aronson M, Redston M, Cotterchio M, Knight J, Gryfe R, Gallinger S. Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst. 2004;96:1631-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 176] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Cheadle JP, Sampson JR. MUTYH-associated polyposis--from defect in base excision repair to clinical genetic testing. DNA Repair (Amst). 2007;6:274-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 99] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Lorca V, Garre P. Current status of the genetic susceptibility in attenuated adenomatous polyposis. World J Gastrointest Oncol. 2019;11:1101-1114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 76. | Theodoratou E, Campbell H, Tenesa A, Houlston R, Webb E, Lubbe S, Broderick P, Gallinger S, Croitoru EM, Jenkins MA, Win AK, Cleary SP, Koessler T, Pharoah PD, Küry S, Bézieau S, Buecher B, Ellis NA, Peterlongo P, Offit K, Aaltonen LA, Enholm S, Lindblom A, Zhou XL, Tomlinson IP, Moreno V, Blanco I, Capellà G, Barnetson R, Porteous ME, Dunlop MG, Farrington SM. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br J Cancer. 2010;103:1875-1884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 77. | Cleary SP, Cotterchio M, Jenkins MA, Kim H, Bristow R, Green R, Haile R, Hopper JL, LeMarchand L, Lindor N, Parfrey P, Potter J, Younghusband B, Gallinger S. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136:1251-1260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 78. | Webb EL, Rudd MF, Houlston RS. Colorectal cancer risk in monoallelic carriers of MYH variants. Am J Hum Genet. 2006;79:768-71; author reply 771-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 79. | Jones N, Vogt S, Nielsen M, Christian D, Wark PA, Eccles D, Edwards E, Evans DG, Maher ER, Vasen HF, Hes FJ, Aretz S, Sampson JR. Increased colorectal cancer incidence in obligate carriers of heterozygous mutations in MUTYH. Gastroenterology. 2009;137:489-494, 494.e1; quiz 725-494.e1; quiz 726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 80. | Jenkins MA, Croitoru ME, Monga N, Cleary SP, Cotterchio M, Hopper JL, Gallinger S. Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population-based case-family study. Cancer Epidemiol Biomarkers Prev. 2006;15:312-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 128] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 81. | Nielsen M, Infante E, Brand R. MUTYH Polyposis. [accessed 2019 Oct 10]. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2020. [PubMed] |
| 82. | Kantor M, Sobrado J, Patel S, Eiseler S, Ochner C. Hereditary Colorectal Tumors: A Literature Review on MUTYH-Associated Polyposis. Gastroenterol Res Pract. 2017;2017:8693182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 83. | Zhang Y, Newcomb PA, Egan KM, Titus-Ernstoff L, Chanock S, Welch R, Brinton LA, Lissowska J, Bardin-Mikolajczak A, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Garcia-Closas M. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 84. | Wasielewski M, Out AA, Vermeulen J, Nielsen M, van den Ouweland A, Tops CM, Wijnen JT, Vasen HF, Weiss MM, Klijn JG, Devilee P, Hes FJ, Schutte M. Increased MUTYH mutation frequency among Dutch families with breast cancer and colorectal cancer. Breast Cancer Res Treat. 2010;124:635-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 85. | Barnetson RA, Devlin L, Miller J, Farrington SM, Slater S, Drake AC, Campbell H, Dunlop MG, Porteous ME. Germline mutation prevalence in the base excision repair gene, MYH, in patients with endometrial cancer. Clin Genet. 2007;72:551-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 86. | Win AK, Cleary SP, Dowty JG, Baron JA, Young JP, Buchanan DD, Southey MC, Burnett T, Parfrey PS, Green RC, Le Marchand L, Newcomb PA, Haile RW, Lindor NM, Hopper JL, Gallinger S, Jenkins MA. Cancer risks for monoallelic MUTYH mutation carriers with a family history of colorectal cancer. Int J Cancer. 2011;129:2256-2262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 87. | Win AK, Reece JC, Dowty JG, Buchanan DD, Clendenning M, Rosty C, Southey MC, Young JP, Cleary SP, Kim H, Cotterchio M, Macrae FA, Tucker KM, Baron JA, Burnett T, Le Marchand L, Casey G, Haile RW, Newcomb PA, Thibodeau SN, Hopper JL, Gallinger S, Winship IM, Lindor NM, Jenkins MA. Risk of extracolonic cancers for people with biallelic and monoallelic mutations in MUTYH. Int J Cancer. 2016;139:1557-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 88. | Beiner ME, Zhang WW, Zhang S, Gallinger S, Sun P, Narod SA. Mutations of the MYH gene do not substantially contribute to the risk of breast cancer. Breast Cancer Res Treat. 2009;114:575-578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 89. | Out AA, Wasielewski M, Huijts PE, van Minderhout IJ, Houwing-Duistermaat JJ, Tops CM, Nielsen M, Seynaeve C, Wijnen JT, Breuning MH, van Asperen CJ, Schutte M, Hes FJ, Devilee P. MUTYH gene variants and breast cancer in a Dutch case–control study. Breast Cancer Res Treat. 2012;134:219-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 90. | Rizzolo P, Silvestri V, Bucalo A, Zelli V, Valentini V, Catucci I, Zanna I, Masala G, Bianchi S, Spinelli AM, Tommasi S, Tibiletti MG, Russo A, Varesco L, Coppa A, Calistri D, Cortesi L, Viel A, Bonanni B, Azzollini J, Manoukian S, Montagna M, Radice P, Palli D, Peterlongo P, Ottini L. Contribution of MUTYH Variants to Male Breast Cancer Risk: Results From a Multicenter Study in Italy. Front Oncol. 2018;8:583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 91. | Rennert G, Lejbkowicz F, Cohen I, Pinchev M, Rennert HS, Barnett-Griness O. MutYH mutation carriers have increased breast cancer risk. Cancer. 2012;118:1989-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 92. | Young LC, Hays JB, Tron VA, Andrew SE. DNA mismatch repair proteins: potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J Invest Dermatol. 2003;121:435-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Cho HG, Kuo KY, Li S, Bailey I, Aasi S, Chang ALS, Oro AE, Tang JY, Sarin KY. Frequent basal cell cancer development is a clinical marker for inherited cancer susceptibility. JCI Insight. 2018;3:e122744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 94. | Pritchard CC, Smith C, Salipante SJ, Lee MK, Thornton AM, Nord AS, Gulden C, Kupfer SS, Swisher EM, Bennett RL, Novetsky AP, Jarvik GP, Olopade OI, Goodfellow PJ, King MC, Tait JF, Walsh T. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 95. | Görgens H, Müller A, Krüger S, Kuhlisch E, König IR, Ziegler A, Schackert HK, Eckelt U. Analysis of the base excision repair genes MTH1, OGG1 and MUTYH in patients with squamous oral carcinomas. Oral Oncol. 2007;43:791-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 96. | Kim CJ, Cho YG, Park CH, Jeong SW, Nam SW, Kim SY, Lee SH, Yoo NJ, Lee JY, Park WS. Inactivating mutations of the Siah-1 gene in gastric cancer. Oncogene. 2004;23:8591-8596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 97. | Shinmura K, Goto M, Suzuki M, Tao H, Yamada H, Igarashi H, Matsuura S, Maeda M, Konno H, Matsuda T, Sugimura H. Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J Pathol. 2011;225:414-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 98. | Gushima M, Hirahashi M, Matsumoto T, Fujita K, Fujisawa R, Mizumoto K, Nakabeppu Y, Iida M, Yao T, Tsuneyoshi M. Altered expression of MUTYH and an increase in 8-hydroxydeoxyguanosine are early events in ulcerative colitis-associated carcinogenesis. J Pathol. 2009;219:77-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 99. | Na K, Hernandez-Prera JC, Lim JY, Woo HY, Yoon SO. Characterization of novel genetic alterations in salivary gland secretory carcinoma. Mod Pathol. 2020;33:541-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 100. | Mills S. Histology for pathologists. 5th ed. Philadelphia PA: Lippincott Williams Wilkins, 2019. |
| 101. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5543] [Article Influence: 461.9] [Reference Citation Analysis (0)] |
| 102. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12135] [Cited by in RCA: 12965] [Article Influence: 1440.6] [Reference Citation Analysis (2)] |
| 103. | Sharbeen G, McCarroll J, Goldstein D, Phillips PA. Exploiting base excision repair to improve therapeutic approaches for pancreatic cancer. Front Nutr. 2015;2:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 1300] [Article Influence: 100.0] [Reference Citation Analysis (0)] |
| 105. | Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513-1530. [PubMed] |
