Published online Jan 27, 2025. doi: 10.4240/wjgs.v17.i1.100790
Revised: October 11, 2024
Accepted: November 18, 2024
Published online: January 27, 2025
Processing time: 112 Days and 5.9 Hours
Anal fistula is increasingly prevalent due to modern lifestyle factors, and surgery remains the primary treatment. However, the rising incidence of antibiotic re
To investigate the relationship between gut microbiota composition and cefu
This study included 30 anal fistula patients categorized into cefuroxime-sensitive (Cefur-S) and cefuroxime-resistant (Cefur-NS) groups. Gut microbiota samples were collected during colonoscopy, and 16S ribosomal DNA sequencing was per
Alpha and beta diversity analyses showed no significant differences in microbial diversity between the Cefur-S and Cefur-NS groups. However, effect size analysis identified Roseburia and Butyricicoccus as dominant genera in the Cefur-S group, with higher butyrate production potentially protecting against cefuroxime resis
This study suggests that specific gut microbiota, particularly Butyricicoccus and Roseburia, may mitigate cefuroxime resistance in anal fistula patients by increasing butyrate production. Probiotic intervention targeting gut microbiota composition presents a promising strategy for reducing antibiotic resistance and improving clinical outcomes.
Core Tip: This study explores the role of gut microbiota in cefuroxime resistance among anal fistula patients, highlighting the protective effects of butyrate-producing bacteria such as Butyricicoccus and Roseburia. The findings demonstrate that modulating gut microbiota through probiotic intervention can significantly reduce cefuroxime resistance, improve stool consistency, and lower bowel movement frequency. This research offers valuable insights into the potential of microbiota-based strategies to manage antibiotic resistance and enhance perioperative care in anal fistula patients.
- Citation: Ling YT, Yao F, Li SJ, Cao CX, Chen ZW, Qiu M, Li BZ, Hu BW, Zhong SY, Hu GL, Li JH. Microbiota in patients with cefuroxime resistance and anal fistula revealed by 16S ribosomal DNA. World J Gastrointest Surg 2025; 17(1): 100790
- URL: https://www.wjgnet.com/1948-9366/full/v17/i1/100790.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v17.i1.100790
Anal fistula is increasingly prevalent due to modern lifestyle and dietary habits, and surgery remains the primary treatment[1,2]. Traditional methods, such as fistulotomy and fistulectomy, though effective, can significantly damage anal sphincter function and surrounding tissues[3]. Newer techniques like the ligation of intersphincteric tract and transanal opening of intersphincteric space procedures aim to preserve sphincter function; however, the open and contaminated nature of surgical wounds still poses a risk for bacterial infections, leading to various postoperative complications. The use of antibiotics during the perioperative period for anal fistula surgery can help prevent infections[4]. However, the overuse of antibiotics has led to significant challenges in clinical treatment due to increasing antibiotic resistance[5]. In our department, we analyzed the antimicrobial susceptibility test results of anal crypt swabs from all anal fistula patients treated between July 2022 and July 2023. We observed a notably high resistance rate to cefuroxime, a commonly used antibiotic. After excluding factors such as lifestyle and dietary habits, we found that patients resistant to cefuroxime often had more frequent bowel movements and looser stools. This observation prompted us to investigate whether gut microbiota influences cefuroxime resistance in anal fistula patients, to better guide the selection and use of antibiotics during the perioperative period.
The 16S ribosomal DNA (16S rDNA) gene plays a crucial role in antimicrobial resistance research due to its highly conserved regions across different bacterial species[6]. By sequencing the 16S rDNA gene, researchers can identify and classify bacteria present in a sample, even those that are difficult to culture[7]. This method allows for the detection of resistant strains within complex microbial communities, facilitating the study of how resistance genes are distributed among different bacterial populations. Additionally, 16S rDNA sequencing can help in understanding the evolution and spread of resistance mechanisms in diverse environments[8]. In this study, we aim to explore the complex relationship between gut microbiota and the prognosis of anal fistula patients with cefuroxime resistance. By utilizing 16S rDNA, we can detect differences in gut microbiota among different patient groups, providing preliminary insights into the potential associations between microbial communities and clinical outcomes. Understanding the interaction between gut mi
This study included 30 patients who underwent surgery for anal fistula at our hospital between July 2023 and April 2024. The diagnosis was based on the latest diagnostic criteria, and all patients signed informed consent forms for treatment. The study protocol was reviewed and approved by the Ethics Committee of Jiaxing Second Hospital, approval Jiaxing Second Hospital Ethics Review, No. 2023-028.
Before the surgery, all patients had samples collected from the anal crypt using specialized bacterial culture swabs. The samples were kept at an appropriate temperature and promptly sent to the Clinical Laboratory of Jiaxing Second Hospital for bacterial culture and antibiotic susceptibility testing. Based on their sensitivity to cefuroxime, patients were categorized into two groups: The cefuroxime-sensitive (Cefur-S) group and the Cefur-NS group.
All patients underwent colonoscopy before surgery. Prior to the procedure, bowel preparation was carried out using a laxative. During the colonoscopy, normal saline was used to irrigate the ascending colon and sigmoid colon, ensuring thorough cleaning (Boston bowel preparation scale: ≥ 3 for both the left and right colon). After washing away residual stool and food debris, 2.5 mL of lavage fluid was aspirated from the ascending colon and 2.5 mL from the sigmoid colon. These samples were combined and stored at -80 °C in our hospital’s freezer for subsequent bacterial DNA extraction.
Patients in the Cefur-NS were administered oral Clostridium butyricum tablets (brand name: Miyairi tablets, produced by Miyarisan Pharmaceutical Co., Ltd., Japan) for 7 consecutive days. After the treatment period, patients were followed up via telephone, and during wound dressing changes, anal crypt swabs were taken again from the area around the wound for antibiotic susceptibility testing. The consistency of stool was assessed by scoring bowel movements over the 7-day period before and after the intervention: Watery stool (4 points), mushy stool (3 points), soft stool (2 points), formed stool (1 point), and hard stool (0 point). Scores were accumulated to generate a stool consistency score (lower scores indicate more formed stools). Additionally, the frequency of daily bowel movements was recorded.
Genomic DNA of the gut microbiota was extracted using the 2 × Phanta Max Master Mix kit (Vazyme Biotech Co., Ltd.). The library products were purified using the QIAquick Gel Extraction Kit (Qiagen) and the AGENCOURT®AMPURE® XP Kit (Beckman Coulter, Inc.). The purity and concentration of the samples were analyzed using the KAPA Library Quantification Kit for Illumina® Platforms. 16S rDNA profiling, targeting the V3-V4 hypervariable region, was performed using the following primers: 341-forward (5’-CCTACGGGNGGCWGCAG-3’) and 785-reverse (5’-GACTACHVGGGTA
Primers were removed from the sequences using cutadapt (version 2.4) software, followed by denoising with the dada2 denoise-paired command in QIIME2 (version 2023.9). The DADA2 pipeline generated amplicon sequence variant (ASV) abundance tables for each sample, with row names corresponding to sample IDs and column names to ASV identifiers. Representative ASV sequences were then aligned with a reference database for taxonomic annotation. The taxonomic annotation of ASVs was performed using the feature-classifier command in QIIME2, which identifies the species with the highest sequence similarity to each ASV. The Silva database (http://www.arb-silva.de) was used as the 16S rDNA reference database. Alpha and beta diversity analyses were conducted using QIIME2. Differential bacterial taxa between the two groups were identified using linear discriminant analysis (LDA) effect size (LEfSe), with significance set at P < 0.05 and an LDA score > 2. R software (version 4.1.2) was used for general data analysis. The Wilcoxon signed-rank test was employed to compare differences in 16S rDNA gene sequences between the two groups. Statistical significance was defined as P < 0.05.
A total of 30 patients were included in this study, comprising 27 males and 3 females, with an age range of 21 to 68 years (mean age: 42.78 ± 13.09 years). The average body mass index of the patients was 25.85 ± 4.98. Postoperative anal swab antibiotic susceptibility testing indicated that 9 patients were resistant to cefuroxime (Cefur-NS group), while the remaining 21 patients were sensitive to this antibiotic (Cefur-S group). There were no significant differences between the two groups in terms of age, gender distribution, height, weight or body mass index (Table 1).
| Features | Cefur-NS (n = 9) | Cefur-S (n = 21) | Statistics | P value |
| Age | 38.22 (12.91) | 44.71 (12.98) | -1.26 | 0.227 |
| Height | 172.33 (6.30) | 171.29 (6.25) | 0.42 | 0.682 |
| Weight | 77.89 (13.05) | 75.76 (17.78) | 0.36 | 0.719 |
| BMI | 25.83 (24.07-29.39) | 24.77 (22.68-25.47) | 109.50 | 0.512 |
| Sex | - | - | - | > 0.999 |
| Female | 1 (11.11) | 2 (9.52) | - | - |
| Male | 8 (88.89) | 19 (90.48) | - | - |
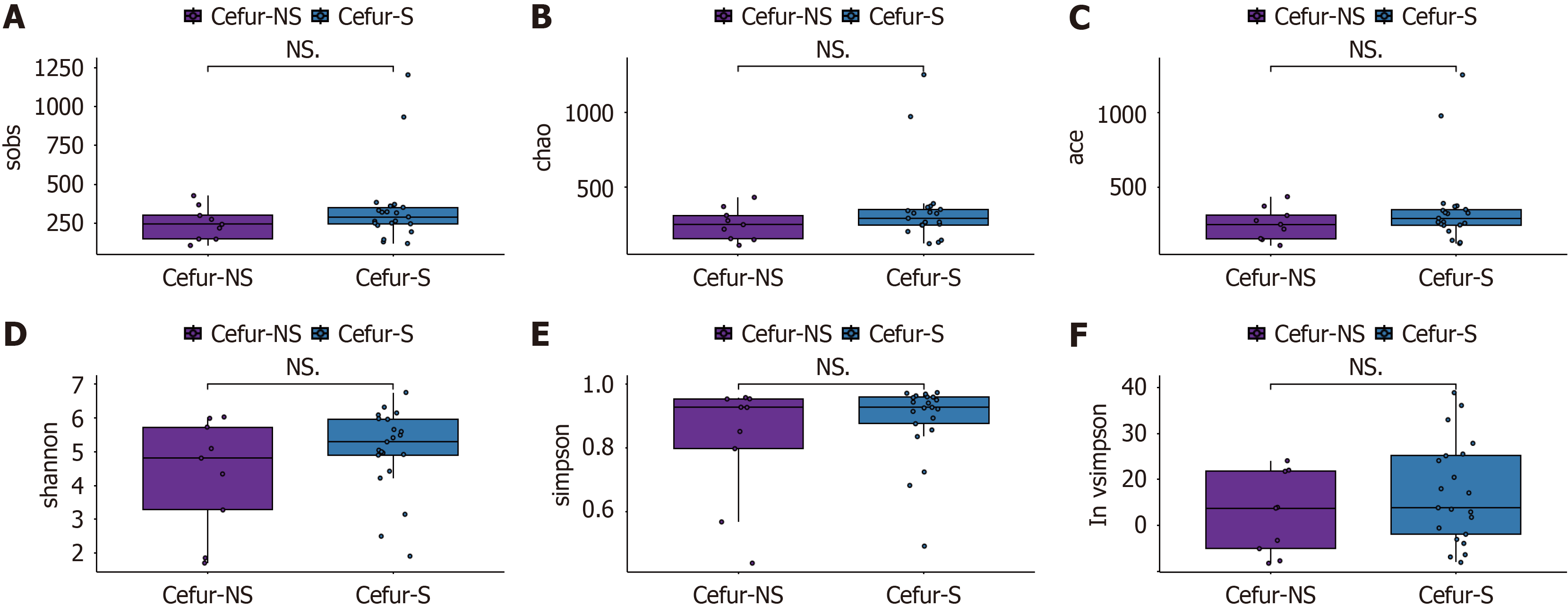
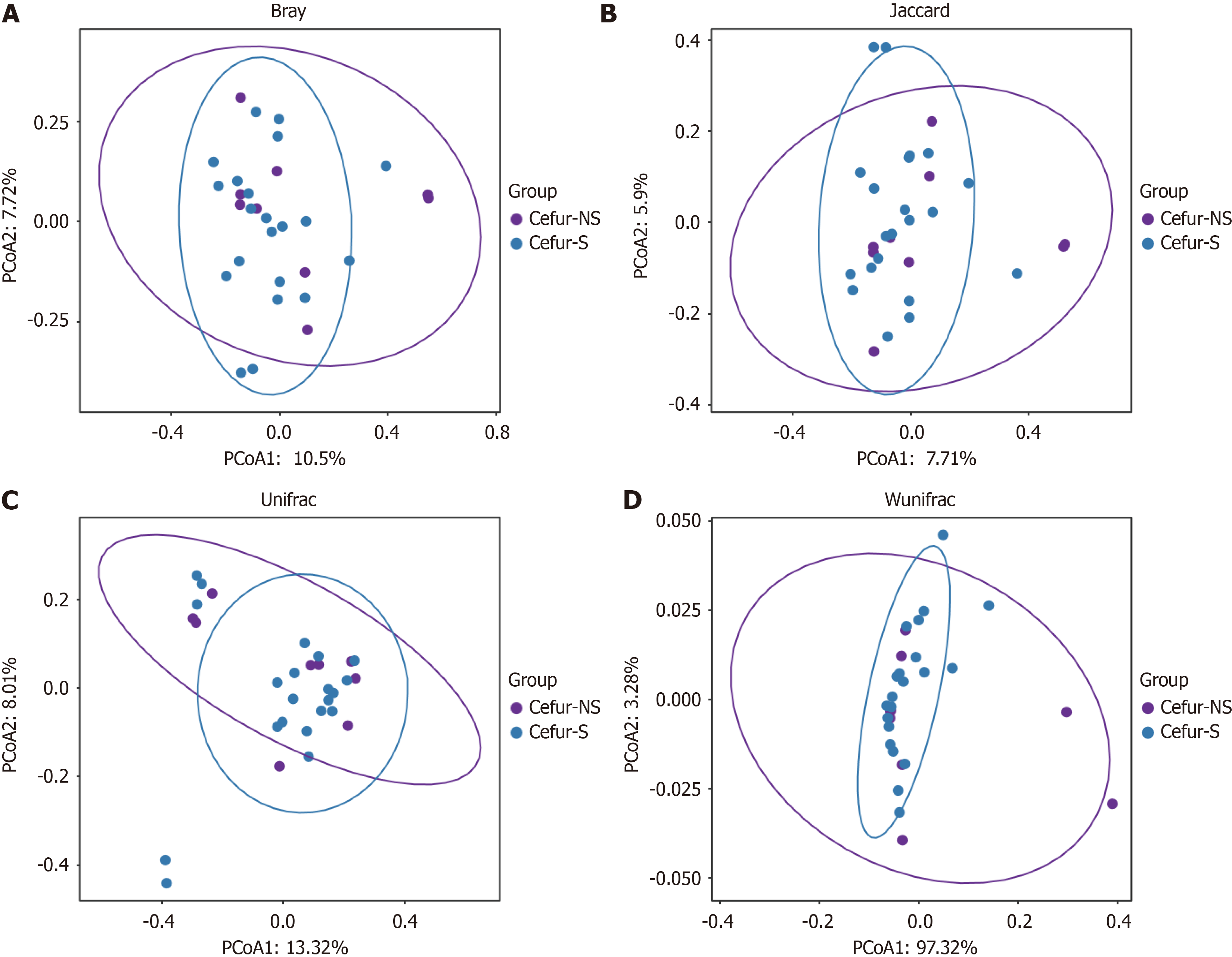
Alpha diversity refers to the analysis of species diversity within individual samples. We calculated the observed number of ASVs using the observed species method, estimated the ASV abundance in the community with the Chao method, and assessed the ASV richness using the abundance-based coverage estimator index. Additionally, we evaluated community diversity using the Shannon, Simpson, and InvSimpson indices. The results showed no significant differences in species richness between the Cefur-NS and Cefur-S groups (Figure 1). Beta diversity involves comparing the microbial com
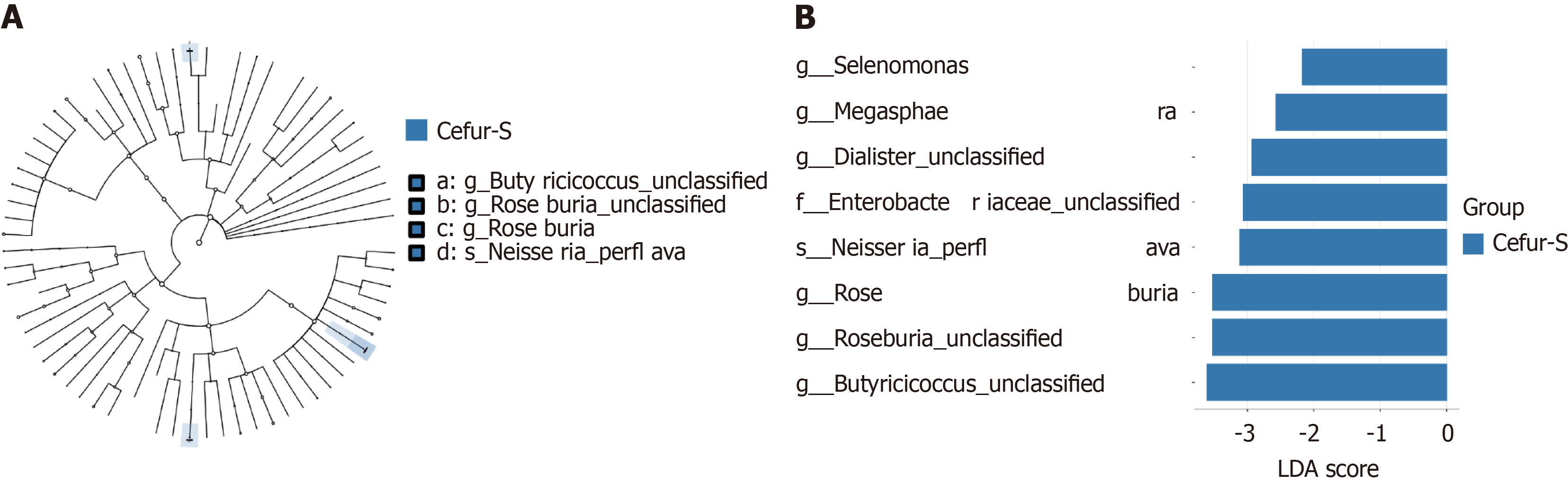
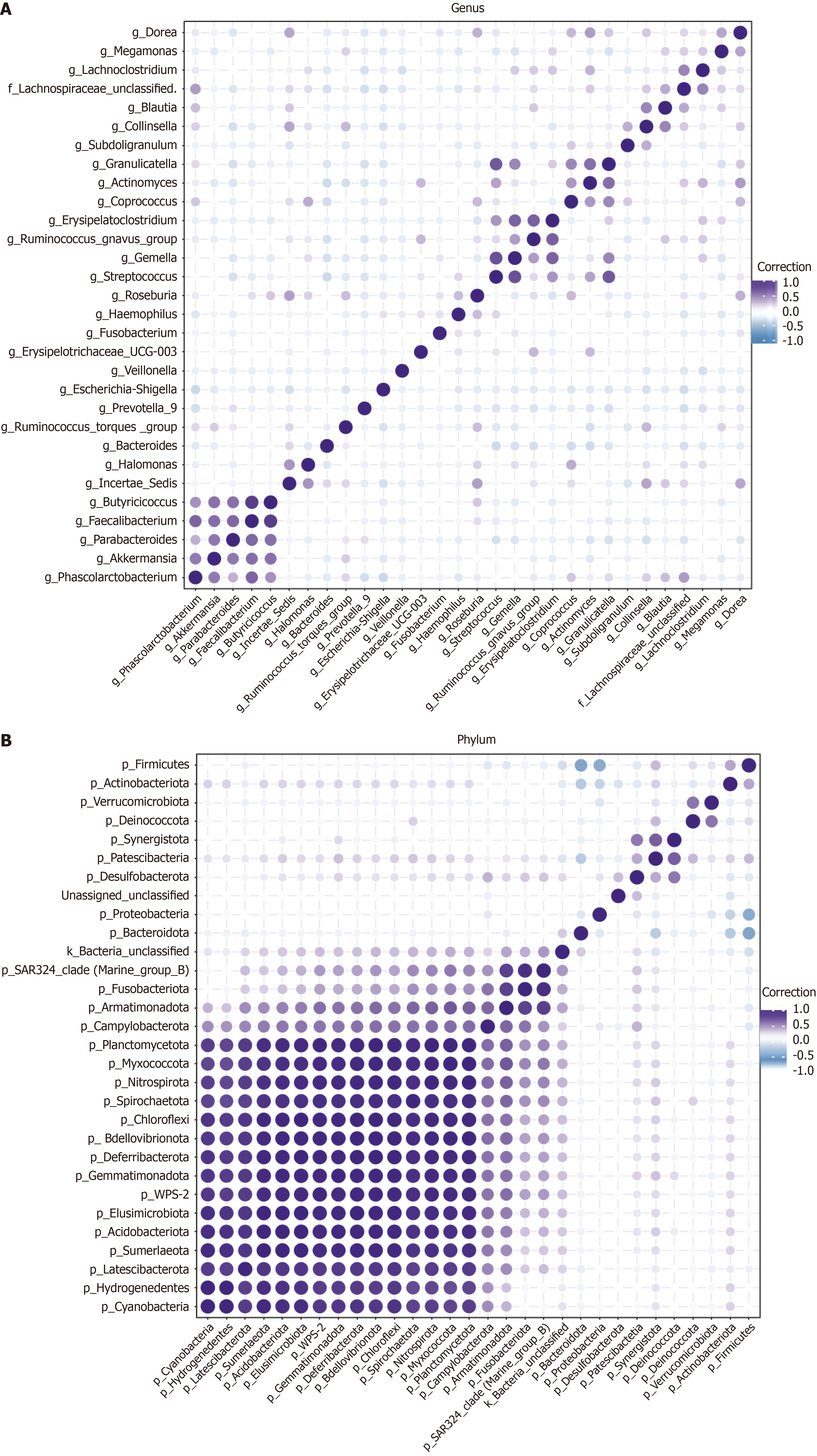
