Copyright
©The Author(s) 2025.
World J Clin Oncol. Apr 24, 2025; 16(4): 104182
Published online Apr 24, 2025. doi: 10.5306/wjco.v16.i4.104182
Published online Apr 24, 2025. doi: 10.5306/wjco.v16.i4.104182
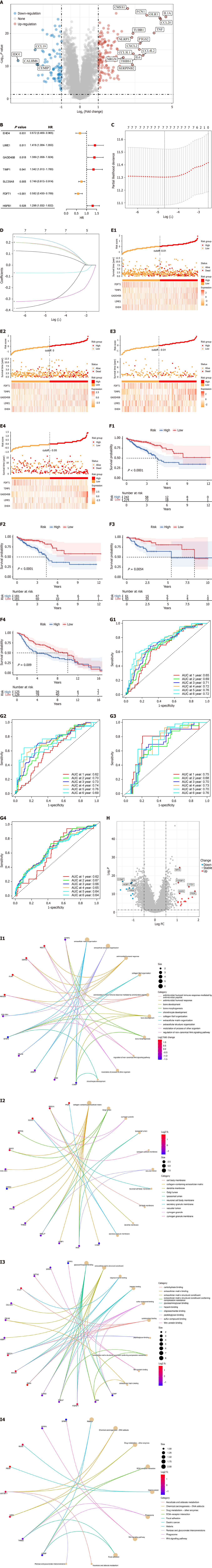
Figure 1 Establishment of prognostic model and enrichment analysis.
A: Volcano plot depicting differential gene expression; B: Forest plot highlighting notable mRNAs; C and D: Least absolute shrinkage and selection operator model selection and feature identification; E: Distribution of risk scores and heatmap of mRNA expression in The Cancer Genome Atlas colorectal cancer dataset. E1: Overall risk score distribution. E2: Training set. E3: Test set. E4: Gene Expression Omnibus (GEO) datasets; F: Kaplan-Meier survival analysis. F1: Total risk score. F2: Training set. F3: Test set. F4: GEO datasets; G: Time-dependent receiver operating characteristic curves. G1: Entire dataset. G2-G4: Training set, test set, and GEO datasets, respectively; H: Volcano plot of differential genes; I: Enrichment results. I1: Biological processes. I2: Molecular function. I3: Cellular component. I4: Kyoto Encyclopedia of Genes and Genomes pathways. HR: Hazard ratio; AUC: Area under the curve.
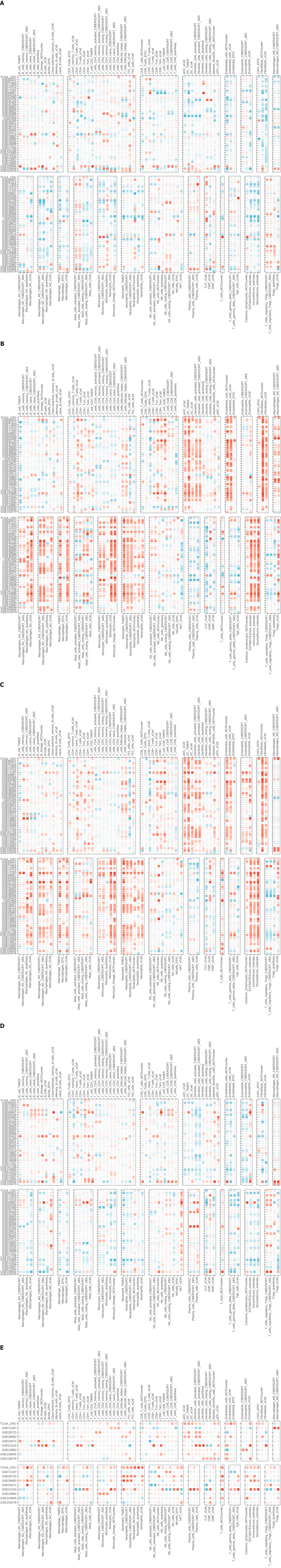
Figure 2 Spearman correlation between FDFT1, TIMP1, GADD45B, LIME1, EHD4 expression and immune cell infiltration in colorectal cancer, analyzed using multiple algorithms.
A: FDFT1; B: TIMP1; C: GADD45B; D: LIME1; E: EHD4.
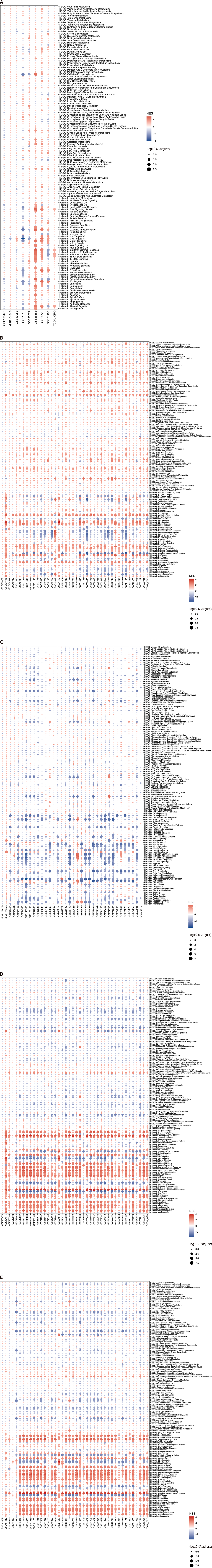
Figure 3 Gene Set Enrichment Analysis.
When enrichment score/Normalized Enrichment Score (NES) is negative, core molecules are primarily concentrated in the low expression group (right side), indicating significant enrichment in the low expression group. When enrichment score/NES is positive, core molecules are primarily concentrated in the high expression group (left side), indicating significant enrichment in the high expression group. A: EHD4; B: FDFT1; C: GADD45B; D: LIME1; E: TIMP1.
Figure 4 Kaplan-Meier survival curves.
The Kaplan-Meier survival curves are presented with the horizontal axis representing survival time (t) and the vertical axis indicating the probability of an individual surviving beyond time t. High expression groups are shown in red, while low expression groups are depicted in blue. Differences between survival curves were evaluated using a log-rank test with a significance threshold of P < 0.05. X represents different EHD4, FDFT1, GADD45B, LIME1, TIMP1 expression levels of the patients. The Y-axis represents proportions, with the total ratio of dead to alive patients in groups with different expression levels being 1, with red representing patients who died and blue representing patients who were alive. A: FDFT1; B: TIMP1; C: GADD45B; D: LIME1; E: EHD4. From left to right are overall survival (OS), disease-specific survival (DSS), progression-free interval, and disease-free interval; F: EHD4, LIME1, GADD45B survival_ OS-status; G1: GSE17537 _GADD45B_Survival_ Chi-Square_test_RFS; G2: GSE17537_ GADD45B_ Survival _Chi-Square_test_OS; H1: GSE17536 _ GADD45B _ Survival_Chi-Square _test _RFS; H2: GSE17536_GADD45B_ Survival_ Chi-Square_ test_ OS; I: TIMP1. CRC. Survival. Chi-Square. Test; J1: GSE87211_ TIMP1_ Survival_ Chi -Square_test_RFS; J2: GSE87211_ TIMP1_Survival_Chi-Square _test_DSS; J3: GSE29621 _TIMP1 _ Survival_ Chi-Square _test_RFS; K1: GSE17536_ TIMP1_ Survival_ Chi-Square _test_RFS; K2: GSE17536_ TIMP1_ Survival_ Chi-Square _test_DSS; L: TIMP1, survival of FDFT1, _OS-Status; M1: GSE72970_FDFT1 _Survival_ Chi-Square_test_PFS; M2: GSE72970 _FDFT1_Survival_Chi-Square_test_OS; N: GSE12945_ FDFT1_ Survival _Chi-Square_test_OS. CRC: Colorectal cancer.