| 106. | Capellá G, Pera G, Sala N, Agudo A, Rico F, Del Giudicce G, Plebani M, Palli D, Boeing H, Bueno-de-Mesquita HB, Carneiro F, Berrino F, Vineis P, Tumino R, Panico S, Berglund G, Simán H, Nyrén O, Hallmans G, Martinez C, Dorronsoro M, Barricarte A, Navarro C, Quirós JR, Allen N, Key T, Bingham S, Caldas C, Linseisen J, Nagel G, Overvad K, Tjonneland A, Boshuizen HC, Peeters PH, Numans ME, Clavel-Chapelon F, Trichopoulou A, Lund E, Jenab M, Kaaks R, Riboli E, González CA. DNA repair polymorphisms and the risk of stomach adenocarcinoma and severe chronic gastritis in the EPIC-EURGAST study. Int J Epidemiol. 2008;37:1316-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 107. | Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q, Lin D, He F. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378-4384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 181] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 108. | Karahalil B, Bohr VA, Wilson DM. Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum Exp Toxicol. 2012;31:981-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 109. | Costa EF, Santos ES, Liutti VT, Leal F, Santos VC, Rinck-Junior JA, Mariano FV, Coutinho-Camillo CM, Altemani A, Lima CS, Lourenço GJ. Association between polymorphisms in genes related to DNA base-excision repair with risk and prognosis of oropharyngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2016;142:1917-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 110. | Wang J, Guo C, Gong X, Ao F, Huang Y, Huang L, Tang Y, Jiang C, Xie X, Dong Q, Huang M, Li J. The impacts of genetic polymorphisms in genes of base excision repair pathway on the efficacy and acute toxicities of (chemo)radiotherapy in patients with nasopharyngeal carcinoma. Oncotarget. 2017;8:78633-78641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 111. | Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 112. | Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 387] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 113. | Miyaishi A, Osawa K, Osawa Y, Inoue N, Yoshida K, Kasahara M, Tsutou A, Tabuchi Y, Sakamoto K, Tsubota N, Takahashi J. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 114. | Singh A, Singh N, Behera D, Sharma S. Genetic Investigation of Polymorphic OGG1 and MUTYH Genes Towards Increased Susceptibility in Lung Adenocarcinoma and its Impact on Overall Survival of Lung Cancer Patients Treated with Platinum Based Chemotherapy. Pathol Oncol Res. 2019;25:1327-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 115. | Raetz AG, Xie Y, Kundu S, Brinkmeyer MK, Chang C, David SS. Cancer-associated variants and a common polymorphism of MUTYH exhibit reduced repair of oxidative DNA damage using a GFP-based assay in mammalian cells. Carcinogenesis. 2012;33:2301-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 116. | Li WQ, Hu N, Hyland PL, Gao Y, Wang ZM, Yu K, Su H, Wang CY, Wang LM, Chanock SJ, Burdett L, Ding T, Qiao YL, Fan JH, Wang Y, Xu Y, Shi JX, Gu F, Wheeler W, Xiong XQ, Giffen C, Tucker MA, Dawsey SM, Freedman ND, Abnet CC, Goldstein AM, Taylor PR. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34:1536-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 117. | Sakurada A, Miyanishi K, Tanaka S, Sato M, Sakamoto H, Kawano Y, Takada K, Nakabeppu Y, Kobune M, Kato J. An intronic single nucleotide polymorphism in the MUTYH gene is associated with increased risk for HCV-induced hepatocellular carcinoma. Free Radic Biol Med. 2018;129:88-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 118. | Chen H, Wang H, Liu J, Cheng Q, Chen X, Ye F. Association of the MUTYH Gln324His (CAG/CAC) variant with cervical carcinoma and HR-HPV infection in a Chinese population. Medicine (Baltimore). 2019;98:e15359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 119. | Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson's disease. J Neurosci Res. 2007;85:919-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 216] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 120. | Arai T, Fukae J, Hatano T, Kubo S, Ohtsubo T, Nakabeppu Y, Mori H, Mizuno Y, Hattori N. Up-regulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson's disease. Acta Neuropathol. 2006;112:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 121. | Sliwinska A, Sitarek P, Toma M, Czarny P, Synowiec E, Krupa R, Wigner P, Bialek K, Kwiatkowski D, Korycinska A, Majsterek I, Szemraj J, Galecki P, Sliwinski T. Decreased expression level of BER genes in Alzheimer's disease patients is not derivative of their DNA methylation status. Prog Neuropsychopharmacol Biol Psychiatry. 2017;79:311-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 122. | Abolhassani N, Leon J, Sheng Z, Oka S, Hamasaki H, Iwaki T, Nakabeppu Y. Molecular pathophysiology of impaired glucose metabolism, mitochondrial dysfunction, and oxidative DNA damage in Alzheimer's disease brain. Mech Ageing Dev. 2017;161:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 123. | Sheng Z, Oka S, Tsuchimoto D, Abolhassani N, Nomaru H, Sakumi K, Yamada H, Nakabeppu Y. 8-Oxoguanine causes neurodegeneration during MUTYH-mediated DNA base excision repair. J Clin Invest. 