INTRODUCTION
Each organ in the human body has unique specialized structures responsible for specific functions. Two investigational approaches are revealing the importance of the organization of molecular constituents in protein structure and function. The first approach focuses on one specific molecule at a time, the structure of the molecule, and the function the molecule is responsible for delivering. The second approach uses a high throughput analyses, capturing molecules in specific locations, performing experiments that enable us to determine their roles, and functions at these locations. The overarching goal of such high throughput experiments is a faster as well as greater understanding of composition, structure, and function. Proteomic analyses of different parts of the eye, in particular the anterior eye structures, involve high throughput methods that help identify proteins and their posttranslational modifications. Proteomics involves all methods that help identify proteins in the anterior eye chamber. The mass spectrometric methods to identify proteins in different locations in the anterior chamber use relatively older techniques and do not properly portray our current state of understanding. We aim to review the current state of advancement in identification of anterior chamber proteins, compared to the data gathered in the earliest era of proteomic mass spectrometry. We will present information on the following areas: cornea, aqueous humor, trabecular meshwork, ciliary body, iris, and lens. As each section of the anterior eye is uniquely different in protein, function and pathology, we have written the review specific to, what we believe, are the key relevant findings in the literature.
MASS SPECTROMETRIC PROTEOMIC ANALYSES OF THE CORNEA
The human cornea is a transparent, avascular, and highly specialized connective tissue which reflects and absorbs light into the lens and retina, and contributes two thirds of the eye’s refractive power. It is the most densely innervated tissue in the body and acts to protect the eye from infection as well as UV light[1]. The cornea also acts as a structural barrier providing the eye with biomechanical stability[2]. It is approximately 530 μm in thickness and is composed of five layers: the epithelium, Bowman’s layer, the stroma, Descemet’s membrane, and the endothelium[3]. The stroma contributes 90% of corneal volume[3]. Diseases of the cornea are commonly infectious, traumatic or genetic in nature and have a tendency to affect certain layers of the cornea[4]. Especially in developing countries, corneal disease often contributes to blindness. The most common etiologies of corneal blindness globally include infectious trachoma (C. trachomatis), oncherciasis (O. volvulus), leprosy (M. lepromatosis), and hypovitaminosis D (xerophthalmia)[5]. Keratoconus and Fuch’s dystrophy, diseases of the stroma and endothelium respectively, are the most common causes of corneal disease resulting in blindness in developed countries[5].
In recent years, our understanding of the identities and functions of the various proteins involved in the cornea has grown immensely. In 2005 just over 140 proteins were identified in the cornea[3]. Since then, over 3000 proteins have been characterized[4]. We have chosen here to focus on a narrow set of 12 proteins that have been identified in multiple studies, and which have important cellular functions.
Transforming growth factor-beta-induced protein (TGFβIp) has been identified in multiple corneal proteome studies[3,6,7] and has been implicated in corneal disease[8]. Numerous isoforms of TGFβIp have been found in the human cornea with 29 isoforms being found in earlier mass spectrometric studies in the mid-2000s[3]. This protein group’s most frequently described isoform, TGFβIp ig-h3, is 683 amino acids in length and has been described in several cellular compartments[7]. These include the membrane, Golgi apparatus, cytoplasm, endoplasmic reticulum, extracellular matrix/space, and the mitochondria[7,8]. Its molecular functions include catalysis, binding of nucleotides, signal transduction, regulation of enzyme activity, protein binding, and cell adhesion[7,8]. The relative abundance of this protein has been shown to be especially high in the stroma and endothelium[4]. In the stroma, it has been characterized as the second most abundant protein (17.6% abundance), and in the endothelium it has been described as the most abundant protein (36.8% abundance)[4]. As mentioned previously, this protein has been implicated in several disease states, including Fuch’s endothelial corneal dystrophy[8]. Simply put, this disease involves the progressive loss of endothelial cells, which is associated with impaired vision[5]. Increased expression and accumulation of TGFβIp ig-h3 has also been associated with other corneal and lattice dystrophies[5]. Overall, more than 50 mutations of this protein have been noted to be involved in disease states[5].
Peroxiredoxins are a group of redox associated proteins[6] which play a role in oxidative stress response in the cornea[9]. These proteins decompose peroxide molecules[10]. It is thought that decreased expression of these and other antioxidant proteins may play a role in Fuch’s dystrophy and keratoconus[8,9]. Peroxiredoxins 1, 2, and 6 have consistently been identified in corneal samples by mass spectrometry[3,6,7]. Peroxiredoxin 1 is 199 amino acids in length, and is found in the membrane, cytoplasm, nucleus, extracellular space, and mitochondria. It is involved in functions such as catalysis, DNA and protein binding, and inhibition of oxidation[7]. Peroxiredoxin 2 is 198 amino acids in length, found in the cytoplasm, nucleus, cytosol, mitochondria, organelle lumena, and chromosomes. It is also involved in catalysis, protein binding, and inhibition of oxidation, as well as metallic ion binding[7]. Peroxiredoxin 6 is 224 amino acids, and is found in similar cellular compartments as Peroxiredoxins 1 and 2, as well as in vacuoles; it also has similar cellular activities as its predecessors[7].
Transketolase is an enzyme involved in the pentose phosphate pathway and is involved in cell transparency[11]. It has been shown to be downregulated in keratoconus[12]. This protein is 623 amino acids in length and is found in the cytoplasm and cytosol. In addition to catalysis, it is involved in protein and metallic ion binding[7].
Mitochondrial ATP synthase subunit alpha has also been found in multiple mass spectrometric corneal proteomic investigations. It is made up of 553 amino acids, and is found in the membrane, cytoplasm, extracellular space, mitochondria, and organelle lumena. In addition to its catalytic function, it also binds proteins, metals, and nucleotides and has transporter actions[7].
At a cellular level, L-lactate dehydrogenase is involved in fermentation of pyruvate to lactate. The protein is upregulated in keratoconus[12]. The beta chain of this protein is 334 amino acids and is found in the cytoplasm, cytosol, nucleus, extracellular space, and mitochondri. It has been found in several corneal proteomics investigations, and in addition to its catalytic activity, it plays a role in transcription regulation, binding of nucleotides and metal ions, and transporter activity. It also regulates other enzymes[7].
F-actin-capping protein subunit alpha-1 is part of a protein which interacts with the fast-growing ends of actin filaments to prevent subunit exchange[13]. Its role in the cornea is not well characterized but it may play some role in colon cancer[14]. The protein is 286 amino acids in length and exists in a wide variety of cellular spaces. In addition to its catalytic activity, it is a structural protein, binds proteins and metals, regulates enzyme activity, and plays a role in redox reactions[7].
Vimentin is a class III intermediate filament protein[15]. It is composed of 466 amino acids and is seen in the cytoskeleton, membrane, cytoplasm, cytosol, and extracellular space. It functions in catalysis, DNA and protein binding, motor and transportation activities, and is involved in structural activities[7]. It has been found to be increased in the epithelium of corneas with keratoconus. As this protein is generally found in mesenchymal cells, it is thought that epithelial to mesenchymal transformation may be a possible characteristic of keratoconus[15].
Annexin A5 is a blood/plasma protein[6] which is thought to be involved in cellular apoptosis and its expression is used to determine cytotoxicity[16]. This protein is found in the cytoplasm and extracellular space. It is 320 amino acids long, and functions in metal and protein binding, as well as in the regulation of enzymes[7].
Keratin, type II cytoskeletal 4 is a protein found in the cytoskeleton. It is 534 amino acids in length, and functions in a wide array of cellular roles including catalysis, binding of nucleotides and proteins, and motor and structural molecular activities[7]. Epidermal fatty acid-binding protein is a small cytoplasmic protein of 135 amino acids, which is primarily involved in catalysis, protein binding, and transporter activity[7].
Understanding cornea proteomics has helped identify key proteins which in turn increased bimolecular understanding of disease and functions of proteins in wound healing[17,18].
MASS SPECTROMETRIC PROTEOMIC ANALYSES OF THE AQUEOUS HUMOR
The aqueous humor plays a substantial role in maintaining homeostasis within the eye. The pigmented and non-pigmented ciliary epithelium is responsible for production of aqueous humor, which is secreted into the posterior chamber. From the posterior chamber a majority of the aqueous humor traverses the trabecular meshwork, (a filter like structure), and flows into the Schlemm’s canal where it continues on to bathe the cornea. A small amount of the aqueous humor follows a less conventional pathway, the uveosceral pathway. The aqueous humor distributes through many sections of the anterior eye and is thus a key component in looking for proteomic biomarkers. A complication in the investigation of these biomarkers is is that there is only 150-200 μL of aqueous humor in an average age individual and this amount decreases with age. There is also a low overall protein concentration present in the aqueous humor. These obstacles can make protein analysis in the aqueous humor challenging and with time, specialized techniques have evolved to provide more accurate analysis. Through the evolution of these specialized techniques, different groups have used specific techniques to analyze the protein make-up of the aqueous humor.
The aqueous humor is abundant in numerous proteins such as antioxidant proteins, immunoregulatory proteins, and anti-angiogenic proteins. These proteins were identified using Multidimensional Protein Identification Technology (MudPIT)[19]. Protein composition of the aqueous humor is intricate as it is a key regulatory component of the eye. Up to 676 nonredundant proteins have been identified in the aqueous humor of patients with no disease. These proteins were identified using nanoflow liquid chromatography electrospray ionization tandem mass spectrometry (nano-LC-ESI-MS/MS). An issue that complicates this type of identification is the high prevalence of albumin, a protein that which makes up 50% of the proteins in the aqueous humor. Its abundance result in the masking of less abundant proteins during analysis. In order to overcome this issue, immunodepletion of several aqueous humor samples of albumin, transferrin, antitrypsin, haploglobin, fibrinogen, IgG, and IgA is commonly performed[13]. The presence of complement regulatory molecules, specifically 23 complement proteins, demonstrates the importance of the aqueous humor in maintaining a healthy environment and protecting against autoimmune disease. Catalytic enzymes crucial for respiratory pathways are also present in the aqueous humor, specifically aldolase and ketolase. Angiogenin, and angiogenic inducer were present along with angiogenic inhibitors, specifically PEDF, type IV collagen, and vitamin D binding protein. Finally, members of the transforming growth factor β (TGFβ), tumor necrosis factor (TNF), fibroblast growth factor, interleukin, and growth differentiation families were also present in the aqueous humor[20]. Taken together, the numerous components present in the aqueous humor make it a powerful regulatory mechanism for maintaining homeostasis in the eye.
The identification of aqueous humor proteins in normal samples provided a baseline for further investigation to take part in diseased counterparts. The study of protein levels in the aqueous humor in diseased individuals provides substantial information for potential biomarkers to possibly identify disease earlier. Analyzing these protein levels also assists in further profiling the protein composition of the aqueous humor. Glaucoma refers to a family of eye optic nerve disorders, some of which are associated with increased intraocular pressure (IOP). The most common form of glaucoma is primary open angle glaucoma. Research has been carried out to analyze alterations in the protein composition of the aqueous humor in patients with increased IOP. Endothelial leukocyte adhesion molecule 1 (ELAM 1) plays a key role in inflammation and is significantly increased in glaucomatous aqueous humor. Interestingly, apolipoprotein B and E are present in increased amounts. Typically, these proteins are responsible for in the delivery of cholesterol to cells. Another set of proteins present are responsible for muscle cell differentiation and function, specifically, myotrophin, myoblast determination protein 1, myogenin, vasodilator-stimulated phosphoprotein, and ankyrin-2. Presence of stress response proteins such as heat shock 60 kilodaltons (kDa) and 90 kDa proteins as well as ubiquitin fusion degradation 1-like are responsible for the removal of damaged protein. Finally, phospholipase C, β, and γ are shown to take part in signal transduction as well as neural development[21]. Similarly, in an investigation performed in patients with primary congenital glaucoma, a select set of proteins was shown to be upregulated and downregulated. Apolipoprotein A-IV (APOA-IV) is a plasma protein commonly involved in lipid absorption and transport. This specific protein is increased in glaucomatous samples. Albumin was also increased in these samples. This protein is crucial for maintenance of colloid osmotic pressure of plasma, antioxidant activity, regulation of normal microvascular permeability as well as fatty acid, and hormone transport. Another protein increased in glaucomatous aqueous humor is antithrombin 3 (ANT3 or SERPINC1), a protease inhibitor belonging to the serpin family. There were several proteins downregulated in glaucomatous samples including Transthyretin (TTR), Glutathione independent prostaglandin D synthase (PTGDS), opticin (OPT), and Retinol binding protein 3 IRBP. TTR is the main iodothyronine-binding protein that transfers T4 from the blood in the brain across the blood-choroid plexus barrier and tends to decrease in serum when acute inflammation is taking place. PTGDS is responsible for converting prostaglandin H2 (PGH2) to prostaglandin D2 (PGD2), common in smooth muscle contraction/relaxation as well as platelet aggregation inhibition. This protein has been demonstrated to bind to retinal and retinoic acid, key players in tissue development/maintenance. OPT, a member of the small leucine-rich repeats proteoglycan (SLRP) gene family is believed to be anti-angiogenic, is present in normal aqueous humor. IRBP is a glycoprotein synthesized by rods and cones. This protein binds to retinoids as well as fatty acids and may act as a retinoid transporter[22]. The presence of these proteins further supports the idea of necessary equilibrium between different elements in the eye that needs to take place in order to maintain a healthy environment.
The profiling of the proteins in the aqueous humor has given insight to its importance as a regulator in many aspects of the eye. Investigating these proteins in the normal state has been as important as investigating those in the diseased state. Overall, the investigations carried out in this area further supports underline the importance of maintaining specific protein levels in the aqueous humor.
MASS SPECTROMETRIC PROTEOMIC ANALYSES OF THE TRABECULAR MESHWORK
The trabecular meshwork (TM) plays a fundamental role in the regulation of intraocular pressure (IOP) and is pathophysiologically involved in the development of glaucoma. The TM can be divided into the uveal, corneoscleral and juxtacanalicular meshworks. It consists of collagen beams, covered by endothelial cells and surrounded by extracellular matrix (ECM)[23,24]. Until recently, the pathogenesis of outflow resistance at the TM was largely unknown. Understanding the pathogenesis that contributes to outflow resistance has recently increased. We now know that TM cell gene expression alters with IOP and mechanical stress[25] which can induce changes in cell proteins. This can lead to altered cell behavior including the increased tendency of the TM to contract with raised IOP[26,27], alterations in metabolic processes, cell adhesion, signal transduction, regulation of transcription, increased stretch activated channels[28], and the remodeling of extracellular matrix of TM in POAG[29-32].
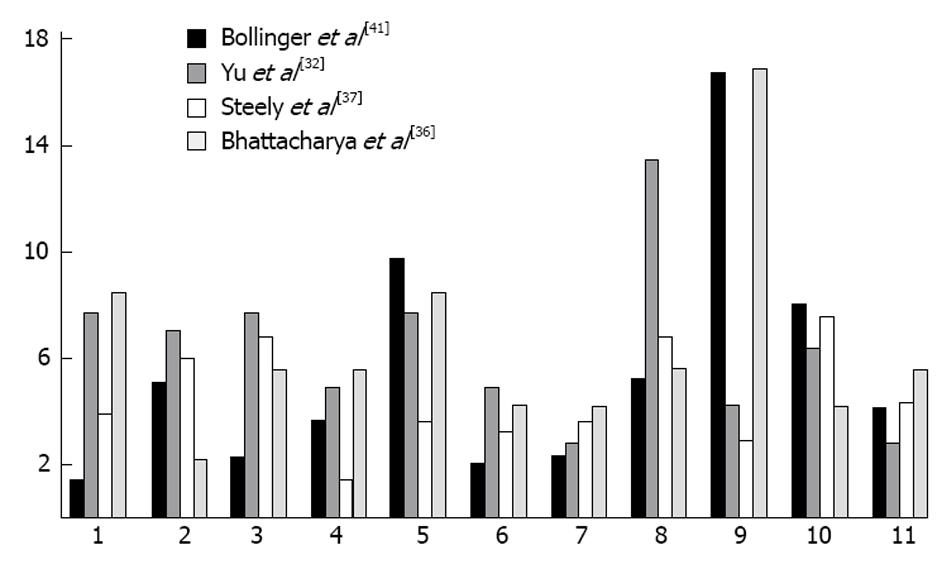
Proteomic analysis of the TM has played a major role in understanding the mechanisms involved in outflow obstruction. Over 850 proteins have been identified in the TM[32] and multiple studies have found alterations in the expression of proteins when IOP is raised[32-34].Multiple proteins are altered in location and quantity with glaucoma. We previously discovered that cochlin, a protein of unknown function is present in conjunction with stretch activated channels, in glaucomatous TM in human eyes but absent in normal samples[35]. Cochlin was also uniquely found in DBA/2J mice with hypertensive IOP but absent in DBA2J with a normal IOP[36]. A study by Yu et al[32] used 2-DE protein-expression, combined gel-spot to identify proteins in the TM of human donors, some of which were cultured in dexamethasone. This study found 877 proteins in human TM, several of which were previously associated with glaucoma. Several proteins belonged to cytoskeletal protein families/extracellular matrix proteins, such as vimentin, lamin, actin, and annexin. The highest proportion of proteins found were involved in metabolic processes (13%), and similar percentages of proteins were involved in anti-apoptosis, motility, carbohydrate metabolism (10%-11%) (Figure 1). In contrast, few proteins were found to play roles in cell division and cell to cell signaling. Another study which grouped protein by their function found the largest number group were in protein folding (16.8%) which was significantly more than what we and Yu et al[32] found (2.9% and 4.2%).
Figure 1 Comparing common protein functions in trabecular meshwork of eyes between fours students.
1: Anti-apoptosis; 2: Carbohydrate metabolic process; 3: Cell adhesion; 4: Cell cycle; 5: Cell motility; 6: Cell proliferation; 7: Lipid metabolic process; 8: Metabolic process; 9: Protein folding/metabolism; 10: Signal transduction; 11: Transport.
Myocilin is a protein found in the TM; mutations in this protein have been associated with glaucoma[38-40]. Myocilin a prominent component of TM exosomes, suggesting that exosomes could contribute to aqueous humour outflow from the trabecular meshwork. As there are few studies which have examined TM exosome proteomics and exosome protein mutation is involved in disease, this is an area of which deserves further investigation.
Transforming growth factor beta 2 (TGFβ2) is often elevated in the TM of patients with POAG. Bollinger et al[41] examined TGFβ2-induced proteomic changes from four donors who were treated with or without TGFβ2. Cellular proteins in the TM were then analyzed by liquid chromatography-mass spectrometry iTRAQ. This study found that TGFβ2 significantly altered 47 proteins. More than half of the elevated proteins induced extracellular matrix remodeling and cytoskeleton interaction. Thirty proteins were elevated and 17 decreased after TGFβ2 treatment. CD9 antigen and mitochondrial superoxide dismutase 2 (SOD2) were the most significantly reduced proteins 64% and 46%, respectively. Interestingly the proteins most greatly decreased were from the mitochondria (40%). Downregulation of mitochondrial proteins may result in mitochondrial dysfunction and reduced ATP production, which may lead to disruption of outflow dynamics.
Overall TM proteomic studies have identified multiple proteins alterations associated with hypertensive IOP. Modulated protein patterns in glaucomatous eyes have emerged through proteomic studies. Future studies may look further into the gene expression of these altered proteins for a better understanding of their occurrence.
MASS SPECTROMETRIC PROTEOMIC ANALYSES OF THE CILIARY BODY
The ciliary body is a circumfirential layer of tissue behind the iris in the anterior chamber of the eye. Its epithelium serves as the main production center of aqueous humor. In recent years, literature regarding the proteome of the ciliary body has been sparse and had utilized immunohistochemistry, immunofluorescence, and Western blot technology, resulting in the characterization of fewer than 50 discrete proteins[42]. However, in 2013 Goel et al[42] profiled the ciliary body proteome utilizing MS/MS analysis on an LTQ-Orbitrap Velos ETD mass spectrometer. In this study, samples from the human ciliary body were processed and run on an SDS-PAGE. The bands were subsequently excised and digested with trypsin prior to LC-MS/MS analysis. MS data was then searched against the NCBI protein database, and 2815 proteins were characterized. Included in these data were proteins previously identified using the aforementioned techniques, including collagen type XVIII alpha 1 (COL18A1), cytochrome P450 family 1 subfamily B polypeptide 1 (CYP1B1), Opticin (OPTC), and aquaporin 1 (AQP1). Several of these proteins have possible implications in ocular disease. OPTC has been investigated as a possible target for primary open angle glaucoma. AQP1 is involved in the production of aqueous humor and its movement into the anterior chamber[42].
Goel et al[42] also identified a large number (> 2000) of proteins which were unknown to exist in the ciliary body. Some of these novel molecules include proteins involved in metabolism and energy pathways such as Neutrophil cytosol factor 2, Myosin-11, Pyruvate kinase isozymes M1/M2, and Alpha-1-antitrypsin. Other proteins such as ER lumen protein retaining receptor 2, Tubulin beta-2A chain, Exportin-1 are involved in transport mechanisms. Exportin-1 is overexpressed in cancer cells. Leukocyte surface antigen CD47 and complement C3 are part of the immune response mechanism. Desmin is an intermediate filament, which when defective is involved in several myopathies.
The Goel et al[42] group further investigated the proteins that were common and disparate between the ciliary body and plasma, and the ciliary body and aqueous humor. The majority of proteins found in the ciliary body (1895 of 2791) were also found in the plasma, which contained a total of 9393 proteins and therefore had 7498 unique proteins. In the comparison of the ciliary body and aqueous humor, 211 of the 2891 ciliary body proteins were also found in the aqueous humor, leaving 321 unique aqueous humor proteins. These comparisons are important to know which proteins are natively found in the ciliary body, and which of them may have originated from elsewhere. In the future, work regarding ciliary body proteomics may explore the proteins now known to be unique in order to investigate further therapeutic targets.
MASS SPECTROMETRIC PROTEOMIC ANALYSES OF THE IRIS
Mass spectrometric analyses of the human iris proteome have not been well-published. Other methods of proteomic analysis have been used on a small number of known iris proteins. One such example includes the immunohistochemical analysis of Opticin (OPTC)[43]. The is protein was identified using an antibody targeting its amino terminal[43,44]. OPTC is the ortholog of a cDNA sequence which has been shown to be expressed abundantly in the iris[42,44]. Mass spectrometric analyses of this and other iris proteins are required to better characterize the more complete human anterior chamber proteome.
Mass spectrometric proteomic analyses of the lens
The Human lens is responsible for the refractive properties of the eye. It is avascular and contains one layer of epithelium found in the anterior capsule and posterior capsule. The lens is mostly acellular, consisting mainly of crystalline proteins with some non-crystalline proteins also present[44]. Its main function is to change shape and thus allow for accommodation of vision. Another function of the lens is to maintain transparency. Loss of accommodation results in presbyopia and loss of transparency results in cataract. There are 3 main types of crystalline proteins in the human body, including type α, β, and γ. Type α-A is a heat shock and chaperone protein and is found mostly in the lens while α-B is ubiquitous throughout the human body. It was also known that the α-crystallines play a role as heat shock proteins and are chaperone proteins. Most recently protein analysis was performed in a mice mouse model in which the genes responsible for the α-crystallin wereas missing. This was carried out to determine what happens with the other proteins inside of the lens giving further insight into the development of cataracts[45]. Wild type and αA/αB knockout mice were compared using two-dimensional gel electrophoresis and mass spectrometry. There was a greater abundance of histones H2A,H4, and H2B fragment, and a low molecular weight β1-catenin in postnatal 2 d of the knockout mice. There was increased abundance of βB2-crystallin and vimentin in 30 d-old lenses of knockout mice. Gel permeation chromatography was able to demonstrate an aggregation of β-crystalline. Therefore, the absence of crystalline type αA and αB resulted in changes of protein expression indicating that lens proteins also result in interactive functions beyond just plain functions the. Aggregation of α crystalline was also found by recent Matrix-assisted laser desorption/ionization (MALDI) studies[4].
Type γ requires the use of post-translational modification in order to maintain its transparency. Given that crystallins are life-long proteins, post-translational modification may play a role in the development of cataracts[46]. Heat and deamidation (a chemical reaction in which an amide functional group is removed from an organic compound and damages the amide-containing side chains of asparagine and glutamine) may play a role in the change of the physical properties of the protein. This study used 2D LC-MS/MS to examine which major lens proteins undergo deamidation and the exact sites of deamidation. It was found that all of the major proteins found in the lens were deamidated. Each crystallin protein differed in the sites and extents of deamidation. Many of the areas of deamidatation were characterized by the presence of a basic amino acid one residue from the glutamine and asparagine.
Although the lens consists mostly of crystalline proteins, the advent of new analytical techniques allowed for analysis of proteins involved in lens besides crystalline. One of the first complete proteomics studies to address the protein inside of the lens was in 2008[47]. The lens from fetal, cataract, and normal lenses were evaluated by 2D LC-MS/MS and PANTHER was used for protein classification. This study identified a total of 231 proteins across all of the lens samples. Fetal samples showed the highest amount of unique proteins compared to cataract and normal lenses. A 5-mm core of lens was used in the adult some of which lacked epithelial and outer cortical fibers which play a role in the metabolic machinery of the lens. The fetal samples were all pooled together. While many studies have shown the crystallin class as the dominant protein, this study showed that many low abundance proteins also existed in the lens.
A more recent study[48] was performed, using MALDI, which concentrated on the major protein differences for identification in order to determine the variances between proteins in age-related cataracts and normal lens nuclei. Observers graded cataracts and total solubilized proteins were compared using gel electrophoresis. MALDI was used to identify the proteins that had different abundances. LC-MS/MS analyses determined the compositions of > 200 kDa molecular weight aggregates found in age related nuclear cataract lens nuclei. It was identified that α, β-A3, βA4, βB1, and γD-crystallin were involving with the higher molecular weight aggregates. An uncharacterized protein found and this protein, along with αA, αB, and γ-D crystallin were more found to be more prone to aggregation. Therefore, aggregation of crystallins may account for the development of cataracts. Also, some enzymes may play a role in the protein aggregation and possibly accelerate the process.
Membrane proteins were purified from young mouse lenses and shotgun proteomics was employed in order to analyze the membrane proteins of the mouse lens cells[49]. These same techniques were then applied to analyze the human lens protein of the membrane[50]. HPLC-mass spectrometry with multidimensional protein identification technology (MudPIT) with and without phosphopeptide enrichment was applied for the study of the proteome of the lens membrane. There were 951 proteins that were identified in which 379 were membrane and membrane-associated proteins. Many of these proteins are responsible for carbohydrate metabolism, proteasome, cell-cell signaling and communication, glutathione metabolism and actin regulation.
LOXL-1 protein and apolipoprotein E, both found in the extracellular matrix, were abnormal in pseudoexfoliation syndrome, a disease of the anterior lens capsule[51]. This study performed mass spectrometry on isolated surgically removed anterior capsules in patients with pseudoexfoliation syndrome. Direct analysis showed LOXL-1 protein and apoliprotein E which shows that these extracellular matrix proteins play a role in pseudoexfoliation[52]. This study employed MALDI imaging on the anterior capsule which showed presence of LOXL-1 protein was more abundant in the iris region and apolipoprotein E in the pseudoexfoliation deposits in anterior capsule in the pupillary area. There could also be significant post-translational modification involved in promoting the aggregation of proteins.
The lens is unique in that it contains many fibers that are acellular and proteins that exist for the lifetime of the individual. The advantage of studying the proteomics of the lens is that it may provide a powerful model for the rest of the human body with regard to understanding the changes involved in proteins that are maintained throughout a lifetime. It is essential that the proteins maintain transparency, and aggregation may result in lack of solubility resulting in cataracts. Proteomic studies have shown that α-crystallins play a role in preventing aggregation and serving as chaperone proteins. α-crystallins are present only in the lens while α-B crystallin is ubiquitous throughout the human body and dysfunction of the α-B protein has been implicated in many degenerative disorders. Post-translational modification also plays a role in the lens protein.
CONCLUSION
Identification of proteins in different regions of the anterior chamber including the: cornea, aqueous humor, trabecular meshwork, ciliary body, and lens has expanded in recent years. Among other proteomic methods, mass spectrometry has enabled rapid protein sequencing while simultaneously determining posttranslational modifications in the amino acid residues. Mass spectrometry has rapidly evolved since 1990, allowing improved identification of proteins. Although the advances in mass spectrometry have been rapid, the identification of proteins from tissue or cell samples often remains unsatisfactory. Currently approximately 5000 proteins from each anterior eye segment tissue or fluid is identified against a theoretical prediction of 20000 proteins. Thus at best approximately 25% of actual proteins are captured compared to theoretical estimates. Part of the reason why protein identification is relatively poor compared to mRNA is due to differences in the chemistry of RNA and proteins. The identification of posttranslational modifications of proteins, remains another frontier in mass spectrometry (or any other suitable high throughput method) that is yet to be conquered. One important issue remaining to be elucidated is the process of natural aging. Several age-related changes that can be easily quantified occur in eyes such as prebyopia and the progressive ability to form sharp images. Several eye diseases are also age associated such as age-related macular degeneration and glaucoma. Important insight into true age related changes, and the result of aging and disease on protein turnover. The Current methods do not allow the juxtaposition of mRNA and protein information together. Modern proteomic methods lack in their ability to juxtapose mRNA and protein information from inactive proteins, deactivated proteins, or proteins undergoing degradation. These are the avenues for future advancement which will expand our insight into how protein-drug interactions keep proteins in their active states. We presented an account of the current state of proteins in different regions of anterior eye chamber and what improvement has occurred compared to that in the previous decade. Further improvements will enable us to address the question of protein turnover in tissues and better enable us to distinguish active, inactive, partially degraded, and degraded states of proteins.