Figure 5 GSVA analysis and cell cycle analysis.
A: Pathway-GSVA-metabolism analysis; B-F: Median expression levels of EHD4, FDFT1, GADD45B, LIME1, and TIMP1 were used as references. The light red area represents the gene expression distribution. Results from left to right correspond to disease-free interval, disease-specific survival, overall survival, and progression-free interval; G-K: Expression differences of EHD4, FDFT1, GADD45B, LIME1, and TIMP1 across cell cycle stages. The upper and lower boundaries of the boxes represent the interquartile range.
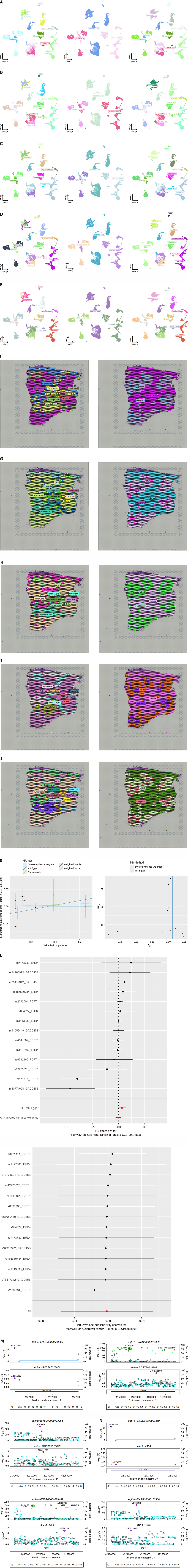
Figure 6 Single-cell transcriptomics, spatial transcriptomics, and genetic analysis.
A-E: Single-cell data distribution after uniform manifold approximation and projection dimensionality reduction. A: EHD4; B: FDFT1; C: GADD45B; D: LIME1; E: TIMP1; F-J: Spatial transcriptome deconvolution and cell localization; F: EHD4; G: FDFT1; H: GADD45B; I: LIME1; J: TIMP1; K and L: Mendelian randomization results; M and N: Genetic variation analysis via expression quantitative trait locus-genome-wide association study colocalization.
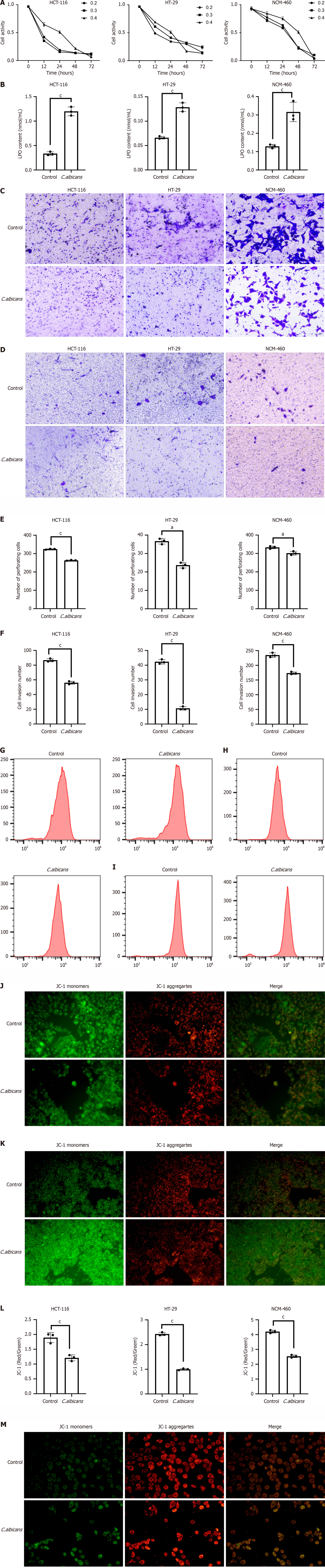
Figure 7 Effects of Candida albicans metabolic mixture on colorectal cancer cell activity, lipid peroxidation content, invasion, migration, reactive oxygen species content, and mitochondrial membrane potential.
A: Candida albicans (C. albicans) metabolite mixture reduced cell viability; B: C. albicans metabolite mixture increased intracellular lipid peroxidation (LPO) levels; C and D: Effects of C. albicans metabolite mixture on cell migration (C) and invasion (D); E and F: Quantified histograms showing effects on cell migration (E) and invasion (F); G-I: Flow cytometry results for reactive oxygen species content. G: HCT-116 cells; H: HT-29 cells; I: NCM-460 cells; J-L: C. albicans metabolite mixture decreased mitochondrial membrane potential. J: HCT-116 cells; K: HT-29 cells; L: Quantitative histogram of mitochondrial membrane potential; M: C. albicans metabolite mixture decreased mitochondrial membrane potential in NCM-460 cells. aP < 0.05, bP < 0.01, cP < 0.001.
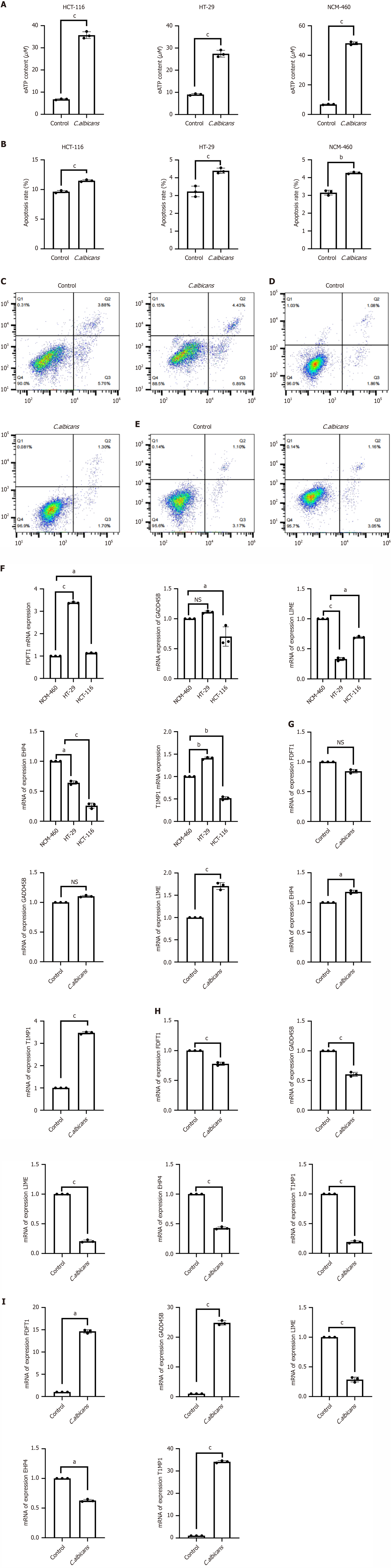
Figure 8 Candida albicans metabolic mixture interferes with extracellular ATP content, apoptosis and PCR detection of prognostic model gene expression in colorectal cancer cells.
A: Detection of extracellular ATP content; B: Histogram depicting cell apoptosis rates; C: Apoptosis rate in HCT-116 cells assessed via flow cytometry; D: Apoptosis rate in HT-29 cells detected by flow cytometry; E: Apoptosis rate in NCM-460 cells evaluated by flow cytometry; F: Expression levels of FDFT1, TIMP1, GADD45B, LIME1, and EHD4 across different cell types measured using real-time quantitative PCR (RT-qPCR); G: RT-qPCR analysis of FDFT1, TIMP1, GADD45B, LIME1, and EHD4 expression in HCT116 cells treated with the Candida albicans (C. albicans) metabolite mixture; H: RT-qPCR analysis of FDFT1, TIMP1, GADD45B, LIME1, and EHD4 expression in HT29 cells treated with the C. albicans metabolite mixture; I: RT-qPCR analysis of FDFT1, TIMP1, GADD45B, LIME1, and EHD4 expression in NCM460 cells treated with the C. albicans metabolite mixture. (aP < 0.05,bP < 0.01, cP < 0.001).
Figure 9 Results of liquid chromatography-mass spectrometry detection of the metabolic mixture of Candida albicans.
A: Superimposed total ion chromatogram for negative [neg-quality control (A1)(QC)] and positive(A2) (pos-QC) samples; B: Extracted ion chromatogram paths for blank samples in negative (B1) and positive (B2) modes; C: Correlation analysis of QC samples in negative (C1) and positive (C2) modes; D: Ring graphs representing metabolite classes in negative (D1) and positive (D2) modes; E: Distribution maps of coefficient of variation values for negative (E1) and positive (E2) modes.The volatility or dispersion of the data is similar, resulting in the two pictures being the same; F: Principal component analysis score plots showing quality spectrum data for samples and QC samples in negative (F1) and positive (F2) groups; G: Cluster analysis results for negative (G1) and positive (G2) modes; H: S-plot from OPLS-DA analysis; I: Dynamic distribution of differential metabolite content; J: Validation of the OPLS-DA model; K: Volcano plot illustrating differential metabolites; L: Scatter plot of differential metabolites; M: Sample-level clustering tree, comparing Candida albicans metabolites mixture (A) and the control group (NC).
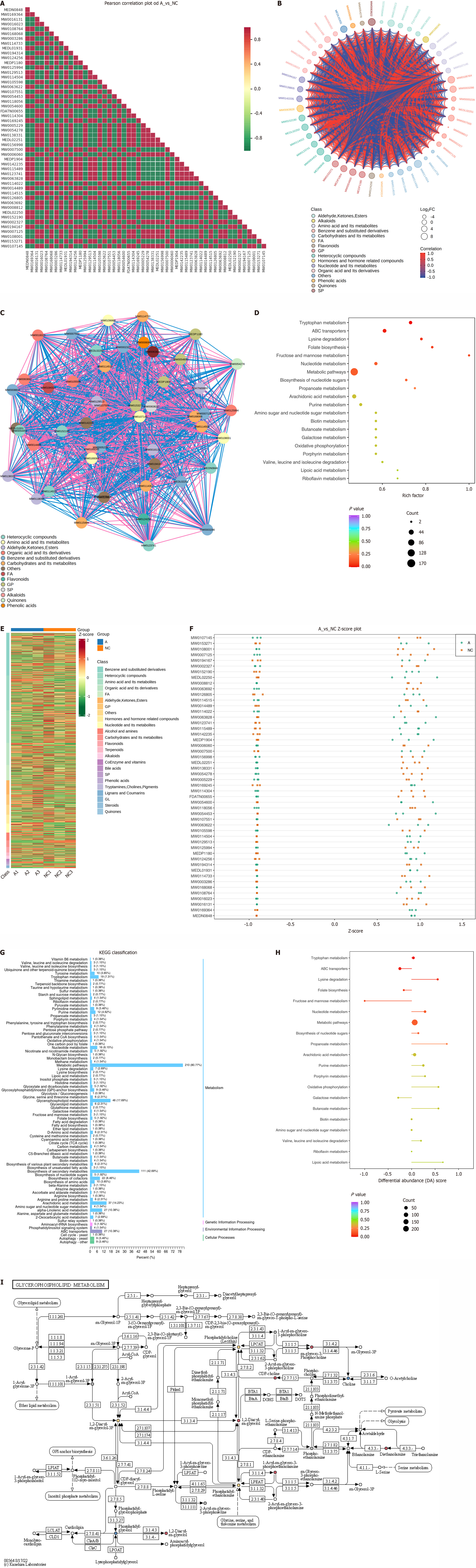
Figure 10 Enrichment analysis of main components of Candida albicans metabolic mixture.
A: Correlation heat map of differential metabolites; B: Chord diagram showing relationships among differential metabolites; C: Correlation network of differential metabolites; D: Kyoto Encyclopedia of Genes and Genomes pathway enrichment map for differential metabolites; E: Cluster heat map of differential metabolites; F: Z-value distribution map for differential metabolites; G: Pathway classification of differential metabolites; H: Abundance score map for differential metabolites; I: Pathway map showing differential metabolite involvement.
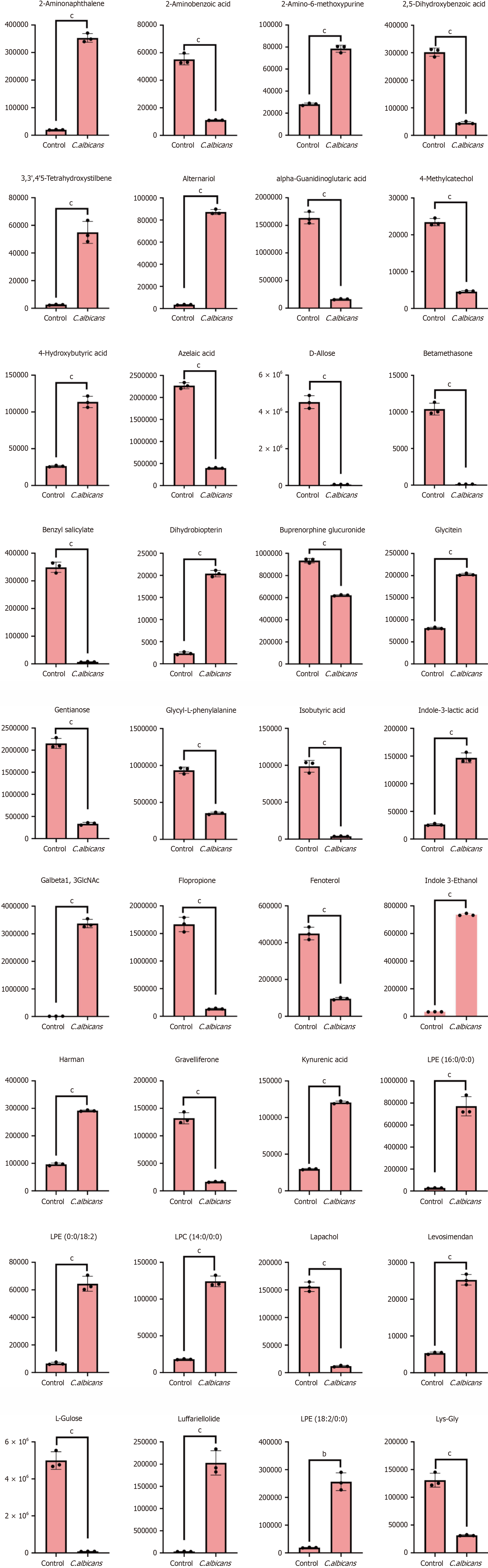
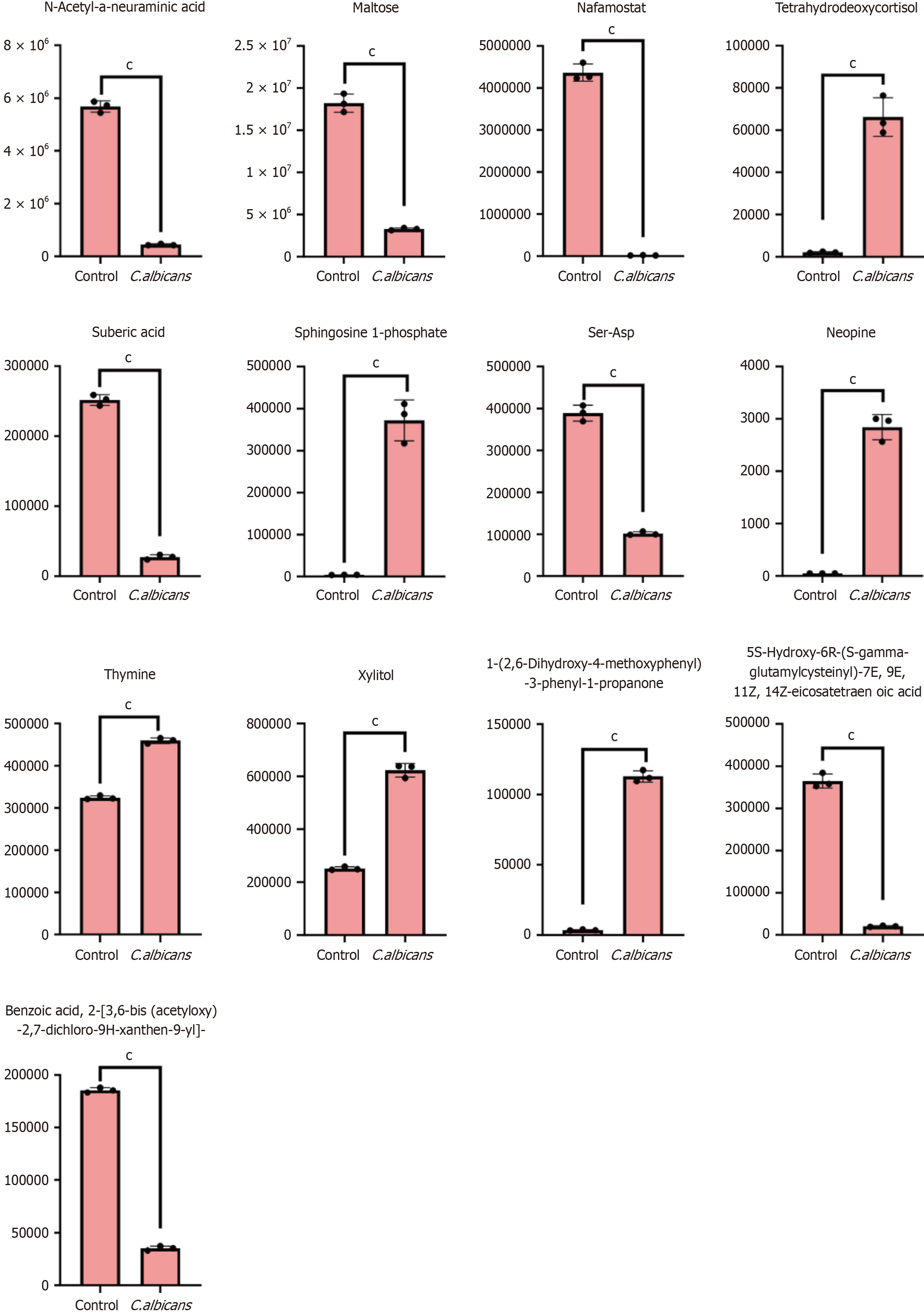
Figure 11 Violin diagram of differential metabolites.
(aP < 0.05, bP < 0.01, cP < 0.001).
Figure 12 Violin diagram of differential metabolites.
(aP < 0.05, bP < 0.01, cP < 0.001).
- Citation: Zhang HL, Zhao R, Wang D, Mohd Sapudin SN, Yahaya BH, Harun MSR, Zhang ZW, Song ZJ, Liu YT, Doblin S, Lu P. Candida albicans and colorectal cancer: A paradoxical role revealed through metabolite profiling and prognostic modeling. World J Clin Oncol 2025; 16(4): 104182
- URL: https://www.wjgnet.com/2218-4333/full/v16/i4/104182.htm
- DOI: https://dx.doi.org/10.5306/wjco.v16.i4.104182