2012;122:4344-4361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 124. | Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 125. | Mantha AK, Sarkar B, Tell G. A short review on the implications of base excision repair pathway for neurons: relevance to neurodegenerative diseases. Mitochondrion. 2014;16:38-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 126. | Shen Y, McMackin MZ, Shan Y, Raetz A, David S, Cortopassi G. Frataxin Deficiency Promotes Excess Microglial DNA Damage and Inflammation that Is Rescued by PJ34. PLoS One. 2016;11:e0151026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 127. | Cossée M, Dürr A, Schmitt M, Dahl N, Trouillas P, Allinson P, Kostrzewa M, Nivelon-Chevallier A, Gustavson KH, Kohlschütter A, Müller U, Mandel JL, Brice A, Koenig M, Cavalcanti F, Tammaro A, De Michele G, Filla A, Cocozza S, Labuda M, Montermini L, Poirier J, Pandolfo M. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 128. | Gomes CM, Santos R. Neurodegeneration in Friedreich's ataxia: from defective frataxin to oxidative stress. Oxid Med Cell Longev. 2013;2013:487534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 129. | Hayashi G, Shen Y, Pedersen TL, Newman JW, Pook M, Cortopassi G. Frataxin deficiency increases cyclooxygenase 2 and prostaglandins in cell and animal models of Friedreich's ataxia. Hum Mol Genet. 2014;23:6838-6847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 130. | Aceto GM, Catalano T, Curia MC. Molecular Aspects of Colorectal Adenomas: The Interplay among Microenvironment, Oxidative Stress, and Predisposition. Biomed Res Int. 2020;2020:1726309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 131. | Talhaoui I, Matkarimov BT, Tchenio T, Zharkov DO, Saparbaev MK. Aberrant base excision repair pathway of oxidatively damaged DNA: Implications for degenerative diseases. Free Radic Biol Med. 2017;107:266-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 132. | Jarem DA, Wilson NR, Schermerhorn KM, Delaney S. Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion. DNA Repair (Amst). 2011;10:887-896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 133. | Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447-452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 347] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 134. | Cilli P, Ventura I, Minoprio A, Meccia E, Martire A, Wilson SH, Bignami M, Mazzei F. Oxidized dNTPs and the OGG1 and MUTYH DNA glycosylases combine to induce CAG/CTG repeat instability. Nucleic Acids Res. 2016;44:5190-5203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 135. | Nakatake S, Murakami Y, Ikeda Y, Morioka N, Tachibana T, Fujiwara K, Yoshida N, Notomi S, Hisatomi T, Yoshida S, Ishibashi T, Nakabeppu Y, Sonoda KH. MUTYH promotes oxidative microglial activation and inherited retinal degeneration. JCI Insight. 2016;1:e87781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 136. | Li Q, Zhao F, Ju Y. Germline mutation of CHEK2 in neurofibromatosis 1 and 2: Two case reports. Medicine (Baltimore). 2018;97:e10894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 137. | Wang W, Shen P, Thiyagarajan S, Lin S, Palm C, Horvath R, Klopstock T, Cutler D, Pique L, Schrijver I, Davis RW, Mindrinos M, Speed TP, Scharfe C. Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing. Nucleic Acids Res. 2011;39:44-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 138. | Kwiatkowski D, Czarny P, Galecki P, Bachurska A, Talarowska M, Orzechowska A, Bobińska K, Bielecka-Kowalska A, Pietras T, Szemraj J, Maes M, Sliwinski T. Variants of Base Excision Repair Genes MUTYH , PARP1 and XRCC1 in Alzheimer's Disease Risk. Neuropsychobiology. 2015;71:176-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 139. | Lillenes MS, Støen M, Günther CC, Selnes P, Stenset VT, Espeseth T, Reinvang I, Fladby T, Tønjum T. Mitochondrial transcription factor A (TFAM) rs1937 and AP endonuclease 1 (APE1) rs1130409 alleles are associated with reduced cognitive performance. Neurosci Lett. 2017;645:46-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 140. | Synowiec E, Wójcik KA, Czubatka A, Polakowski P, Izdebska J, Szaflik J, Błasiak J, Szaflik JP. Lack of association between polymorphisms of the DNA base excision repair genes MUTYH and hOGG1 and keratoconus in a Polish subpopulation. Arch Med Sci. 2015;11:1101-1110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 141. | Cuchra M, Markiewicz L, Mucha B, Pytel D, Szymanek K, Szemraj J, Szaflik J, Szaflik JP, Majsterek I. The role of base excision repair in the development of primary open angle glaucoma in the Polish population. Mutat Res. 2015;778:26-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 142. | Szaflik JP, Cuchra M, Przybylowska-Sygut K, Dziki L, Kurowska AK, Gacek M, Drzewoski J, Szaflik J, Majsterek I. Association of the 399Arg/Gln XRCC1, the 194 Arg/Trp XRCC1, the 326Ser/Cys OGG1, and the 324Gln/His MUTYH gene polymorphisms with clinical parameters and the risk for development of primary open-angle glaucoma. Mutat Res. 2013;753:12-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 143. | Synowiec E, Blasiak J, Zaras M, Szaflik J, Szaflik JP. Association between polymorphisms of the DNA base excision repair genes MUTYH and hOGG1 and age-related macular degeneration. Exp Eye Res. 2012;98:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 144. | Czarny P, Kwiatkowski D, Galecki P, Talarowska M, Orzechowska A, Bobinska K, Bielecka-Kowalska A, Szemraj J, Maes M, Su KP, Sliwinski T. Association between single nucleotide polymorphisms of MUTYH, hOGG1 and NEIL1 genes, and depression. J Affect Disord. 2015;184:90-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 145. | Czarny P, Kwiatkowski D, Toma M, Kubiak J, Sliwinska A, Talarowska M, Szemraj J, Maes M, Galecki P, Sliwinski T. Impact of Single Nucleotide Polymorphisms of Base Excision Repair Genes on DNA Damage and Efficiency of DNA Repair in Recurrent Depression Disorder. Mol Neurobiol. 2017;54:4150-4159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 146. | Zhang S, Guo H, Chen D, Chen X, Jin Q. MUTYH and ORAI1 polymorphisms are associated with susceptibility to osteoarthritis in the Chinese Han population. J Int Med Res. 2018;46:2292-2300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 147. | Kung YJ, Tsai KS, Huang CM, Lin HJ, Chen TH, Hsu YA, Chang CY, Huang YS, Wan L. MUTYH Gene Polymorphisms as Risk Factors for Rheumatoid Arthritis. Biomed Res Int. 2015;2015:893796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 148. | Chen SY, Chen HH, Huang YC, Liu SP, Lin YJ, Lo SF, Chang YY, Lin HW, Huang CM, Tsai FJ. Polymorphism and protein expression of MUTYH gene for risk of rheumatoid arthritis. BMC Musculoskelet Disord. 2017;18:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 149. | Sun C, Chen H, Guo W, Zhang K, Qi Q, Gu X, Zhu D, Wang Y. A common mutation of the MYH gene is associated with increased DNA oxidation and age-related diseases. Free Radic Biol Med. 2010;48:430-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 150. | Cao L, Zhou W, Zhu Y, Guo W, Cai Z, He X, Xie Y, Li X, Zhu D, Wang Y. Combined analysis of polymorphism variants in hMTH1, hOGG1 and MUTYH genes on the risk of type 2 diabetes in the Chinese population. Gene. 2013;519:50-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 151. | Chen H, Sun C, Guo W, Meng R, Du H, Qi Q, Gu X, Li L, Zhang K, Zhu D, Wang Y. AluYb8 insertion in the MUTYH gene is related to increased 8-OHdG in genomic DNA and could be a risk factor for type 2 diabetes in a Chinese population. Mol Cell Endocrinol. 2011;332:301-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 152. | Corredor Z, Filho MIDS, Rodríguez-Ribera L, Velázquez A, Hernández A, Catalano C, Hemminki K, Coll E, Silva I, Diaz JM, Ballarin J, Vallés Prats M, Calabia Martínez J, Försti A, Marcos R, Pastor S. Genetic Variants Associated with Chronic Kidney Disease in a Spanish Population. Sci Rep. 2020;10:144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 153. | Corredor Z, da Silva Filho MI, Rodríguez-Ribera L, Catalano C, Hemminki K, Coll E, Silva I, Diaz JM, Ballarin JA, Henández A, Försti A, Marcos R, Pastor S. Loci associated with genomic damage levels in chronic kidney disease patients and controls. Mutat Res. 2020;852:503167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 154. | Cai Z, Guo W, Chen H, Tao J, Cao L, Sun W, Wang Y. Base excision repair gene polymorphisms are associated with inflammation in patients undergoing chronic hemodialysis. Biochem Biophys Res Commun. 2012;424:611-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 155. | Guo W, Zheng B, Cai Z, Xu L, Guo D, Cao L, Wang Y. The polymorphic AluYb8 insertion in the MUTYH gene is associated with reduced type 1 protein expression and reduced mitochondrial DNA content. PLoS One. 2013;8:e70718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |