Published online Aug 15, 2014. doi: 10.4291/wjgp.v5.i3.213
Revised: April 8, 2014
Accepted: May 29, 2014
Published online: August 15, 2014
Processing time: 218 Days and 13.7 Hours
Escherichia coli (E. coli), and particularly the adherent invasive E. coli (AIEC) pathotype, has been increasingly implicated in the ethiopathogenesis of Crohn’s disease (CD). E. coli strains with similar pathogenic features to AIEC have been associated with other intestinal disorders such as ulcerative colitis, colorectal cancer, and coeliac disease, but AIEC prevalence in these diseases remains largely unexplored. Since AIEC was described one decade ago, substantial progress has been made in deciphering its mechanisms of pathogenicity. However, the molecular bases that characterize the phenotypic properties of this pathotype are still not well resolved. A review of studies focused on E. coli populations in inflammatory bowel disease (IBD) is presented here and we discuss about the putative role of this species on each IBD subtype. Given the relevance of AIEC in CD pathogenesis, we present the latest research findings concerning AIEC host-microbe interactions and pathogenicity. We also review the existing data regarding the prevalence and abundance of AIEC in CD and its association with other intestinal diseases from humans and animals, in order to discuss the AIEC disease- and host-specificity. Finally, we highlight the fact that dietary components frequently found in industrialized countries may enhance AIEC colonization in the gut, which merits further investigation and the implementation of preventative measures.
Core tip: In this review we critically revise the findings on Escherichia coli (E. coli) populations associated with Crohn’s disease and ulcerative colitis. Then we focus on adherent invasive E. coli (AIEC), especially in its mechanisms of pathogenicity and epidemiology. We discuss about AIEC disease- and host-specificity and we underline the importance of discovering specific molecular tools to detect AIEC for further epidemiologic studies. Finally we point out to a putative role of diet on AIEC gut colonization.
-
Citation: Martinez-Medina M, Garcia-Gil LJ.
Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasiveEscherichia coli pathogenicity. World J Gastrointest Pathophysiol 2014; 5(3): 213-227 - URL: https://www.wjgnet.com/2150-5330/full/v5/i3/213.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v5.i3.213
The intestinal microbiota has been implicated in the pathogenesis of Crohn’s disease (CD) and ulcerative colitis (UC), the main idiopathic inflammatory bowel diseases (IBDs)[1]. CD patients demonstrate an altered intestinal microbial community, and the dysbioses present in colonic CD and ileal CD are different[2]. In contrast, a specific dysbiosis in UC is starting to be defined, although differences between studies have hampered attempts to reach a clear consensus to date[2-5]. A number of culture-based and molecular-based studies support the theory that Escherichia coli (E. coli) is a microbiological factor implicated in CD, but some controversy exists regarding its role in UC[2,6-17]. In this section, we examine data on E. coli populations in CD and UC related to abundance, association with disease activity, translocation of the gut mucosa, and pathogenic features of the strains to highlight the evidence that supports or refutes putative implications for this bacterium in each IBD subtype.
Several independent studies based on quantitative Polymerase Chain Reaction (PCR) have indicated that E. coli is augmented in CD patients in comparison with controls[2,6,11,13]. However, differences are especially significant for CD patients with ileal disease, and no clear association with colonic or ileocolonic CD has been demonstrated. On average, in our cohort, E. coli 16S rRNA gene copies accounted for 14% and 33% of total bacteria 16S rRNA gene copies in healthy subjects and ileal CD patients, respectively (P < 0.001)[13]. Of note, a higher abundance of E. coli was observed in active CD patients than in patients in remission[6,11,18]. Accordingly, a previous study using Fluorescent In Situ Hybridization (FISH) demonstrated increased E. coli numbers in the epithelium and within the lamina propria in active CD patients compared to inactive CD patients[14]. In addition, we determined that higher numbers of this species correlated with a reduced amount of time before relapse[11]. These findings are in agreement with previous data reporting that the higher numbers of E. coli isolated from the neoterminal ileum of CD patients are associated with early recurrence of the disease[7], and that high levels of antibodies against the E. coli outer membrane protein C (OmpC) correlate with disease progression, longer duration, and greater need for surgery among CD patients[19-21].
There is substantial controversy regarding the abundance of E. coli in the colonic mucosa of UC patients (Table 1). Several works have consistently reported no increase with respect to healthy subjects[2,6,7,11-13], arguing against a putative role for E. coli in UC, while others have reported increased E. coli abundance in UC patients[8,10,14,16,18,22,23]. As in the majority of these studies both CD and UC patients were analyzed, these controversial observations can not be explained by differences in methodology between studies. We postulate that they can be attributable to differences in the disease severity of the patients included in the studies, as increased numbers of E. coli have been associated with activity status in UC patients. Using FISH, epithelium-associated E. coli were found to be more abundant in active UC compared to inactive UC or controls[14], and quantitative PCR indicated that increased numbers of E. coli were present in active UC patients compared to inactive UC patients[22] as well as in inflamed vs non-inflamed UC tissue[23].
| Ref. | Method | Samples | Comments |
| Increased E. coli abundance in CD but not UC | |||
| Martin et al[12] | Culture | Biopsies | Specially hemagglutinin-positive strains |
| Martinez-Medina et al[13] | qPCR | Biopsies | Specially in ileal CD |
| Lopez-Siles et al[11] | qPCR | Biopsies | Specially in active CD |
| Darfeuille-Michaud et al[7]1 | culture | Biopsies | Specially in ileal lesions |
| Baumgart et al[6]1 | qPCR | Biopsies | Specially in ileal CD |
| Willing et al[2]1 | qPCR | Biopsies | Specially in ileal CD |
| Increased E. coli abundance in CD and UC | |||
| Mylonaki et al[14] | FISH | Biopsies | Specially in active UC patients |
| Kotlowski et al[10] | culture | Biopsies | |
| Rehman et al[16] | cloning | Biopsies | |
| Fujita et al[8] | qPCR | Biopsies | |
| Schwiertz et al[18] | qPCR | Feces | Specially in active CD patients |
| Sha et al[22] | qPCR | Feces | Specially in active UC and CD patients |
| Pilarczyk-Zurek et al[23]2 | qPCR | Biopsies | Specially in inflamed UC tissue |
Altogether, substantial evidence supports an overgrowth of E. coli in ileal CD patients, while there is still no convincing data that exists for other IBD subtypes. Further studies aimed at comparing the abundance of E. coli in IBD patients categorized by disease subtype and assessing any correlation with activity status of the disease would shed light on the role of this bacterium in each IBD subtype and its putative application as a diagnostic and/or prognostic tool.
E. coli has been found in the mucus layer, close to the intestinal epithelial cells and in ulcers of both CD and UC patients[24,25]. Translocation of the intestinal mucosa has been primarily observed in CD[6] and higher amounts of intracellular E. coli were detected in inflamed compared to non-inflamed mucosa[6,26]. With FISH and immunohistochemistry, E. coli has been detected scattered within the lamina propria, either in the extracellular space or inside macrophages, as well as in the subserosal layer, the perivascular areas of the submucosa, the muscle layer, and in germinal centers of lymph follicles of CD patients[8,14,27]. A recent study using high throughput sequencing indicated a greater proportion of E. coli reads in the lymph nodes of ileal CD patients than other CD patients[28]. Interestingly, E. coli DNA was also more frequently found in the granulomas of CD patients (80%) than in non-CD control patients (10%) in a study that used Laser Capture Microdissection and PCR[29]. In contrast, E. coli has not been frequently found to translocate the mucosa of UC patients[8,24,25], although some controversy exists as some authors have detected E. coli in the lamina propria of UC patients[14,27].
The majority of the aforementioned studies are based on techniques that do not distinguish viable bacteria from dead bacteria. Further studies should study the viability of translocated E. coli, particularly in lymph nodes and granulomas, as these locations would be more relevant to establish a link between this bacterium and CD pathogenesis. These studies should also focus on UC patients to clarify the existing controversial data. A lack of E. coli translocation in UC would suggest that E. coli does not play a primary role in UC pathogenesis or that it plays a different role than in CD.
E. coli strains isolated from IBD patients are clonally diverse[6,13,17] and belong to distinct serotypes[6,13,30] and to different sequence types[6,31-33]. Although a close genetic relationship was detected in a study of IBD pediatric patients[34], the hypothesis that there is a particular clone associated with IBD has largely been ruled out.
In turn, E. coli strains isolated from IBD patients primarily belong to the B2 and D phylogroups in conjunction with extraintestinal pathogenic E. coli (ExPEC). Some works demonstrate major colonization by B2+D phylogroups in IBD patients in comparison with healthy controls[10,31], but in other studies, a similar distribution of phylogroups exist between IBD and healthy subjects[13,29,30,33-36]. Differences between studies could be based on the types of samples analyzed, as it has been reported in healthy individuals that transient E. coli (more likely to be found in feces) are principally A and B1, whereas resident E. coli (more likely to be found in the mucosa) mainly belong to the B2 and D phylogroups[35]. Therefore, studies based on mucosal samples tend to indicate enrichment of B2 and D strains, even in healthy controls. Another factor that could influence the distribution of phylogroups in IBD is the disease severity of patients analyzed, as an increased proportion of B2 and D isolates has been found in active IBD patients[32], which was significantly associated with the inflammation state of IBD tissues[30]. This denotes a shift in E. coli populations to isolates that are better adapted to the inflamed tissue in IBD and/or that are involved in the inflammation itself. Of note, no differences in phylogroup distribution between CD and UC have ever been reported.
E. coli isolated from IBD patients carry different sets of virulence genes that are characteristic of ExPEC strains, whereas intestinal pathogenic E. coli are rare or absent[6,10,13,30,32,34,36-39]. These virulence factors are also frequent in E. coli from healthy subjects and are considered “colonization factors” necessary for successful establishment in the intestinal mucosa[40]. Virulence gene profiles are inexorably linked with the phylogenetic origins of the strains. Based on the distribution of phylogenetic groups, virulence-associated genes characteristic of ExPEC were more frequently found in IBD patients than in healthy subjects in those studies where B2+D predominated in IBD[10,31], whereas no differences were found in other works[13,36,37,41,42]. A shift in the phylogroup distribution would then lead to an increased proportion of E. coli equipped with colonization factors that would facilitate establishment and persistence in IBD patients. However, it is not clear whether this shift occurs specifically in IBD patients or is a general trend taking place in industrialized countries[43]. Although no particular genetic traits distinguish E. coli from the intestinal mucosa of CD or UC, some virulence factors have been found to be differentially distributed between these IBDs. For example, a diarrhea-associated hemolytic E. coli strain called cell-detaching E. coli (CDEC), which commonly harbors hemolysin, cytotoxic necrotizing factor 1, pilus P and S-fimbria genes, was found in 24% of UC E. coli and only in 4.7% of CD E. coli[44]. The gene usp encoding for the uropathogenic-specific protein was also more frequently found in UC E. coli than in CD E. coli[30]. Recently, E. coli carrying the iroN gene, which encodes for a receptor for iron-chelating siderophores, was more frequently isolated from inflammatory and unchanged mucosa of active-phase UC patients[23].
On the other hand, approximately one decade ago, Darfeuille-Michaud et al[45] discovered a new pathotype of E. coli with distinctive phenotypic pathogenic traits that was associated with CD, but not with UC, named adherent invasive E. coli (AIEC). Altogether, these observations suggest that specific E. coli types could be involved in each IBD. We further discuss this issue in the section dedicated to AIEC prevalence in ulcerative colitis.
To date, AIEC is the most likely candidate to cause specific damage to people who are genetically susceptible to the development of CD, and therefore the following sections will focus on discussing the most recent research findings on this pathovar. We review (1) the latest research regarding AIEC pathogenicity; (2) the prevalence and abundance of the pathotype in several intestinal disorders, discussing its putative contribution to other intestinal diseases in addition to CD; (3) the evidence that supports a lack of host-specificity and thus a risk for zoonosis; and (4) recent research that points to a putative role for environmental factors in the fate of AIEC development in the intestine.
The AIEC pathotype was defined as E. coli strains that (1) are able to adhere to differentiated Caco-2 and/or undifferentiated I-407 intestinal epithelial cells with an adhesion index equal or superior to 1 bacteria per cell; (2) are able to invade I-407 cells with an invasion index equal or superior to 0.1% of the original inoculum; (3) involve host cell actin polymerization and microtubule recruitment in bacterial uptake; (4) do not have known invasive determinants; and (5) are able to survive and replicate within J774-A1 macrophages[45]. Since its definition, invasive determinants characteristic from ExPEC have been detected in some AIEC, but not consistently in all AIEC, and thus are not a particularity of the AIEC pathotype[6,13,36,46-48].
Pathogenicity mechanisms of AIEC have mainly been studied in the reference AIEC strain LF82, and its features have been comprehensively linked to many characteristics of CD pathogenesis.
Adhesion to intestinal epithelial cells is in part mediated by type 1 pili, which interact with the glycoprotein CEACAM6 in a mannose-associated manner[49,50]. CEACAM6 is overexpressed in CD patients with ileal disease, which makes them more susceptible to overcolonization by AIEC. Although type 1 pili is present in almost all E. coli, including non-pathogenic strains, we have recently demonstrated that AIEC strains usually present FimH adhesin variants that allow them to more efficiently bind intestinal epithelial cells[31]. Some non-AIEC strains carry these mutations as well, but they do not express type 1 pili. Flagella are also important for adhesion to and invasion of intestinal epithelial cells and elicit the secretion of the pro-inflammatory cytokine IL-8 and chemokine CCL20 in polarized intestinal epithelial cells, which in turn leads to the recruitment of macrophages and dendritic cells to the site of infection[51,52]. The further secretion of INFγ and TNFα by macrophages and lymphocytes leads to CEACAM6 expression, which enhances AIEC colonization. The binding of LF82 type 1 pili to CEACAM6 and flagella to TLR5 in intestinal epithelial cells induces the production of HIF-1a and activation of the classical NF-κB pathway[53]. In turn, these molecules cooperatively control the transcription of IL-8 and pro-angiogenic factors contributing to inflammation and vascularization.
The intermediate filament vimentin, expressed on the host cell surface of mesenchymal cells, has been recently proposed to act as a receptor for AIEC[54]. At the intracellular side, vimentin leucine-rich repeats interact with NOD2 leading to the recruitment of these proteins at the plasma membrane. This is necessary for a proper function of NOD2 in terms of antigen detection, NF-κB activation and autophagy induction. CD patients have specific NOD2 variants (L1007fs and R702W) that are unable to interact with vimentin and, in turn, they localize in the cytosol. That leads to a defective inflammatory response, autophagy induction and handling of CD-associated AIEC. Altogether, NOD2 and vimentin appear to play an important role in AIEC recognition and polymorphisms in these two proteins may have an impact on the ability of AIEC to colonize the host.
A new host-microbe interaction that mediates adhesion of LF82 to intestinal epithelial cells and involves a bacterial and a human chitinase has recently been proposed[55]. Chitinases are enzymes that hydrolyze chitin, a long-chain polymer of an N-acetylglucosamine. The authors demonstrate that specific polymorphisms in two chitin binding domains characteristic of LF82 and other pathogenic E. coli are required to interact with an N-glycosylated asparagine of the human chitinase CHI3L1. Interestingly, human chitinases are overexpressed in intestinal epithelial cells and moderately expressed in cells of the lamina propria during inflammation.
Outer membrane vesicles (OMVs) containing the transmembrane protein OmpA play a role in LF82 invasion of intestinal epithelial cells[48]. OmpA binds the endoplasmic reticulum-localized stress response chaperone Gp96 that is overexpressed on the apical surface of ileal epithelial cells in patients with CD. OMVs fuse with host cells, and it is thought that the release of bacterial effectors that are still undefined is involved in the actin polymerization and microtubule recruitment that occurs during invasion. Point mutations in the ompA sequence of LF82 and other B2 strains mediate better interactions with Gp96[56]. In turn, Gp96 is overexpressed in the ileum of CD patients, which renders them more susceptible to AIEC infection.
Once inside the host cell, LF82 bacteria can be found in several types of intracellular compartments: individually or in groups within single membrane vacuoles, within damaged vacuoles, or within LC3-positive autophagosomes, which indicates that autophagy restricts a subpopulation of intracellular LF82 bacteria[57]. Nevertheless, it was recently demonstrated that AIEC can abrogate the autophagic process[58]. Intracellular LF82 activates NF-κB, leading to the increased expression of MIR30C and MIR130A in T84 cells and in mouse enterocytes, and the upregulation of these microRNAs reduces levels of ATG5 and ATG16L1, inhibiting autophagy and enhancing the inflammatory response. In turn, defects in autophagy mechanisms related to the ATG16L1 and IRGM genes have been associated with CD patients, and these defects confer an advantage for AIEC to survive inside human cells[57]. Therefore, it is a combination of host deficiency factors and AIEC pathogenicity that determines the fate of intracellular E. coli survival.
In addition to adhesion and invasion capacity, LF82 is also able of translocating via the M cells of the Peyer’s patches, gaining access to the lamina propria. This interaction is mediated by type 1 pili and long polar fimbriae (Lpf), which can interact independently with GP2, a surface protein specific to M cells. It is of note that the sites of initial inflammation in CD are the Peyer’s patches and colonic lymphoid follicles; thus, this mechanism of translocation is consistent with early clinical signs of the disease[59].
Another mechanism that can facilitate bacterial translocation is the ability of LF82 to alter intestinal permeability by inducing the expression of the pore-forming protein claudin-2[60] and by displacing ZO-1 and E-cadherin from apical tight junctions, leading to decreased transepithelial resistance and loss of barrier function[17,61]. Besides, pro-inflammatory cytokines like TNFα can drive alterations in intestinal permeability[62]. As AIEC infection induces the secretion of large amounts of TNFα and IL-8[17]; thus, the loss of barrier function induced by LF82 can in part be mediated by the induction of TNFα secretion.
A novel mechanism of pathogenicity observed in LF82 and two other AIEC strains (O83:H1 and UM146) is the evasion of host immune responses via subversion of the IFNγ pathway in intestinal epithelial cells[63]. Phosphorylation of the Signal Transducer and Activator of Transcription STAT-1 is blocked, thus preventing the transcription of IFNγ-dependent genes, which reduces host immune responses and results in an inability to mount an appropriate anti-microbiocidal response. Enterohemorrhagic E. coli (EHEC) strain O157:H7, in part through its Shiga toxin, is also able to block tyrosine phosphorylation and activation of STAT1 after IFNγ stimulation, in contrast with enteropathogenic E. coli E2348/69 or commensal E. coli HB101 which do not present this mechanism of pathogenicity. However, AIEC do not present Shiga toxins. Presumably a small secreted peptide may be responsible for this pathogenic mechanism in AIEC[63].
Once AIEC has gained access to the lamina propria, these bacteria can be engulfed by macrophages. Intramacrophage LF82 do not escape into the cytoplasm but induce the formation of a large vacuole (phagosome) that fuses with lysosomes[64], suggesting that AIEC bacteria have the ability to replicate in an environment with acidic pH, oxidative stress, active proteolytic enzymes, and antimicrobial compounds. Indeed, it was demonstrated in vitro that an acidic environment is necessary for replication of AIEC LF82 bacteria[64]. The protease HtrA and the thiol-disulfide oxidoreductase DsbA have been reported to be important for survival and replication within macrophages[65,66]. The authors linked these proteins to the ability of LF82 to resist the stress conditions of the phagolysosomes, as isogenic mutants for these proteins were less efficient in growing in acidic and nutrient-poor medium, and these proteins were overexpressed not only in LF82 during macrophage infection but also in acidic nutrient-poor medium. Interestingly, the overexpression of HtrA is dependent on the LF82 background, as non-pathogenic E. coli do not overexpress that protein under similar growth conditions. The RNA-binding protein Hfq, which functions as a global posttranscriptional regulator of gene expression, has also been implicated in survival and replication within macrophages and in stress tolerance but also other aspects of LF82 pathogenicity, such as adhesion and invasion capability[67]. Hfq binds small regulatory RNA molecules, facilitating their interaction with mRNA, but the target genes are still unknown.
Continuous replication of LF82 within macrophages results in the secretion of high levels of TNFα without inducing host cell death[68]. This can explain inflammation and granuloma formation in the gut of CD patients, which has been demonstrated in vitro[50,69,70]. A direct role for LF82 in delaying apoptosis of infected macrophages and dendritic cells has recently been reported[71]. LF82 infection was found to alter the function of caspase-3, a protease that plays an essential role in apoptosis, and to increase degradation of this molecule in the proteasome.
Also supporting AIEC capability to replicate within immune cells, strain LF82 was able to replicate within monocytes isolated from CD patients for the first 20 h after infection but then CD monocytes started to clear intracellular bacteria[72]. Interestingly, those patients with polymorphisms in CARD15 gene (R702W, G908R and 1007fs) showed reduced early inflammatory response towards AIEC infection with decreased levels of IL-1β, IL-6 and IL-10. In contrast, Asp299Gly mutation in TLR4 had no effect on monocyte response to AIEC. Besides, a recent study revealed that CD monocyte-derived dendritic cells stimulated with lipopolysaccharide show an attenuated inflammatory response with decreased levels of IL-6 and IL-1β, as well as an impaired autophagy with reduced LC3 expression[73]. Moreover, these cells had a reduced capacity to support the expansion of allogeneic Th17 cells from CD4+ memory T cells. The authors propose that mucosal Th17 activation in CD patients is a secondary event in response of poor bacterial clearance due to defects in innate immunity. Further studies showing AIEC effects on CD defective dendritic cells regarding, not only cytokine release, but also autophagy function and the level of IL-17A response induction in T cells, are necessary to decipher whether the alterations observed in lipopolysaccharide-stimulated dendritic cells equally occur after AIEC exposure.
AIEC LF82 bacteria are also able to invade and replicate within human neutrophils, but in contrast to its behavior inside macrophages and intestinal epithelial cells, LF82 induces the autophagic death of infected neutrophils, which later undergo an alternative cell death process called NETosis[74]. In neutrophils, LF82 are localized inside autolysosomes, as observed by the colocalization of phagosome and lysosome markers, but there is no acidification, which suggests that LF82 avoids autolysosome maturation. Infected neutrophils secrete cytokines, in particular IL-8, contributing to mucosal inflammation.
The ability to form biofilms is a pathogenic feature frequently found among AIEC strains. We found that 17 out of 27 AIEC strains and only 9 out of 38 intestinal non-AIEC strains were biofilm producers[75]. Motility and flagellar type are of relevance in biofilm production, as non-motile strains were not able to form biofilms, and all strains with the H1 flagellar antigen were strong biofilm producers. Recently, Chassaing et al[76] have demonstrated the ability of the LF82 strain to form biofilms on intestinal epithelial cells using cell culture and animal models.
Despite all the research conducted on AIEC pathogenicity, we still do not know the genetic factors that are characteristic of the AIEC pathotype. The majority of genes related to its pathogenicity are not AIEC-specific, as is the case for fimH, ompA, dsbA or htrA, and are present in the majority of E. coli strains, including non-pathogenic strains[31,48,65,66]. Point mutations or differential gene expression are involved in the increased fitness and/or virulence of AIEC strains. Unfortunately, these genetic factors have been studied in very few strains or exclusively in the prototype strain LF82. Conversely, virulence genes that are not usually present in non-pathogenic E. coli, such as afaC, pks or lpf, have been found frequently, but not consistently, in AIEC strains[13,59,77]. AIEC strains are clonally diverse, belong to different serotypes and carry different sets of virulence genes that are characteristic of ExPEC strains; these features also describe non-AIEC ExPEC-like strains inhabiting the intestinal mucosa[13]. The AIEC pathotype comprises high genotype variability, which complicates the identification of specific genetic factors of the pathotype.
It is of note that despite the genetic similarity between AIEC and ExPEC, the latter generally does not exhibit the AIEC phenotype. We determined that only 4 out of 63 ExPEC strains of different origins were AIEC-like[78], conferring a particular identity on the pathotype. Identification of additional genetic elements or the differential expression of key genes that must be involved in AIEC pathogenicity represents an important milestone that can be achieved through genome- and transcriptome-based studies.
Four AIEC genomes belonging to B2 strains have been sequenced and published to date[46,47,79,80], and comparative genomics have been carried out for the LF82 and NRG857c strains[47]. Although novel virulence factors not previously found in AIEC by PCR genotyping, such as a type-6 secretion system, have been detected in genomic islands of the sequenced strains, genomic studies have corroborated the notion that AIEC resembles ExPEC. Unique sequences for AIEC were found in common between the LF82 and NRG857 strains. However, both strains belong to the same phylogroup and serotype (B2 O83:H1), which indicates they are genetically very close. Given the high variability of AIEC seropathotypes, studying the distribution of these genes in other AIEC strains is essential to confirm whether these elements are common features of the pathotype or are strain-specific. Comparative genomics of phylogenetically distant AIEC strains would presumably reveal a significantly greater number of genetic differences. Although it will complicate the situation, sequencing additional AIEC strains from different phylogenetic origins is crucial to determine the common genetic features involved in the AIEC phenotype.
AIEC have generally been isolated from tissue samples, but there is no evidence regarding its exact localization within the intestinal mucosa. Although AIEC are invasive bacteria, they have not been convincingly observed within intestinal epithelial cells or in the lamina propria in resected tissue or mucosal biopsies. The studies conducted by Martin et al[12] and Eliott et al[36] addressed whether E. coli are intraepithelial or mucosa-associated by treating biopsies with gentamicin. This approach has brought indirect evidence of E. coli invasion of intestinal epithelial cells in CD but not in UC. However, the complete AIEC phenotype was not studied in the intracellular E. coli strains obtained from these studies.