LEfSe analysis was conducted to identify differences in gut microbiota abundance between the two groups at the phylum level (Figure 3A). The results revealed that the Cefur-S had eight dominant bacterial taxa compared to the Cefur-NS (LDA score > 2, P < 0.05) (Figure 3B). Notably, Roseburia and Butyricicoccus unclassified were identified as the most prominent taxa, with LDA scores exceeding 3.5. To further elucidate the relationships between different taxa at the phylum and genus levels, we conducted a Pearson correlation analysis on the top 30 abundant taxa and visualized the results using a heatmap. At the genus level, a negative correlation was observed between Escherichia and Collinsella (-0.14), as well as unclassified Helicobacter (-0.24). Conversely, Erysipelatoclostridium was positively correlated with Ruminococcus (0.29) and Actinomyces (0.3) (Figure 4A). At the phylum level, Firmicutes showed a negative correlation with Bacteroidetes (-0.65) and Proteobacteria (-0.6), while it was positively correlated with Acinetobacter (0.38) (Figure 4B).
To explore whether modulating the gut microbiota could reduce cefuroxime resistance, the Cefur-NS group (9 patients) was treated with Clostridium butyricum tablets for one week. Upon follow-up, anal crypt swabs were taken again for bacterial culture and antibiotic susceptibility testing. The results showed that after the intervention, only 2 out of the 9 patients remained resistant to cefuroxime, a statistically significant reduction (P < 0.05). Additionally, these 9 patients experienced a notable improvement in stool consistency, with reduced bowel movement frequency and better-formed stools (Table 2).
| Features | Before taking CBT | After taking CBT | Statistics | P value |
| Cefuroxime resistance (n) | 9 | 2 | -1.56 | < 0.01 |
| Defection frequency (times/day), mean ± SD | 2.3 ± 1.2 | 1.7 ± 0.7 | 1.78 | 0.02 |
| Stool consistency score, mean ± SD | 19.3 ± 7.8 | 11.9 ± 8.1 | -3.73 | 0.02 |
Anal fistula is a common disease in colorectal region, with an increasing incidence due to rising living standards and the fast-paced lifestyle prevalent today[9]. Patients often require surgical treatment due to recurrent perianal pain and discharge that significantly impact their quality of life[10]. However, the precise pathogenesis of anal fistulas remains unclear. The most widely accepted theory is that anal fistulas form due to infections in the anal crypt glands, though this theory does not fully explain all aspects of fistula formation[11]. Given the uncertain etiology, the choice of antibiotics during the perioperative period is crucial. Current guidelines recommend the use of first-generation cephalosporins, such as cefazolin, or second-generation cephalosporins, like cefuroxime[12]. Unfortunately, our department’s data, collected from July 2022 to July 2023, involving 149 anal fistula patients, revealed high resistance rates to cefazolin (37.3%) and cefuroxime (35.4%) based on bacterial cultures from anal crypt swabs. Furthermore, patients resistant to these antibiotics exhibited notable differences in bowel movement frequency and stool consistency compared to those who were sensitive to the antibiotics. These findings underscore the importance of further investigating the relationship between gut microbiota changes and antibiotic resistance. Understanding this relationship could inform better antibiotic selection during the perioperative period, potentially reducing the intensity of antibiotic use and improving patient outcomes.
The gut microbiota, comprising an enormous number of microorganisms, maintains a dynamic balance between beneficial and harmful bacteria[13]. This balance is crucial for various functions, including nutrient absorption, intestinal immunity, and the metabolism of certain drugs, thereby establishing a symbiotic relationship between the gut microbiota and the host to maintain overall health. Consequently, changes in gut microbiota can significantly impact health[14]. Existing literature has demonstrated that there are significant differences in the abundance and diversity of gut microbiota between healthy individuals and those with anal fistulas[15]. Research by Yang et al[15] on the gut microbiota of anal fistula patients also suggests that alterations in the gut microbiota may have a potential influence on the pro
At the phylum and genus levels, the analysis of bacterial composition between the two patient groups revealed significant differences. At the phylum level, Firmicutes was the dominant phylum in both groups, followed by Bacteroidetes, Proteobacteria, and Actinobacteria. Notably, there was a statistically significant difference in the proportions of Firmicutes and Proteobacteria between the two groups. Correlation analysis further indicated that Firmicutes was negatively correlated with Bacteroidetes and Proteobacteria, while positively correlated with Actinobacteria. At the genus level, the dominant genera in both groups included Escherichia, Prevotella, unclassified Helicobacter, Collinsella, Ruminococcus, Erysipelatoclostridium, and Megamonas (each accounting for more than 1% of the total composition). When comparing the Cefur-S group with the resistant group, there was a notable increase in the proportions of unclassified Helicobacter and Collinsella in the sensitive group, while the proportions of Escherichia and Erysipelatoclostridium were significantly lower. Related studies, such as the research by Bahar-Tokman et al[16] on the gut microbiota of type 2 diabetes patients, found that an increased Firmicutes-to-Bacteroidetes ratio can indirectly promote the upregulation of interleukin-1β (IL-1β), IL-6, IL-8, toll-like receptor 4, and toll-like receptor 5 gene expression, providing a protective effect against intestinal inflammation[16]. In our study, the higher proportion of Escherichia in the gut microbiota of the resistant group is significant, as literature suggests that Escherichia can enhance antibiotic resistance by sharing genetic material. Moreover, when resistance genes are transferred to plasmids, they can be horizontally transferred between different bacteria, including across genera. This may explain why an increased proportion of Escherichia correlates with a higher rate of resistance to cephalosporins.
LEfSe analysis at the genus level identified eight bacterial taxa with significant differences between the Cefur-S and Cefur-NS groups[17]. Among these, the genera with an LDA score greater than 3.5 included Roseburia, unclassified Roseburia, and unclassified Butyricicoccus. Roseburia (including unclassified Roseburia) is a genus of gram-positive anaerobes with flagella, belonging to the phylum Firmicutes. This genus can utilize carbon sources such as xylose and β-mannan in the gut to produce short-chain fatty acids (SCFAs), particularly butyrate[18]. Roseburia has been associated with several systemic diseases, including inflammatory bowel disease, diabetes, and fatty liver disease. It is thought to exert anti-inflammatory effects and protect the gut through mechanisms involving butyrate, flagellin, immune cell regulation, tryptophan metabolism, and the gut-brain axis[19,20]. Butyricicoccus and Roseburia both belong to the phylum Firmicutes and are key butyrate-producing bacteria in the gut. Butyricicoccus and its metabolic products, such as lipopolysaccharides, increase the concentration of bile acids and SCFAs, thereby improving pathways related to inflammation, lipid metabolism, and energy metabolism[21]. Research has shown that butyrate can reduce the damage to mitochondrial morphology and barrier function in colonic epithelial cells caused by adherent-invasive Escherichia coli (AIEC) through the expression of the free fatty acid receptor[22]. The research of Recharla et al[23] on the protective mechanisms of butyrate in inflammatory bowel disease revealed that butyrate acts on the G protein-coupled receptors on cell walls to upregulate the expression of glucagon-like peptide-1, peptide tyrosine-tyrosine, and leptin, thereby enhancing glucose and lipid metabolism. Additionally, butyrate influences the immune system by acting on naive T cells, promoting the upregulation of regulatory T cells, increasing the secretion of cytokines IL-10 and IL-2L, and inhibiting the differentiation of T helper cells, thus suppressing intestinal inflammation[22].
Moreover, studies suggest that Roseburia can prevent colorectal tumor development by producing butyrate, which induces functional CD8 T cells and enhances the efficacy of anti-programmed death 1 therapy[24]. Interestingly, while high concentrations of SCFAs like butyrate can inhibit gut inflammation, low concentrations may increase the expression of adhesion and virulence genes in invasive AIEC[25]. Therefore, this study suggests that the dominance of Roseburia and Butyricicoccus in the gut microbiota of Cefur-S patients represents beneficial bacteria that, through increasing butyrate levels, reduce the expression of intestinal inflammatory factors and minimize the damage to the intestinal mucosa caused by AIEC. These findings highlight the important role of Roseburia and Butyricicoccus in the etiology of anal fistula by increasing butyrate levels, inhibiting inflammatory factors, and protecting the colonic epithelial barrier. In the study, we observed that patients in the Cefur-NS group exhibited a decreased proportion of Firmicutes and an increased proportion of Proteobacteria in their gut microbiota, with a noted negative correlation between these two phyla. At the genus level, the increased proportion of Escherichia, which belongs to the phylum Proteobacteria, suggests that Escherichia may play a significant role in cefuroxime resistance. To address this, we administered Clostridium butyricum tablets, a probiotic belonging to the phylum Firmicutes, as an intervention for patients resistant to cefuroxime. Butyric acid Bacillus is well-tolerated by gastric acid, easy for patients to take, and shares 78% homology with AIEC, differing by only 332 nu
Despite the promising findings, this study has several limitations that should be acknowledged. First, the sample size was relatively small, with only 30 patients included, which may limit the generalizability of the results. A larger cohort would provide more robust data and could help to validate the observed relationships between gut microbiota com
Although we provided an insight into butyrate-producing bacteria safeguarding cattle from cefuroxime resistance, further investigation to examine the probiotic intervention duration in the long term will be required. Longitudinal studies are needed for further evaluation to assess if the reduction in antibiotic resistance observed in the short term persists over longer times. This will help to clarify the durability and clinical relevance of probiotics treatment for antibiotic resistance management. Moreover, future studies should further broaden the patient cohort and sample size, including various demographics of patients with whom antibiotic resistance is a major burden. Wider studies in many cohorts may help to describe the variation between gut microbiota community and antibiotic resistance in diverse populations. Further, using metagenomics and metabolomics may elucidate the particular mechanisms by which certain bacterial taxa and their metabolites (e.g., butyrate) contribute to antibiotic resistance protection. This comprehensive strategy will facilitate the individualization of probiotic therapy, with a scope that could likely be expanded to not only those treated with cefuroxime but also other specific classes of antibiotics. Our study showed reduced extended-spectrum β-lactamase producing Enterobacteriaceae cefuroxime resistance rates in anal crypt swab cultures when Clostridium butyricum was used. Patients also saw significant reductions in stool consistency and bowel movements per week post-treatment compared with baseline. This provides evidence for the development of strategies to effectively alter antibiotic resistance through manipulation of gut microbiota composition. Finally, increasing the prevalence of beneficial Firmicutes, especially Clostridium butyricum, seems to be a critical strategy to attenuate cefuroxime resistances. This further highlights the potential of targeted probiotic interventions as a promising measure in combatting antibiotic resistance in the clinic.
In conclusion, our study suggests that specific gut microbiota, particularly Butyricicoccus and Roseburia, may help mitigate cefuroxime resistance in anal fistula patients by producing butyrate, which protects the intestinal mucosa. Moreover, probiotic intervention targeting gut microbiota composition shows potential in reducing antibiotic resistance, high
| 1. | Chen B, Liu Y, Wang Y, Wang Q. Causal relationship between body mass index and anal fistula: a two-sample Mendelian randomization study. Front Genet. 2024;15:1406231. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Liu Y, Zhao W, Hu W, Xu J, Zhang H, Huang T, Wu C, Yang J, Mao W, Yao X, Lu Y, Wang Q. Exploring the relationship between anal fistula and colorectal cancer based on Mendelian randomization and bioinformatics. J Cell Mol Med. 2024;28:e18537. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 3. | Xu J, Mei Z, Wang Q. Integrating multidisciplinary perspectives in complex anal fistula management: a blueprint for future research and precision surgery. Int J Surg. 2024;110:1810-1812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Mocanu V, Dang JT, Ladak F, Tian C, Wang H, Birch DW, Karmali S. Antibiotic use in prevention of anal fistulas following incision and drainage of anorectal abscesses: A systematic review and meta-analysis. Am J Surg. 2019;217:910-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 5. | Kesler M, Koch A, Rychlik M, Bierca J, Kołodziejczak M. Assessment of the rationale for the use of preventive antibiotic (clindamycin) therapy in patients operated on for anal fistula based on authors’ own experience. Nowa Medycyna. 2021;. [DOI] [Full Text] |
| 6. | Zothanpuia, Passari AK, Gupta VK, Singh BP. Detection of antibiotic-resistant bacteria endowed with antimicrobial activity from a freshwater lake and their phylogenetic affiliation. PeerJ. 2016;4:e2103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Peck K, Stedtfeld R, Rosefigura J, Reed B, Mcusic D, Irish J, Etchebarne B, Johnson T, Kurihara L, Makarov V. Abstract 1484: Microbial sequencing using a single-pool target enrichment of multiple variable regions of the 16S rRNA gene, the nuclear ribosomal internal transcribed spacer (ITS) region, and antimicrobial resistance genes. Cancer Res. 2019;79:1484. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 8. | Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45:88-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 387] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 9. | Ye Q, Han Y, Du P, Yang M, Zheng D, Mei Z, Wang Q. Clinical efficacy of the bared external anal sphincter (BEAS) in high horseshoe-shaped anal fistulas: Protocol for a real-world, prospective cohort study. Heliyon. 2024;10:e35024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Micallef J, Barns M. High perianal fistula: a narrative review of common management techniques. Int Surg J. 2022;10:186-194. [DOI] [Full Text] |
| 11. | Gottesman L. Classical Cryptoglandular Theory for Anorectal Infection: Reconsidered. Dis Colon Rectum. 2021;64:259-261. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 12. | Trailokya A, Sastry GL, Nandi M, Mukhopadhyay M, Dumbre R, Bhattacharjee S, Sukumar G, Pawar R. Role of cefuroxime as antibiotic prophylaxis for general surgery: An expert opinion. IP J Surg Allied Sci. 2021;3:58-71. [DOI] [Full Text] |
| 13. | Dahiya D, Nigam PS. The Gut Microbiota Influenced by the Intake of Probiotics and Functional Foods with Prebiotics Can Sustain Wellness and Alleviate Certain Ailments like Gut-Inflammation and Colon-Cancer. Microorganisms. 2022;10:665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 14. | Zhang YJ, Li S, Gan RY, Zhou T, Xu DP, Li HB. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16:7493-7519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 442] [Cited by in RCA: 599] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 15. | Yang J, Li L, Su W, Zhang S, Xu H, Wang M, Shen W. Microbiomic signatures of anal fistula and putative sources of microbes. Front Cell Infect Microbiol. 2024;14:1332490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Bahar-Tokman H, Demirci M, Keskin FE, Cagatay P, Taner Z, Ozturk-Bakar Y, Ozyazar M, Kiraz N, Kocazeybek BS. Firmicutes/Bacteroidetes Ratio in the Gut Microbiota and IL-1β, IL-6, IL-8, TLR2, TLR4, TLR5 Gene Expressions in Type 2 Diabetes. Clin Lab. 2022;68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Källman O, Giske CG, Samuelsen Ø, Wretlind B, Kalin M, Olsson-Liljequist B. Interplay of efflux, impermeability, and AmpC activity contributes to cefuroxime resistance in clinical, non-ESBL-producing isolates of Escherichia coli. Microb Drug Resist. 2009;15:91-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Singh V, Lee G, Son H, Koh H, Kim ES, Unno T, Shin JH. Butyrate producers, "The Sentinel of Gut": Their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front Microbiol. 2022;13:1103836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 242] [Article Influence: 121.0] [Reference Citation Analysis (0)] |
| 19. | Nie K, Ma K, Luo W, Shen Z, Yang Z, Xiao M, Tong T, Yang Y, Wang X. Roseburia intestinalis: A Beneficial Gut Organism From the Discoveries in Genus and Species. Front Cell Infect Microbiol. 2021;11:757718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 283] [Article Influence: 70.8] [Reference Citation Analysis (1)] |
| 20. | Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 482] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 21. | Chen CY, Ho HC. Roles of gut microbes in metabolic-associated fatty liver disease. Tzu Chi Med J. 2023;35:279-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Yu Q, Yu F, Li Q, Zhang J, Peng Y, Wang X, Li T, Yin N, Sun G, Ouyang H, Chen Y, Mine Y, Tsao R, Zhang H. Anthocyanin-Rich Butterfly Pea Flower Extract Ameliorating Low-Grade Inflammation in a High-Fat-Diet and Lipopolysaccharide-Induced Mouse Model. J Agric Food Chem. 2023;71:11941-11956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Recharla N, Geesala R, Shi XZ. Gut Microbial Metabolite Butyrate and Its Therapeutic Role in Inflammatory Bowel Disease: A Literature Review. Nutrients. 2023;15:2275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 62] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 24. | Kang X, Liu C, Ding Y, Ni Y, Ji F, Lau HCH, Jiang L, Sung JJ, Wong SH, Yu J. Roseburia intestinalis generated butyrate boosts anti-PD-1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. 2023;72:2112-2122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 129] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 25. | Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology (Reading). 2009;155:521-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |