Copyright
©The Author(s) 2023.
World J Biol Chem. Oct 17, 2023; 14(5): 84-98
Published online Oct 17, 2023. doi: 10.4331/wjbc.v14.i5.84
Published online Oct 17, 2023. doi: 10.4331/wjbc.v14.i5.84
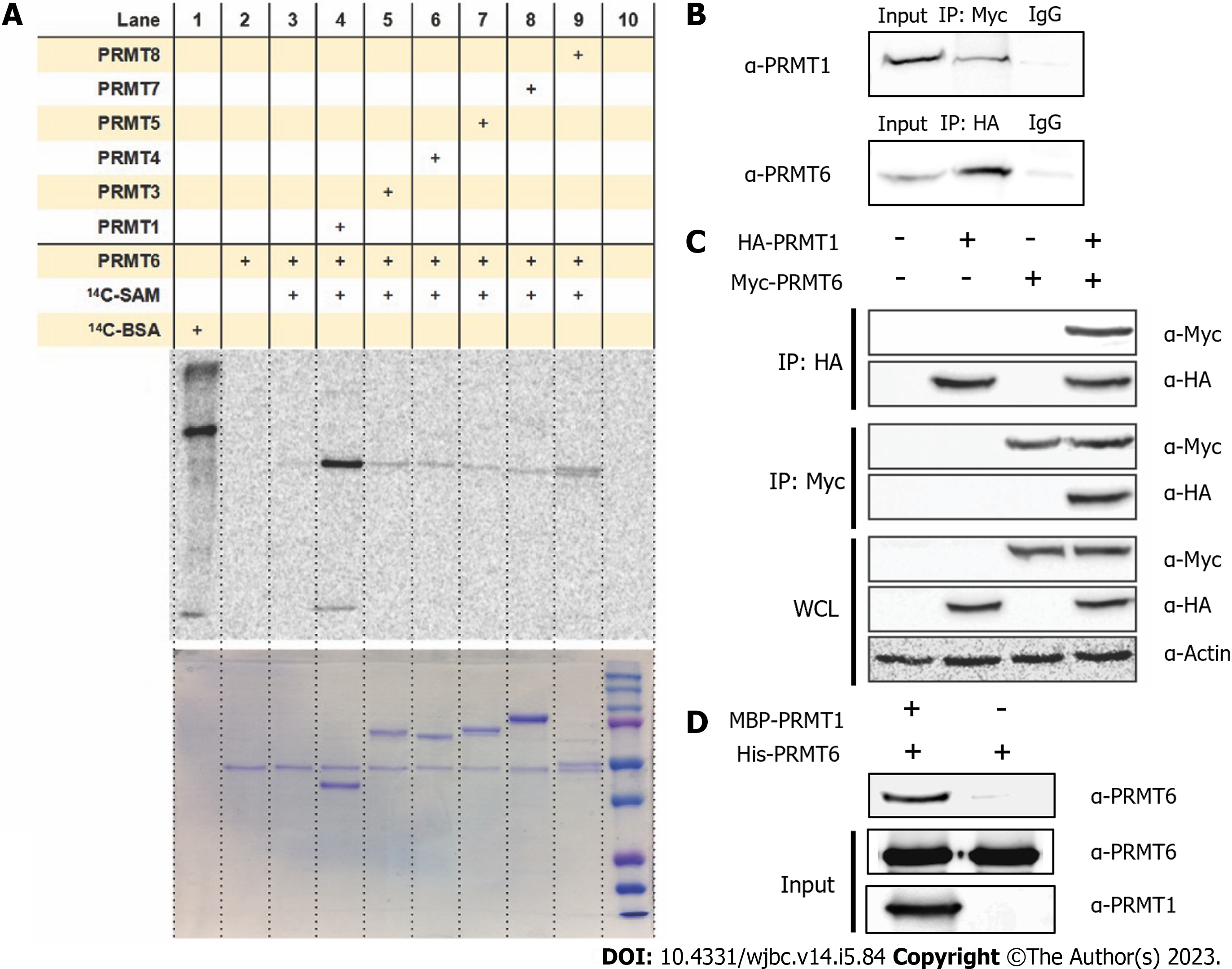
Figure 1 Protein arginine methyltransferase 1 interacts with protein arginine methyltransferase 6 in vitro and in vivo.
A: Gel electrophoresis-autoradiographic image analysis of protein arginine methyltransferase (PRMT) 6 methylation incubated with other PRMT family members. 0.5 µM of PRMT6 was incubated with 1 µM of PRMT1, PRMT3, coactivator associated arginine methyltransferase (PRMT4), PRMT5, PRMT7 and PRMT8. Reactions were held for 30 min at 30 ℃, the samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 12% polyacrylamide gel; B: Co-immunoprecipitation assay was performed between HA-PRMT1 and endogenous PRMT6 or Myc-PRMT6 and endogenous PRMT1 in HEK293T cells. Co-transfected HEK293T cell lysate was immunoprecipitated with anti-HA antibody or anti-Myc antibody followed by protein A/G plus agarose beads, proteins on the beads were eluted and separated on 12% SDS-PAGE and were detected by anti-PRMT1 or anti-PRMT6 antibody; C: Immunoprecipitation assay was performed between HA-PRMT1 and Myc-PRMT6. Single transfected HEK293T Cell lysate was immunoprecipitated with anti-HA antibody or anti-Myc antibody followed by protein A/G plus agarose beads, proteins on the beads were eluted and separated on 12% SDS-PAGE and were detected by anti-Myc or anti-HA antibody; D: Western blot analysis of maltose-binding protein (MBP) pull down assay. PRMT6 binds to purified Amylose beads or MBP-PRMT1 was detected. PRMT1 was incubated with the amylose beads for 30 min, followed by washing by phosphate-buffered saline and incubated with PRMT6 for 4h. The resin was washed and the pulled-down proteins were eluted and separated by 12% SDS-PAGE and detected by anti-PRMT1 or anti-PRMT6. IgG: Immunoglobulin G; PRMT: Protein arginine methyltransferase; SAM: S-adenosyl methionine.
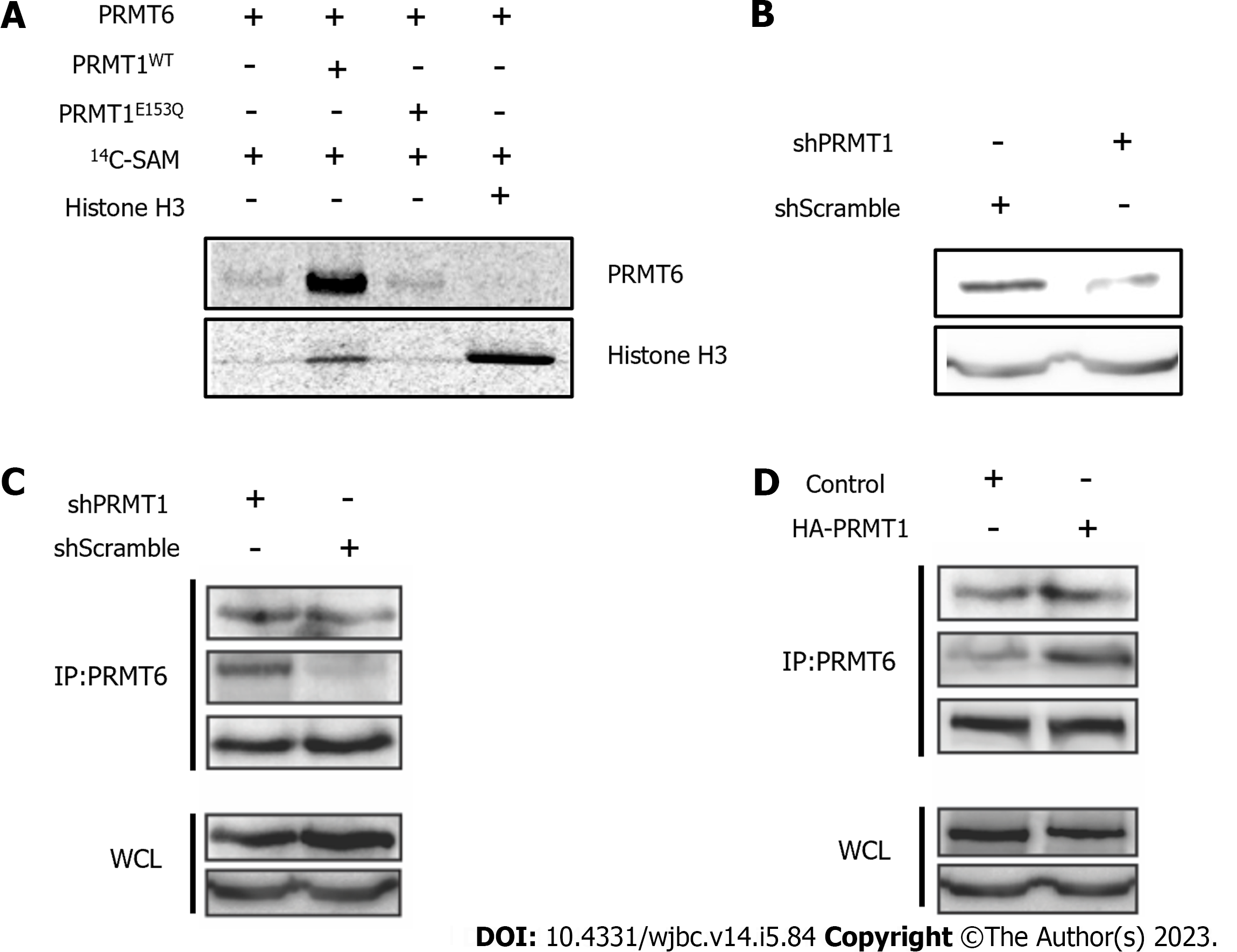
Figure 2 Protein arginine methyltransferase 6 is methylated by protein arginine methyltransferase 1.
A: Gel electrophoresis-autoradiographic image analysis of histone H3 methylation level change in the presence of protein arginine methyltransferase (PRMT) 1WT or PRMT1E153Q. 1 µM PRMT6 and 20 µM 14C-S-adenosyl methionine were incubated with or without 3 µM PRMT1WT, PRMT1E153Q or H3 protein respectively at 30 ℃ in the reaction buffer for 1h. The samples were separated on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis polyacrylamide gel. The radioactive SDS gel was dried and stored for two days and then scanned for the phosphor image; B: Western blot was performed to analysis PRMT1 level in HEK293T cells transfected by hPRMT1 or shScramble; C and D: Immunoprecipitation was performed to precipitate PRMT6 in HEK293T cell lysate transfected by shPRMT1 or shScramble or HA-PRMT1, followed by western blot to analysis monomethyl arginine and asymmetric dimethylarginine level of precipitated PRMT6. PRMT: Protein arginine methyltransferase.
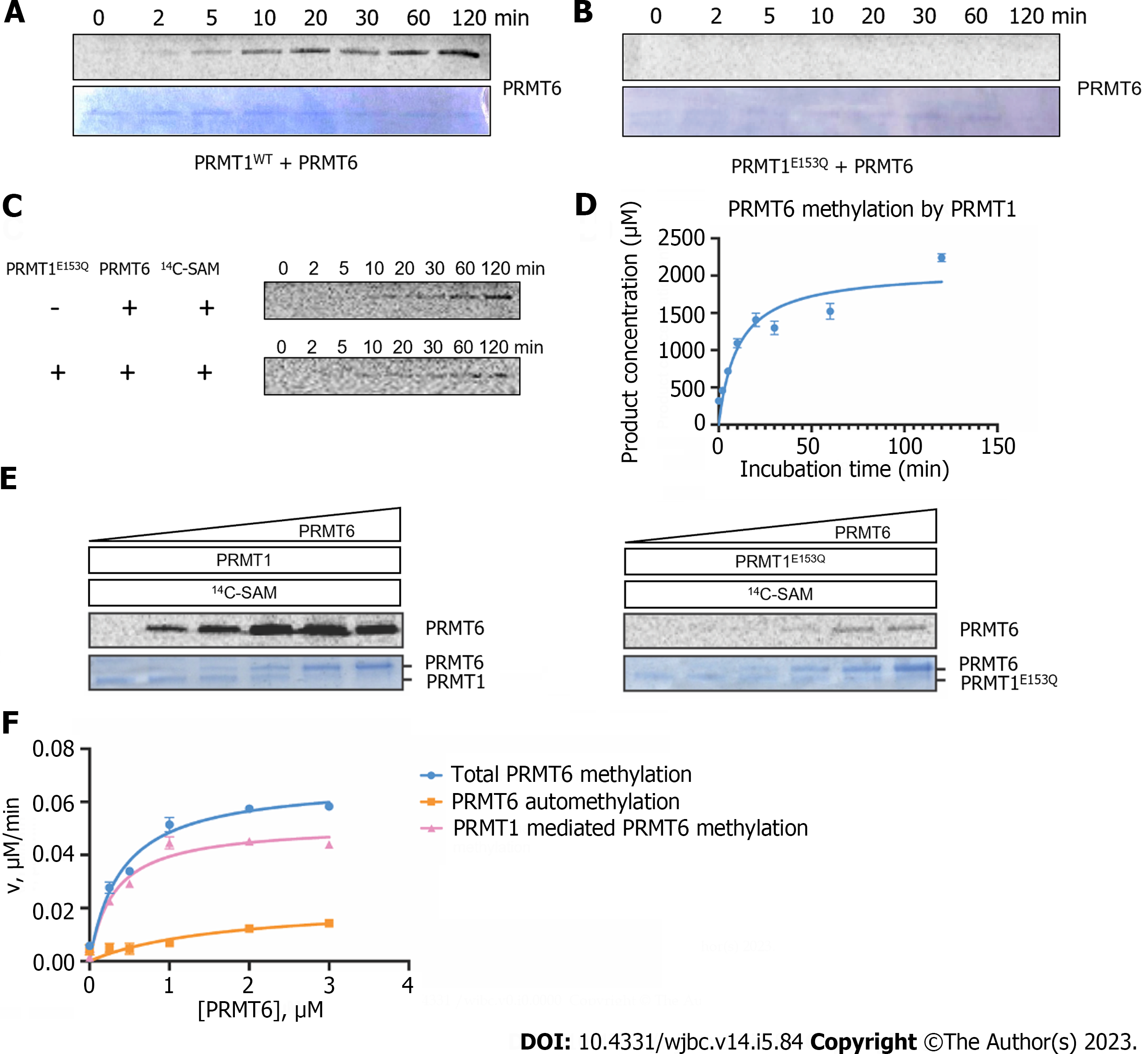
Figure 3 Time-dependent and concentration-dependent protein arginine methyltransferase 6 methylation catalyzed by protein arginine methyltransferase 1.
A and B: Time-dependent radioactive gel methyltransferase assays were carried out at 30 ℃ in an incubation system of 30 µL. 1.5 µM of protein arginine methyltransferase (PRMT) 6 and 10 µM of 14C-S-adenosyl methionine (SAM) were incubated with 0.4 µM of PRMT1WT or PRMT1E153Q, and the reactions were quenched at different time points (0-120 min) by sodium dodecyl sulfate (SDS) loading buffer. The reaction mixture was separated on 12% SDS-polyacrylamide gel electrophoresis (PAGE) and methylated PRMT6 were visualized by Coomassie blue staining and phosphor image scanning, respectively; C: Gel electrophoresis autoradiography assays were performed at 30 ℃ in an incubation system of 30 µL. 4.5 µM PRMT6 was incubated with 10 µM 14C-SAM with or without 0.4 µM of PRMT1E153Q; D: Time course of PRMT6 methylation with PRMT1, the linear part of the curve was used to calculate the steady-state rate; E: Concentration-dependent radioactive gel methyltransferase assay was performed. Different concentration of PRMT6 (0, 0.25, 0.5, 1, 2, 4 M) was incubated with 10 M 14C-SAM in the presence of 0.2 M of PRMT1WT or PRMT1E153Q in the reaction buffer at 30 ℃ for 1h. The reaction mixture was separated on 12% SDS-PAGE and the phosphor image was quantified by ImageJ. Data were normalized according to the reading of 5 M 14C-BSA and the reaction yield was calculated according to the reading of 5 M reaction mixture in liquid scintillation; F: Comparison of the total PRMT6 methylation in the presence of PRMT1, PRMT6 automethylation, and PRMT1-mediated PRMT6 methylation at different concentration of PRMT6. SAM: S-adenosyl methionine; PRMT: Protein arginine methyltransferase.
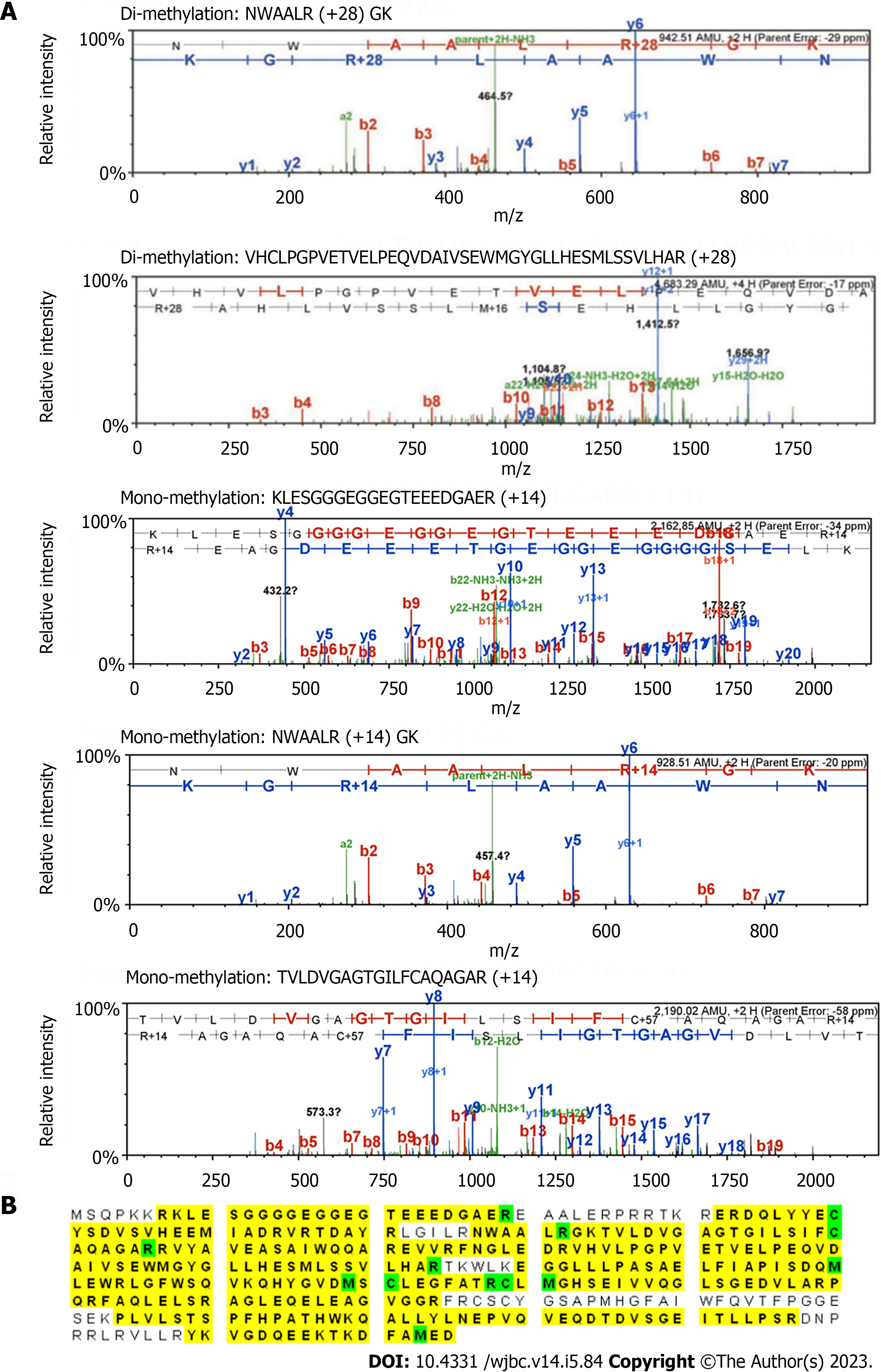
Figure 4 Liquid chromatography with tandem mass spectrometry analysis of the methylation sites on protein arginine methyltransferase 6.
A: 5.5 M protein arginine methyltransferase (PRMT) 6 and 200 M S-adenosyl methionine were incubated with 1 M PRMT1 at 30 ℃ for 3 h in the reaction buffer. The methylated PRMT6 band was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, cut from the gel, washed and vacuum-dried. The sample was sent to UAB proteomics and mass spectrometry facility (Birmingham, AL 35294) for in-gel digestion and liquid chromatography-tandem mass spectrometry (MS/MS) detection. Tandem MS spectra showing the arginine-dimethylated peptides; B: PRMT6 protein sequence coverage (yellow) and the modified residues (green) detected by tandem MS analysis. Green "C" is with carboamidomethylation, and green "M" is with oxidation.
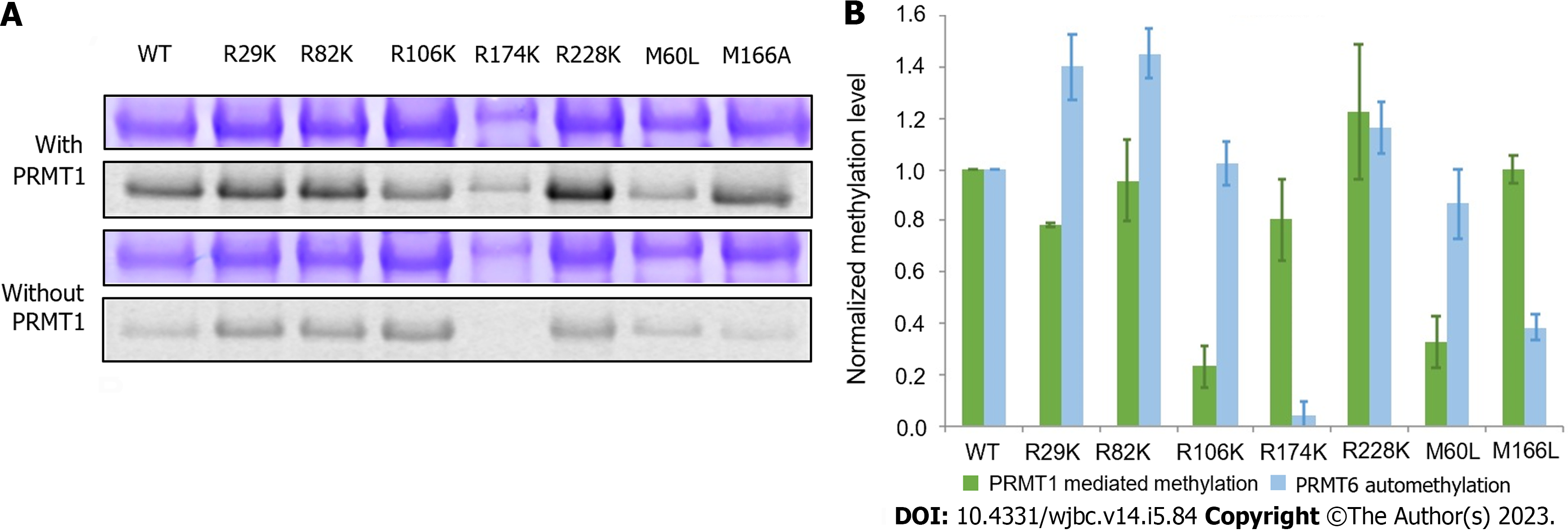
Figure 5 Comparison of protein arginine methyltransferase 1-mediated methylation and automethylation among different protein arginine methyltransferase 6 mutants.
A: Radioactive methyltransferase assay was performed to compare protein arginine methyltransferase (PRMT) 1-mediated methylation level and automethylation level of PRMT6 mutants. 5 M of each PRMT6 mutant was incubated with 30 M 14C-S-adenosyl methionine in the presence or absence of 1 M PRMT1 in the reaction buffer at 30 ℃ for 2 h. The products were separated on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and visualized by Coomassie blue staining and phosphor image scanning. The methylation intensity of each mutant was quantified with Quantity One and normalized against PRMT6WT. The assay was repeated at least three times. Coomassie blue staining and phosphor image of methylated PRMT6 mutants in the presence or absence of PRMT1; B: Column graph comparing the methylation of PRMT6 m. PRMT: Protein arginine methyltransferase.
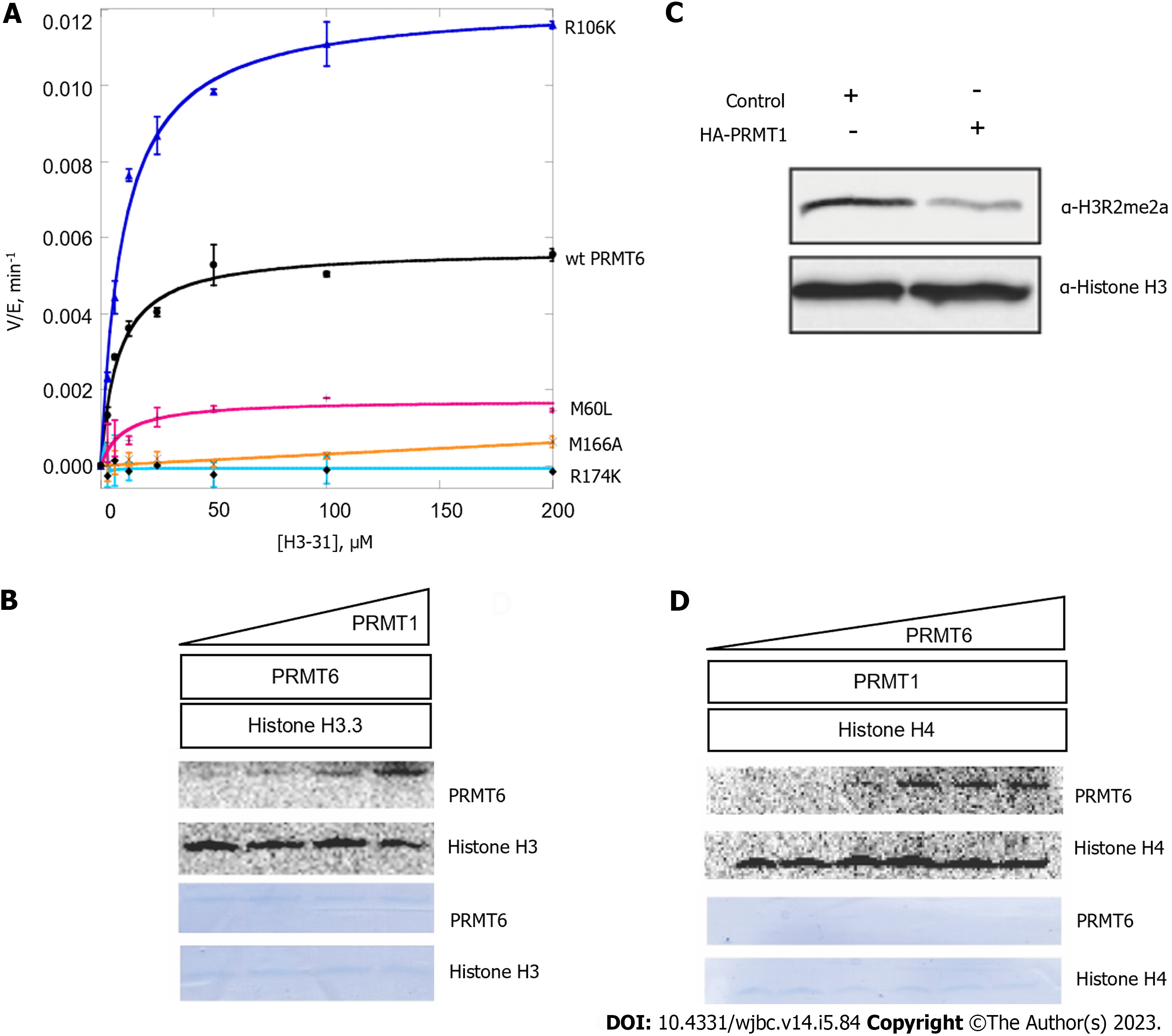
Figure 6 Methylation by protein arginine methyltransferase 1 may regulate the enzymatic activity of protein arginine methyltransferase 6.
A: Radioactive methyltransferase assay was performed to show activity of protein arginine methyltransferase (PRMT) 6 wild type and PRMT6 mutants on histone H3 (1-31). 0.6 M PRMT6 mutant and 30 µM 14C-S-adenosyl methionine (SAM) were incubated with 0 to 200 µM of H3 (1-31). After the enzyme was added, the reaction was continuously incubated for 60 min and then quenched by spreading the mixture onto P81 filter paper. Following washing and drying the paper, liquid scintillation was performed to quantify the methylated products. The data were fit to the Michaelis-Menten curve; B: Radioactive methyltransferase assay was performed to show histone H3 methylation level change as PRMT1 concentration change. Different concentration of PRMT1 was incubated with 30 µM 14C-SAM and 1 µM PRMT6 for 1 h, histone H3 was added and incubated for another 1 h. The systems were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and the phosphor image was captured after 72 h; C: Histone H3 methylation of HEK293T cell lysate was detected by western blot using anti-H3R2me2a antibody. HEK293T cell was transfected with HA-PRMT1 or control and cell lysate was extracted; D: Radioactive methyltransferase assay was performed to show histone H4 methylation level change as PRMT6 concentration change. Different concentration of PRMT6 was incubated with PRMT1 and 30 µM 14C-SAM, then 1 µM histone H4 was added and incubated for another 1 h. The systems were separated by 12% SDS-PAGE gel and the phosphor image was captured after 72 h. PRMT: Protein arginine methyltransferase.
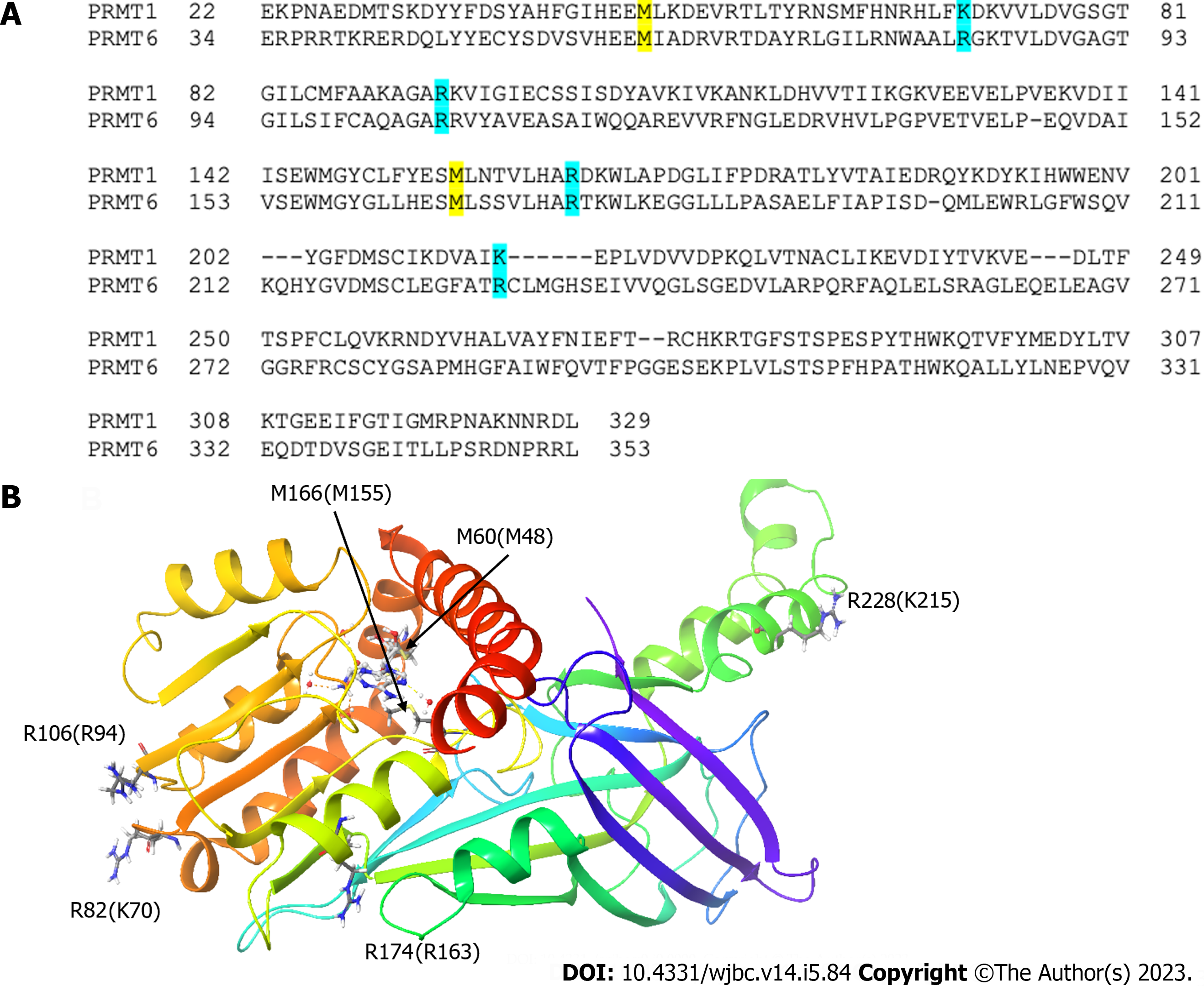
Figure 7 The mutation sites of protein arginine methyltransferase 6 and corresponding residues on protein arginine methyltransferase 1.
A: Sequence alignment of protein arginine methyltransferase (PRMT) 1 and PRMT6 showing the corresponding mutation sites; B: The corresponding mutation sites are highlighted on PRMT6 structure. hPRMT6 monomer (PDB: 4Y30) is shown in a cartoon model. The mutation sites of PRMT6 with their corresponding residues on PRMT1 and the S-Adenosyl-L-homocysteine molecule are shown in a stick model.
- Citation: Cao MT, Feng Y, Zheng YG. Protein arginine methyltransferase 6 is a novel substrate of protein arginine methyltransferase 1. World J Biol Chem 2023; 14(5): 84-98
- URL: https://www.wjgnet.com/1949-8454/full/v14/i5/84.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v14.i5.84