Currently, identifying the exact localization of AIEC strains in the mucosa is nearly impossible to do, as no molecular tools that specifically target the AIEC pathovar are available. Some evidence has been obtained using animal models infected with known reference strains. For example, by staining for the O83 antigen, it has recently been demonstrated that the LF82 and NRG857c strains colonize the ileum, cecum and colon of several mouse models and that they are located at the base of the crypts and within goblet cells[81]. Engineered LF82 with a plasmid containing the GFP protein permitted fluorescence-microscopic examination of the localization of LF82 in the nematode C. elegans. In this situation, there was robust gut colonization, but bacteria remained in the lumen and were not attached to intestinal epithelial cells[67]. To visualize the extent of bacterial adhesion and invasion in in vivo infection, Low et al[55] stained E. coli lipopolysaccharides with specific antibodies and compared basal levels of fluorescence in uninfected mice (corresponding to indigenous bacteria) with levels in infected mice. They found higher counts of stained bacteria in the intestinal epithelial cells and lamina propria of infected mice, suggesting AIEC intestinal epithelial cell invasion and translocation.
Despite the virulence genes that are encoded in the genome of many AIEC strains and the mechanisms of pathogenicity reported for the prototype strain LF82, AIEC are generally considered pathobionts. This assumption is supported by the fact that, although at a lower frequency than in CD, healthy subjects can carry AIEC in their intestinal mucosa[6,13,17,45,82]. The prevalence varies between studies, ranging from the absence to 15.8% of colonic samples with AIEC and from 6.2% to 18% in ileal samples. Although AIEC bacteria may colonize the intestinal mucosa of non-IBD patients, these bacteria usually do not translocate a healthy mucosal barrier, as bacterial invasion of the mucosa has not frequently been observed in control patients[14] and intracellular E. coli was rarely cultivated from the colonic mucosa of healthy subjects (from the absence to 9%)[6,26,83]. AIEC strains are more abundant, and consequently more frequently found, in the ileum than in the colon of healthy subjects. We found that AIEC accounted for 3.58% and 0.95%, respectively, of the ileal and colonic E. coli populations[13]. Accordingly, a larger number of AIEC LF82 bacteria were attached to ileal than to colon tissue in ex vivo samples from healthy subjects infected with this AIEC strain[84]. Altogether, these data suggest that AIEC can more easily colonize the ileum with respect to other E. coli, and at least approximately 1 out of 6 healthy individuals can be considered “asymptomatic carriers”.
Genetic or environment-derived host defects at the intestinal barrier may determine the ability of AIEC to colonize and translocate the gut. A number of host deficiencies frequently found in CD patients have been linked with the increased ability of AIEC LF82 to cause infection. For example, these defects include the overexpression of the CEACAM6 and Gp96 receptors in the apical membrane of intestinal epithelial cells, which facilitates AIEC adhesion and invasion[48,49], or defects in autophagy related to NOD2, ATG16L1 and IRGM function and expression, which impair the ability of host cells to resolve infections[57,85]. Additionally, it has been suggested that the altered bile salts metabolism in CD patients could enhance the expression of long polar fimbriae in AIEC, which could permit better translocation via M cells[86]. Moreover, decreased levels of the protease meprin, which are characteristic of severe inflammation in IBD patients, have been proposed to determine the fate of AIEC in terms of their ability to colonize the host, as these proteases degrade type 1 pili[87].
Intraepithelial E. coli with adherent and invasive properties were isolated from the sigmoid colon mucosa in 29% of CD patients[12] and in 90% of CD patients in a cohort composed of ileal, ileocolonic and colonic disease phenotypes[36]. Differences between studies could be explained by the disease activity status of the cohort of patients, who were mainly in the relapse stage in the latter study.
In the last decade, several independent laboratories have reported a higher prevalence of AIEC in CD patients than in healthy subjects[6,13,17,45,82]. Unfortunately, not all of these studies categorized CD patients by their disease subtype or analyzed prevalence based on anatomic location in the gut. The first study was conducted by Darfeuille-Michaud et al[45] in 2004 and revealed that 22% of CD patients with ileal involvement harbored AIEC strains in ileal chronic lesions and at a similar frequency in healthy mucosa. However, AIEC bacteria were more likely to be found in the early ileal lesions that occurred in patients after ileostomy (36.4%). AIEC strains were only isolated from the colon of 3.7% of CD patients with a colonic disease phenotype. The authors proposed an association between AIEC and ileal CD and suggested that the pathovar could be involved in the initiation of the inflammatory process. Conversely, Baumgart et al[6] reported a prevalence of AIEC strains in the ileum of 38.5% of CD patients with ileal involvement and 37.5% with colonic CD, indicating that AIEC is associated both with ileal and colonic disease phenotypes. Sasaki et al[17] demonstrated that 24.3% of CD patients exhibited AIEC strains, but neither the localization of these strains in the gut nor the disease phenotypes of the positive patients were detailed. A similar prevalence was reported by Dogan et al[82] in the ileum of CD patients with ileal disease. We detected AIEC strains at a higher frequency in comparison with previous studies, most likely due to the methodological approach used. Whereas other studies analyzed from 1 to 15 E. coli colonies per patient, we searched for AIEC strains in a collection of 95 - 150 E. coli colonies per patient. This approach not only enabled us to obtain a more accurate prevalence value but also to study the abundance of AIEC strains within the E. coli population. We detected AIEC strains in the ileum of 54.5% of CD patients and in the colon of 50% of CD patients[13]. Although data depicted by disease subtype were not reported in the original work, we also found a higher prevalence in CD patients with ileal involvement (66.7% of ileal and 58.3% of colonic samples) than those with colonic disease (50% of ileal and 25% of colonic samples). Colonic CD patients denoted also a high prevalence of AIEC, what supports the observations of Baumgart et al[6], but the pathotype was more frequently found in the ileum than in the colon of CD patients, in line with the findings of Darfeuille-Michaud et al[45] The abundance of AIEC, defined as the percentage of AIEC within the E. coli population, was low and variable, ranging from 1% to 50%. On average, AIEC isolates represented 9.3%, 3.7% and 3.1% of E. coli isolates in ileal, ileocolonic and colonic CD patients, respectively. Jensen et al[84] supported these data using quantitative PCR targeting indigenous LF82 bacteria. The increased expression of CEACAM6 in the ileum of ileal CD patients may explain the higher prevalence and abundance of AIEC in CD patients with ileal involvement. However, additional host-microbial interactions or environmental factors may be involved in colonization of the colonic mucosa, as no differences in CEACAM6 expression exist at the level of the colon between CD patients and control subjects[49]. Our work demonstrates that AIEC are more prevalent than expected in all CD disease subtypes, reinforces the hypothesis that the microenvironment of ileal CD specifically favors AIEC expansion, and suggests that the colon is also a niche effectively colonized by AIEC.
More than two decades ago, the adhesion capabilities of E. coli from both UC and CD patients were assessed. Mannose-resistant adhesion was characteristic of E. coli from both IBDs, which raised the question of whether adhesive E. coli could also be involved in UC pathogenesis[88,89]. Recent studies have confirmed that, in UC patients, adherent E. coli strains are found as frequently as[90] or even more frequently than[34,91] in CD patients. An undefined adhesion pattern was most prevalent in E. coli from both UC and CD patients[42], although aggregative adherence was particularly frequent in UC patients[42,90]. Molecular tools to detect adhesive determinants of IBD E. coli did not demonstrate specific adhesion factors in UC E. coli in comparison to CD E. coli[10,37,41,42], whereas in other studies UC E. coli carried some adhesion factors more frequently than CD E. coli[30,34,44]. Some of these studies are based on pediatric or newly diagnosed patients, which provides supporting arguments for the early contribution of adherent E. coli to IBD rather than being its development a consequence of inflammation. Moreover, the higher frequency of E. coli B2 strains with at least one positive adhesion-related gene was correlated with disease activity in UC patients (86% in active vs. 13% in non-active patents)[32]. Therefore, there is substantial agreement among studies regarding the adhesion capacity of E. coli strains from UC patients.
Intracellular E. coli were cultured from 47%[36] and 19%[12] of UC patients in two studies using gentamicin protection assay. However, few works have sought to identify the AIEC pathovar in UC patients, and some controversial results have been obtained. In the first study that searched for AIEC in UC any of UC patients had AIEC bacteria in their colon[45], and similar results were obtained in a later study[92]. In contrast, in studies with larger cohorts, one of them based on pediatric patients, AIEC were detected in 7.2% to 10% of UC patients[17,93]. Other investigators that studied the invasion ability of IBD E. coli, but did not study the complete AIEC phenotype, detected a high prevalence of invasive strains in UC patients (37.5%)[44]. Moreover, similar invasion rates in I407 cells were observed for E. coli from pediatric UC and CD patients[42], whereas in a previous study the invasion index using differentiated Caco-2 cells was lower in E. coli from UC than CD patients[17]. Noteworthy, the intra-macrophage survival capacity of E. coli strains was found to be highest in UC patients from a cohort of newly diagnosed IBD patients. Unfortunately, no information about adhesion and invasion abilities was provided[30].
Sasaki et al[17] observed that although AIEC from UC were less invasive than CD E. coli, they induced the secretion of similar amounts of TNFα and higher amounts of IL-8, suggesting that UC-associated E. coli are distinct from those associated with CD. Accordingly, a recent study reported that CD E. coli are frequently lpf+ afaC+, whereas UC E. coli do not possess lpf gene and frequently harbor the afaC and pks genes together[77]. Lpf mediate translocation of bacteria via M cells, while the afimbrial adhesin AfaC mediates a diffuse adherence to and invasion of intestinal epithelial cells and also induces vascular endothelial growth factor expression. The polyketide synthase gene complex (pks) contains the genes to synthesize the metabolite colibactin, a genotoxin with the ability to cause epithelial DNA damage.
The evidence collected to date suggests that E. coli strains with adhesive and other virulence properties could be involved in UC pathogenesis, but further work clarifying the role of these strains in conjunction with host defects in the mucosal barrier is needed. Furthermore, in view of the few studies and conflicting results regarding AIEC prevalence in UC, additional studies characterizing E. coli populations from different anatomical sites, and for both affected and unaffected tissue, in active and inactive UC patients would be of relevance to elucidate the possible role of AIEC in UC.
An analysis of fecal bacterial diversity by pyrosequencing demonstrated that the Escherichia/Shigella genus was enriched in colorectal cancer (CRC) patients[94]. In contrast, studies conducting quantitative PCR did not find an increase in the E. coli population in CRC[8,91]. However, intracellular E. coli has frequently been found in CRC patients. Swidsinski and collaborators detected intracellular E. coli in 87% of patients with CRC and not in controls using a gentamicin protection assay[95]. Similarly, Martin et al[12] isolated intramucosal E. coli from 33% of tumors in CRC patients and 9% of control subjects, surpassing the prevalence found among IBD patients, and Bonnet et al [96] isolated intramucosal E. coli in 86% of colon cancer tumor specimens and 48% of diverticulosis samples. Moreover, high levels of mucosa-associated E. coli correlated with poor colorectal carcinoma prognostic factors and a higher proliferative index of epithelial cells, suggesting a role for these bacteria in tumor progression.
E. coli strains isolated from the study by Prorok-Hamon et al[77] were hemagglutination-positive, adherent to HT29 and I407 intestinal epithelial cells and frequently able to invade I407 cells, all characteristics that resemble the AIEC pathotype. A recent study conducted by the same research group showed that at least one of the isolates obtained from a patient with CRC shared the complete AIEC phenotype. In addition, E. coli isolated from a pediatric cohort with polyposis, who were included as a healthy control group, showed the highest invasion efficiency compared with E. coli strains isolated from IBD children[42]. However, as far as we know, there is no data regarding the prevalence of AIEC in patients with CRC.
Several studies have demonstrated that E. coli associated with CRC are frequently colibactin-producing[72,93-95]. Not only is the pks genomic island encoding for the genotoxin colibactin frequent in CRC, but other cyclomodulins such as CNF, CDT and CIF. Buc et al[97] found that cyclomodulin-encoding genes were over-represented among E. coli from CRC patients (65.8%), particularly distal colon cancer (76.5%), compared with diverticulosis samples (19.54%). These molecules can be genotoxic and/or modulate cellular differentiation, apoptosis, and proliferation. Prorok-Hamon et al[77] observed that CRC E. coli frequently harbored the pks gene but also the adhesins AfaC and LpfA, partially resembling those E. coli isolated from CD and UC. These factors confer the ability to adhere to and invade I407 cells, to upregulate vascular endothelial growth factor expression in intestinal epithelial cells, and presumably, to translocate via M cells and cause genotoxicity to host cells. Recently, pathogenic cyclomodulin-positive E. coli strains were found to be more prevalent in the mucosa of patients with advanced stages of the disease[96].
Few studies have been focused on E. coli populations in CRC patients do date, and the results obtained point to a putative role for a subset of E. coli with pathogenic features relevant to CRC pathogenesis. Given that AIEC possessing virulence factors relevant to enterocyte adhesion and invasion, vascular endothelial growth factor expression and carcinogenesis have been detected in CRC patients and the fact that intramucosal E. coli with features similar to AIEC have been more frequently found in CRC than in IBD patients, further studies determining the prevalence of AIEC in CRC are needed to corroborate or refute the hypothesis for a putative role for AIEC in CRC.
Coeliac disease is a chronic inflammatory disorder exclusively affecting the small intestine, in which genetically predisposed individuals feature a permanent intolerance to dietary gluten. Several studies have provided evidence that coeliac patients exhibit intestinal microbial dysbiosis, similar to what occurs in IBD patients. In a study based on PCR-TGGE of duodenal samples, E. coli was found more frequently in coeliac children (92.1%) than in healthy children (20%)[98]. Quantification of E. coli by FISH showed also that this species was more abundant in active coeliac patients than in inactive patients and controls[99], but this was not observed in fecal samples[100]. Another study found changes in Enterobacteriaceae diversity and increased virulence-gene carriage in E. coli isolates from coeliac children[101]. In particular, E. coli strains largely belonging to the B2 and D phylogenetic groups and carrying ExPEC-like features, e.g., pilus P and hemolysin A, were found to be more abundant in celiac patients when compared to healthy controls. This dysbiosis of the E. coli population is similar to that found in CD patients.
Given the association between E. coli and coeliac disease in terms of abundance and the correlation with disease activity, as well as the genetic similarities between isolates from the intestinal mucosa of coeliac patients and CD patients, further studies aimed at identifying the AIEC phenotype amongst coeliac E. coli isolates are of interest to better define the disease specificity of the AIEC pathotype.
AIEC strains isolated from CD patients genetically resemble avian pathogenic E. coli and other animal ExPEC. We studied the AIEC phenotype in a strain collection obtained from animals with extraintestinal infection and intestinal disease to determine the disease and host specificity of the AIEC pathotype. All these strains were classified as ExPEC in terms of their phylogenetic origin and virulence genotype. ExPEC strains of extraintestinal origin rarely shared the AIEC phenotype, whereas ExPEC-like strains of intestinal origin were frequently AIEC-like in cats (82%), dogs (35%) and swine (32%) with intestinal disease[102]. The high prevalence of AIEC in companion and farm animals highlights a putative risk of zoonosis between humans and animals. In a previous study, Simpson et al[103] detected AIEC in boxer dogs. Interestingly, these dogs suffered from granulomatous colitis, a disease with pathological features that overlap with CD, which supports the role of AIEC in human CD and analogous diseases in animals.
Altogether, these results suggest that the AIEC pathotype is disease-specific rather than host-specific and raises the question of whether there is a possible route of transmission between animals and humans. Further studies examining the distribution of AIEC strains in different healthy and diseased animals and in the environment would contribute to our understanding of the epidemiology, putative reservoirs, host-specificity and possible routes of transmission of AIEC.
Recent studies have implicated some emulsifiers and food stabilizers frequently used in developed countries as having a role in AIEC colonization. Maltodextrin, a polysaccharide derived from starch hydrolysis that is used as food additive, has been shown to markedly enhance AIEC biofilm formation and adhesion to intestinal epithelial cells and macrophages[104]. Maltodextrin favors type 1 pili expression, which is required for biofilm formation and adhesion. Moreover, a higher prevalence of the gene malX, which is essential for maltodextrin metabolism, was found in bacteria isolated from ileal CD patients than from healthy controls (71% vs 18%, respectively). These observations suggest that a diet rich in maltodextrin would aid maltodextrin-utilizing bacteria, would enhance E. coli gut colonization, and thus contribute to dysbiosis. Furthermore, polysorbate-80, an emulsifier commonly used in processed foods, was found to enhance translocation of the AIEC HM605 strain across M cells and intestinal epithelial cells[105]. Using animal models, we also observed that a diet enriched in fat and sugar induced dysbiosis and low grade-inflammation[106]. In this work we also showed that dysbiosis and low-grade inflammation in susceptible individuals lead to increased AIEC colonization, what in turn exacerbated the inflammatory response and epithelial barrier disruption.
The type of dietary fiber intake may influence bile acid metabolism. For example, daily dietary supplementation for four weeks with the purified fiber components pectin and cellulose in humans leads to differential bile acid composition. In cellulose-treated volunteers, cholic acid increased whereas deoxycholic acid decreased, which inversely occurred in pectin-treated individuals[107]. Increased concentrations of cholic acid and chenodeoxycholic acid have been reported in CD patients[108], and lithocholic acid has been reported particularly in ileal CD patients[109]. Interestingly, all of these bile salts induced the expression of the lpf operon in AIEC LF82 strain[86]. Therefore, dietary fiber consumption could also influence the tropism of AIEC for CD ileal tissue by altering bile acid composition and thus the expression of lpf in AIEC in the gut.
These studies demonstrate that dietary components may impact the success of AIEC in colonizing the host and therefore contribute to disease susceptibility. For that reason, intervention studies are needed to evaluate the effects of diet, probiotics, and/or prebiotics on the intestinal microbial community, including the AIEC population with respect to CD activity status and disease progression.
Several studies based on animal models have shown that there is a need of microbial dysbiosis and/or intestinal inflammation to succeed with AIEC infection. An effective colonization only occurs in mice that have been treated with antibiotics[50,81,110], dextran sodium sulfate[69] or high-fat/high-sugar diet[106] before infection, having these treatments an effect on gut bacteria composition and mucosal homeostasis. Moreover, Craven et al[111], nicely showed that moderate to severe ileitis produced by protozoan infection in mice models induced dysbiosis and proliferation of endogenous mucosally invasive E. coli. These works suggest that inflammation and dysbiosis favors AIEC proliferation. Therefore, AIEC overgrowth in the intestine can be seen as a consequence of inflammation.
On the other hand, it has been recently shown that AIEC infection itself induced lasting changes in the intestinal microbiota[112]. This study was conducted on mice lacking flagellin receptor TLR5 (T5KO) which are prone to develope spontaneous colitis. The authors hypothesized that transient colonization of T5KO mice by AIEC results in an altered gut microbiota community with greater proinflammatory potential, which can persist in the host and induce chronic inflammation due to its increased levels of lipopolysaccharide and flagellin. The effects of AIEC infection on host mucosal immunity, barrier integrity and inflammation induction have been demonstrated in multiple animal models[50,60,69,81,106,110] but the work of Chassaing et al[112] is the first showing that AIEC infection contribute to intestinal dysbiosis. Overall, these studies suggest that AIEC overgrowth in the intestine can be seen as a cause of inflammation.
Therefore, inflammation can instigate imbalances in E. coli, especially the AIEC pathotype and, in turn, these bacteria can be involved in a further dysbiosis and increased intestinal inflammation.
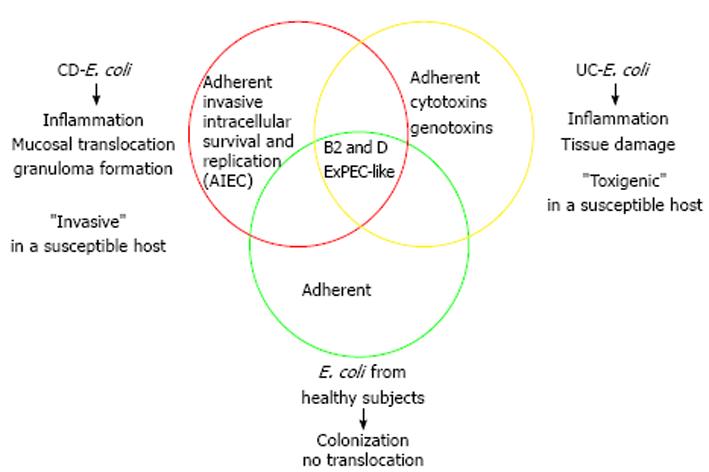
Substantial evidence indicates that E. coli is involved in CD and growing data suggest that this species is also a contributing factor in UC pathogenesis (Figure 1). Studies focused on defining virulence gene profiles of E. coli populations have shown that E. coli associated to the mucosa of healthy subjects resemble those of IBD patients. Genes related with adhesion, iron transport, capsule formation and toxins are present in E. coli from both healthy subjects and IBD patients. These features are thought to be necessary for an effective colonization of the intestinal tract. However, the intestinal microenvironment in IBD patients, especially those in relapse, would predispose to E. coli proliferation. Moreover, E. coli from CD patients have probably evolved towards the AIEC pathotype, which has the capacity to adhere to and to invade intestinal epithelial cells, as well as to survive and replicate within a number of cell types. Virulence properties of AIEC described to date can explain several features of CD pathophysiology such as inflammation, mucosal bacterial translocation and granuloma formation. Conversely, E. coli strains from UC patients appear to present a “toxigenic” behavior rather than the “invasive” pathogenic mechanism of CD- E. coli. Recent research has pointed out that E. coli from UC patients frequently carry virulence genes related to cytotoxicity and genotoxicity, which can contribute to mucosal inflammation and tissue damage. This is in accordance with previous works that did not found E. coli translocating the epithelial barrier of UC patients, and could be linked with some aspects of UC pathophysiology.
Since the AIEC pathotype was defined one decade ago, substantial research has been conducted focusing on the identification of the mechanisms of pathogenicity and also in the field of epidemiology with regard to CD. However, additional epidemiologic studies are still needed to corroborate the role of AIEC in CD and to clarify the AIEC disease- and host-specificity. An important limitation to epidemiological studies is the absence of specific molecular tools to detect and quantify this pathotype, as the current available techniques to identify the AIEC pathotype are based exclusively on phenotypic screening of cultured bacteria, which is highly time-consuming. The execution of large-scale epidemiologic studies would also provide new insights into its distribution, putative reservoirs and transmission pathways. Moreover, the molecular bases of AIEC pathogenicity are still not fully understood, as only a few model strains have been studied and there is a wide variety of seropathotypes and phylotypes within the AIEC pathotype. Genomic and transcriptomic studies including wider and more diverse AIEC strain collections could assist in identifying new genetic elements associated with the AIEC phenotype, which may help us to gain a better understanding of the mechanisms of pathogenicity and could result in significant advances in the detection of new therapeutic targets for CD.
P- Reviewer: Gassler N, Silva MA S- Editor: Song XX L- Editor: A E- Editor: Lu YJ
| 1. | Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1339] [Cited by in RCA: 1376] [Article Influence: 80.9] [Reference Citation Analysis (1)] |
| 2. | Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 362] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 3. | Andoh A, Imaeda H, Aomatsu T, Inatomi O, Bamba S, Sasaki M, Saito Y, Tsujikawa T, Fujiyama Y. Comparison of the fecal microbiota profiles between ulcerative colitis and Crohn’s disease using terminal restriction fragment length polymorphism analysis. J Gastroenterol. 2011;46:479-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 4. | Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1036] [Cited by in RCA: 1333] [Article Influence: 121.2] [Reference Citation Analysis (3)] |
| 5. | Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn’s disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 198] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 6. | Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 2007;1:403-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 483] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 7. | Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405-1413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 609] [Cited by in RCA: 638] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 8. | Fujita H, Eishi Y, Ishige I, Saitoh K, Takizawa T, Arima T, Koike M. Quantitative analysis of bacterial DNA from Mycobacteria spp., Bacteroides vulgatus, and Escherichia coli in tissue samples from patients with inflammatory bowel diseases. J Gastroenterol. 2002;37:509-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 9. | Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136-4141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 425] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 10. | Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 323] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 11. | Lopez-Siles M, Martinez-Medina M, Busquets D, Sabat-Mir M, Duncan S, Flint H, Aldeguer X, Garcia-Gil L. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish Irritable Bowel Syndrome and Inflammatory Bowel Disease phenotypes. Int J Med Microbiol. 2014;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 103] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 12. | Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology. 2004;127:80-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 558] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 13. | Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn’s disease. Inflamm Bowel Dis. 2009;15:872-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 305] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 14. | Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 184] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 15. | Neut C, Bulois P, Desreumaux P, Membré JM, Lederman E, Gambiez L, Cortot A, Quandalle P, van Kruiningen H, Colombel JF. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am J Gastroenterol. 2002;97:939-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 201] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 16. | Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, Ott SJ. Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J Med Microbiol. 2010;59:1114-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A. Invasive Escherichia coli are a feature of Crohn’s disease. Lab Invest. 2007;87:1042-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 188] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 18. | Schwiertz A, Jacobi M, Frick JS, Richter M, Rusch K, Köhler H. Microbiota in pediatric inflammatory bowel disease. J Pediatr. 2010;157:240-244.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 19. | Beaven SW, Abreu MT. Biomarkers in inflammatory bowel disease. Curr Opin Gastroenterol. 2004;20:318-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J, Targan SR. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 338] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 21. | Mow WS, Vasiliauskas EA, Lin YC, Fleshner PR, Papadakis KA, Taylor KD, Landers CJ, Abreu-Martin MT, Rotter JI, Yang H. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 375] [Cited by in RCA: 372] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 22. | Sha S, Xu B, Wang X, Zhang Y, Wang H, Kong X, Zhu H, Wu K. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis. 2013;75:245-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 23. | Pilarczyk-Zurek M, Chmielarczyk A, Gosiewski T, Tomusiak A, Adamski P, Zwolinska-Wcislo M, Mach T, Heczko PB, Strus M. Possible role of Escherichia coli in propagation and perpetuation of chronic inflammation in ulcerative colitis. BMC Gastroenterol. 2013;13:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 664] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 25. | Walmsley RS, Anthony A, Sim R, Pounder RE, Wakefield AJ. Absence of Escherichia coli, Listeria monocytogenes, and Klebsiella pneumoniae antigens within inflammatory bowel disease tissues. J Clin Pathol. 1998;51:657-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Vasquez N, Mangin I, Lepage P, Seksik P, Duong JP, Blum S, Schiffrin E, Suau A, Allez M, Vernier G. Patchy distribution of mucosal lesions in ileal Crohn’s disease is not linked to differences in the dominant mucosa-associated bacteria: a study using fluorescence in situ hybridization and temporal temperature gradient gel electrophoresis. Inflamm Bowel Dis. 2007;13:684-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37:1034-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 240] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 28. | O'Brien CL, Pavli P, Gordon DM, Allison GE. Detection of bacterial DNA in lymph nodes of Crohn’s disease patients using high throughput sequencing. Gut. 2014;Jan 15; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Ryan P, Bennett MW, Aarons S, Lee G, Collins JK, O’Sullivan GC, O’Connell J, Shanahan F. PCR detection of Mycobacterium paratuberculosis in Crohn’s disease granulomas isolated by laser capture microdissection. Gut. 2002;51:665-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, Ziebell K, Krause DO. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1451-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 31. | Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C. Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9:e1003141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 32. | Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9:171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 33. | Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Phylogenetic analysis of inflammatory bowel disease associated Escherichia coli and the fimH virulence determinant. Inflamm Bowel Dis. 2009;15:1737-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Schippa S, Conte MP, Borrelli O, Iebba V, Aleandri M, Seganti L, Longhi C, Chiarini F, Osborn J, Cucchiara S. Dominant genotypes in mucosa-associated Escherichia coli strains from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:661-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Nowrouzian FL, Adlerberth I, Wold AE. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 2006;8:834-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 36. | Elliott TR, Hudspith BN, Wu G, Cooley M, Parkes G, Quiñones B, Randall L, Mandrell RE, Fagerquist CK, Brostoff J. Quantification and characterization of mucosa-associated and intracellular Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2013;19:2326-2338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 37. | Gombošová L, Lazúrová I, Zakuciová M, Curová K, Kmeťová M, Petrášová D, Siegfried L. Genes of intestinal Escherichia coli and their relation to the inflammatory activity in patients with ulcerative colitis and Crohn’s disease. Folia Microbiol (Praha). 2011;56:367-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 38. | Schultsz C, Moussa M, van Ketel R, Tytgat GN, Dankert J. Frequency of pathogenic and enteroadherent Escherichia coli in patients with inflammatory bowel disease and controls. J Clin Pathol. 1997;50:573-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Vejborg RM, Hancock V, Petersen AM, Krogfelt KA, Klemm P. Comparative genomics of Escherichia coli isolated from patients with inflammatory bowel disease. BMC Genomics. 2011;12:316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 40. | Nowrouzian F, Adlerberth I, Wold AE. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect. 2001;126:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 41. | de Souza HL, de Carvalho VR, Romeiro FG, Sassaki LY, Keller R, Rodrigues J. Mucosa-associated but not luminal Escherichia coli is augmented in Crohn’s disease and ulcerative colitis. Gut Pathog. 2012;4:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Sobieszczańska BA, Duda-Madej AB, Turniak MB, Franiczek R, Kasprzykowska U, Duda AK, Rzeszutko M, Iwańczak B. Invasive properties, adhesion patterns and phylogroup profiles among Escherichia coli strains isolated from children with inflammatory bowel disease. Adv Clin Exp Med. 2012;21:591-599. [PubMed] |
| 43. | Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 950] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 44. | Curová K, Kmetová M, Sabol M, Gombosová L, Lazúrová I, Siegfried L. Enterovirulent E. coli in inflammatory and noninflammatory bowel diseases. Folia Microbiol (Praha). 2009;54:81-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 45. | Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1075] [Cited by in RCA: 1123] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 46. | Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb el H, Segurens B, Barbe V, Sauvanet P. Complete genome sequence of Crohn’s disease-associated adherent-invasive E. coli strain LF82. PLoS One. 2010;5:e12714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 137] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 47. | Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics. 2010;11:667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 48. | Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hébuterne X, Hofman P, Darfeuille-Michaud A. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355-1362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 49. | Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566-1574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 435] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 50. | Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179-2189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 243] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 51. | Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Jezek GE, Islas-Islas M, Torres AG. Escherichia coli isolated from a Crohn’s disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 142] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 52. | Subramanian S, Rhodes JM, Hart CA, Tam B, Roberts CL, Smith SL, Corkill JE, Winstanley C, Virji M, Campbell BJ. Characterization of epithelial IL-8 response to inflammatory bowel disease mucosal E. coli and its inhibition by mesalamine. Inflamm Bowel Dis. 2008;14:162-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 53. | Mimouna S, Gonçalvès D, Barnich N, Darfeuille-Michaud A, Hofman P, Vouret-Craviari V. Crohn disease-associated Escherichia coli promote gastrointestinal inflammatory disorders by activation of HIF-dependent responses. Gut Microbes. 2011;2:335-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 54. | Stevens C, Henderson P, Nimmo ER, Soares DC, Dogan B, Simpson KW, Barrett JC, Wilson DC, Satsangi J. The intermediate filament protein, vimentin, is a regulator of NOD2 activity. Gut. 2013;62:695-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 55. | Low D, Tran HT, Lee IA, Dreux N, Kamba A, Reinecker HC, Darfeuille-Michaud A, Barnich N, Mizoguchi E. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology. 2013;145:602-612.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 56. | Miquel S, Claret L, Bonnet R, Dorboz I, Barnich N, Darfeuille-Michaud A. Role of decreased levels of Fis histone-like protein in Crohn’s disease-associated adherent invasive Escherichia coli LF82 bacteria interacting with intestinal epithelial cells. J Bacteriol. 2010;192:1832-1843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 247] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 58. | Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146:508-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 194] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 59. | Chassaing B, Rolhion N, de Vallée A, Salim SY, Prorok-Hamon M, Neut C, Campbell BJ, Söderholm JD, Hugot JP, Colombel JF. Crohn disease--associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J Clin Invest. 2011;121:966-975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 201] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 60. | Denizot J, Sivignon A, Barreau F, Darcha C, Chan HF, Stanners CP, Hofman P, Darfeuille-Michaud A, Barnich N. Adherent-invasive Escherichia coli induce claudin-2 expression and barrier defect in CEABAC10 mice and Crohn’s disease patients. Inflamm Bowel Dis. 2012;18:294-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 61. | Wine E, Ossa JC, Gray-Owen SD, Sherman PM. Adherent-invasive Escherichia coli, strain LF82 disrupts apical junctional complexes in polarized epithelia. BMC Microbiol. 2009;9:180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 62. | Mankertz J, Amasheh M, Krug SM, Fromm A, Amasheh S, Hillenbrand B, Tavalali S, Fromm M, Schulzke JD. TNFalpha up-regulates claudin-2 expression in epithelial HT-29/B6 cells via phosphatidylinositol-3-kinase signaling. Cell Tissue Res. 2009;336:67-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 129] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 63. | Ossa JC, Ho NK, Wine E, Leung N, Gray-Owen SD, Sherman PM. Adherent-invasive Escherichia coli blocks interferon-γ-induced signal transducer and activator of transcription (STAT)-1 in human intestinal epithelial cells. Cell Microbiol. 2013;15:446-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Bringer MA, Glasser AL, Tung CH, Méresse S, Darfeuille-Michaud A. The Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell Microbiol. 2006;8:471-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 113] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 65. | Bringer MA, Barnich N, Glasser AL, Bardot O, Darfeuille-Michaud A. HtrA stress protein is involved in intramacrophagic replication of adherent and invasive Escherichia coli strain LF82 isolated from a patient with Crohn’s disease. Infect Immun. 2005;73:712-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 66. | Bringer MA, Rolhion N, Glasser AL, Darfeuille-Michaud A. The oxidoreductase DsbA plays a key role in the ability of the Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82 to resist macrophage killing. J Bacteriol. 2007;189:4860-4871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 67. | Simonsen KT, Nielsen G, Bjerrum JV, Kruse T, Kallipolitis BH, Møller-Jensen J. A role for the RNA chaperone Hfq in controlling adherent-invasive Escherichia coli colonization and virulence. PLoS One. 2011;6:e16387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 68. | Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn’s disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529-5537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 348] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 69. | Carvalho FA, Barnich N, Sauvanet P, Darcha C, Gelot A, Darfeuille-Michaud A. Crohn’s disease-associated Escherichia coli LF82 aggravates colitis in injured mouse colon via signaling by flagellin. Inflamm Bowel Dis. 2008;14:1051-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 70. | Meconi S, Vercellone A, Levillain F, Payré B, Al Saati T, Capilla F, Desreumaux P, Darfeuille-Michaud A, Altare F. Adherent-invasive Escherichia coli isolated from Crohn’s disease patients induce granulomas in vitro. Cell Microbiol. 2007;9:1252-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 71. | Dunne KA, Allam A, McIntosh A, Houston SA, Cerovic V, Goodyear CS, Roe AJ, Beatson SA, Milling SW, Walker D. Increased S-nitrosylation and proteasomal degradation of caspase-3 during infection contribute to the persistence of adherent invasive Escherichia coli (AIEC) in immune cells. PLoS One. 2013;8:e68386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 72. | Peeters H, Bogaert S, Laukens D, Rottiers P, De Keyser F, Darfeuille-Michaud A, Glasser AL, Elewaut D, De Vos M. CARD15 variants determine a disturbed early response of monocytes to adherent-invasive Escherichia coli strain LF82 in Crohn’s disease. Int J Immunogenet. 2007;34:181-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 73. | Nieminen JK, Sipponen T, Färkkilä M, Vaarala O. Monocyte-derived dendritic cells from Crohn’s disease patients exhibit decreased ability to activate T helper type 17 responses in memory cells. Clin Exp Immunol. 2014;177:190-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 74. | Chargui A, Cesaro A, Mimouna S, Fareh M, Brest P, Naquet P, Darfeuille-Michaud A, Hébuterne X, Mograbi B, Vouret-Craviari V. Subversion of autophagy in adherent invasive Escherichia coli-infected neutrophils induces inflammation and cell death. PLoS One. 2012;7:e51727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 75. | Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 2009;9:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 76. | Chassaing B, Darfeuille-Michaud A. The σE pathway is involved in biofilm formation by Crohn’s disease-associated adherent-invasive Escherichia coli. J Bacteriol. 2013;195:76-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 77. | Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, Knight P, Codling C, Marchesi JR, Winstanley C. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 78. | Martinez-Medina M, Mora A, Blanco M, López C, Alonso MP, Bonacorsi S, Nicolas-Chanoine MH, Darfeuille-Michaud A, Garcia-Gil J, Blanco J. Similarity and divergence among adherent-invasive Escherichia coli and extraintestinal pathogenic E. coli strains. J Clin Microbiol. 2009;47:3968-3979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 77] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 79. | Clarke DJ, Chaudhuri RR, Martin HM, Campbell BJ, Rhodes JM, Constantinidou C, Pallen MJ, Loman NJ, Cunningham AF, Browning DF. Complete genome sequence of the Crohn’s disease-associated adherent-invasive Escherichia coli strain HM605. J Bacteriol. 2011;193:4540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 80. | Krause DO, Little AC, Dowd SE, Bernstein CN. Complete genome sequence of adherent invasive Escherichia coli UM146 isolated from Ileal Crohn’s disease biopsy tissue. J Bacteriol. 2011;193:583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 81. | Small CL, Reid-Yu SA, McPhee JB, Coombes BK. Persistent infection with Crohn’s disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat Commun. 2013;4:1957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 133] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 82. | Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang ZD, Dupont HL, Garneau P, Harel J. Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm Bowel Dis. 2013;19:141-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 83. | Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 532] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 84. | Jensen SR, Fink LN, Nielsen OH, Brynskov J, Brix S. Ex vivo intestinal adhesion of Escherichia coli LF82 in Crohn’s disease. Microb Pathog. 2011;51:426-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 85. | Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 149] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 86. | Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. Bile salts induce long polar fimbriae expression favouring Crohn’s disease-associated adherent-invasive Escherichia coli interaction with Peyer’s patches. Environ Microbiol. 2013;15:355-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 87. | Vazeille E, Bringer MA, Gardarin A, Chambon C, Becker-Pauly C, Pender SL, Jakob C, Müller S, Lottaz D, Darfeuille-Michaud A. Role of meprins to protect ileal mucosa of Crohn’s disease patients from colonization by adherent-invasive E. coli. PLoS One. 2011;6:e21199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 88. | Burke DA, Axon AT. Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhoea. BMJ. 1988;297:102-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 96] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 89. | Giaffer MH, Holdsworth CD, Duerden BI. Virulence properties of Escherichia coli strains isolated from patients with inflammatory bowel disease. Gut. 1992;33:646-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 82] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 90. | Thomazini CM, Samegima DA, Rodrigues MA, Victoria CR, Rodrigues J. High prevalence of aggregative adherent Escherichia coli strains in the mucosa-associated microbiota of patients with inflammatory bowel diseases. Int J Med Microbiol. 2011;301:475-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 91. | Iebba V, Conte MP, Lepanto MS, Di Nardo G, Santangelo F, Aloi M, Totino V, Checchi MP, Longhi C, Cucchiara S. Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease. Infect Immun. 2012;80:1408-1417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 92. | Raso T, Crivellaro S, Chirillo MG, Pais P, Gaia E, Savoia D. Analysis of Escherichia coli isolated from patients affected by Crohn’s disease. Curr Microbiol. 2011;63:131-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Negroni A, Costanzo M, Vitali R, Superti F, Bertuccini L, Tinari A, Minelli F, Di Nardo G, Nuti F, Pierdomenico M. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:913-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 94. | Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 738] [Cited by in RCA: 949] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 95. | Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, Lochs H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 219] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 96. | Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 330] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 97. | Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8:e56964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 401] [Cited by in RCA: 405] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 98. | Schippa S, Iebba V, Barbato M, Di Nardo G, Totino V, Checchi MP, Longhi C, Maiella G, Cucchiara S, Conte MP. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 2010;10:175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 99. | Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 288] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 100. | De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 253] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 101. | Sánchez E, Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 2008;8:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 102. | Martinez-Medina M, Garcia-Gil J, Barnich N, Wieler LH, Ewers C. Adherent-invasive Escherichia coli phenotype displayed by intestinal pathogenic E. coli strains from cats, dogs, and swine. Appl Environ Microbiol. 2011;77:5813-5817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 103. | Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL, German AJ, Yates RM, Russell DG, Johnson SE. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immun. 2006;74:4778-4792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 195] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 104. | Nickerson KP, McDonald C. Crohn’s disease-associated adherent-invasive Escherichia coli adhesion is enhanced by exposure to the ubiquitous dietary polysaccharide maltodextrin. PLoS One. 2012;7:e52132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 105. | Roberts CL, Keita AV, Duncan SH, O’Kennedy N, Söderholm JD, Rhodes JM, Campbell BJ. Translocation of Crohn’s disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut. 2010;59:1331-1339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 106. | Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 397] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 107. | Hillman LC, Peters SG, Fisher CA, Pomare EW. Effects of the fibre components pectin, cellulose, and lignin on bile salt metabolism and biliary lipid composition in man. Gut. 1986;27:29-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 31] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 108. | Rutgeerts P, Ghoos Y, Vantrappen G, Fevery J. Biliary lipid composition in patients with nonoperated Crohn’s disease. Dig Dis Sci. 1986;31:27-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 109. | Lapidus A, Akerlund JE, Einarsson C. Gallbladder bile composition in patients with Crohn ‘s disease. World J Gastroenterol. 2006;12:70-74. [PubMed] |
| 110. | Drouet M, Vignal C, Singer E, Djouina M, Dubreuil L, Cortot A, Desreumaux P, Neut C. AIEC colonization and pathogenicity: influence of previous antibiotic treatment and preexisting inflammation. Inflamm Bowel Dis. 2012;18:1923-1931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 111. | Craven M, Egan CE, Dowd SE, McDonough SP, Dogan B, Denkers EY, Bowman D, Scherl EJ, Simpson KW. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS One. 2012;7:e41594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 112. | Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 168] [Article Influence: 15.3] [Reference Citation Analysis (0)] |