Published online May 27, 2024. doi: 10.4240/wjgs.v16.i5.1328
Revised: April 11, 2024
Accepted: April 12, 2024
Published online: May 27, 2024
Processing time: 91 Days and 8 Hours
Few studies have investigated the expression of GLI1 and PTTG1 in patients undergoing radical surgery for colorectal carcinoma (CRC) and their association with lymph node metastasis (LNM). Therefore, more relevant studies and ana
To explore GLI1 and PTTG1 expression in patients undergoing radical surgery for CRC and their correlation with LNM.
This study selected 103 patients with CRC admitted to our hospital between April 2020 and April 2023. Sample specimens of CRC and adjacent tissues were co
Significantly higher positive rates and expression levels of GLI1 and PTTG1 were observed in CRC tissue samples compared with adjacent tissues. GLI1 and PTTG1 were strongly linked to LNM in patients undergoing radical surgery for CRC, with higher GLI1 and PTTG1 levels found in patients with LNM than in those without. The areas under the ROC curve of GLI1 and PTTG1 in assessing LNM in patients with CRC were 0.824 and 0.811, respectively.
GLI1 and PTTG1 expression was upregulated in patients undergoing radical surgery for CRC and are significantly related to LNM in these patients. Moreover, high GLI1 and PTTG1 expression can indicate LNM in patients with CRC undergoing radical surgery. The expression of both genes has certain diagnostic and therapeutic significance.
Core Tip: The main goal of radical surgery for colorectal carcinoma (CRC) is to completely remove the primary tumor and regional lymph nodes. Lymph nodes are essential in predicting patient prognosis and for determining the need for adjuvant chemotherapy after surgical resection. In this study, the expression of GLI1 and PTTG1 in patients with CRC undergoing radical surgery and their correlation with lymph node metastasis (LNM) were examined. Aberrant overexpression of GLI1 and PTTG1 was confirmed in patients undergoing radical surgery for CRC. In addition, both GLI1 and PTTG1 were sig
- Citation: Cao F, Chen YY, Wang HC. GLI1 and PTTG1 expression in colorectal carcinoma patients undergoing radical surgery and their correlation with lymph node metastasis. World J Gastrointest Surg 2024; 16(5): 1328-1335
- URL: https://www.wjgnet.com/1948-9366/full/v16/i5/1328.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v16.i5.1328
Colorectal carcinoma (CRC), which originates from the colon or rectum, is the third most common malignancy in both men and women worldwide[1]. Overweight, family history, tall stature, excessive intake of alcoholic beverages, and lack of regular exercise are the predisposing factors[2]. Etiologically, CRC is associated with excessive mucosal epithelial cell proliferation, distant tumor metastasis, long-term intestinal inflammatory responses, and abnormal changes in the gut microbiota[3,4]. The global number of new CRC cases in 2018 was 1.8 million, with nearly 900000 deaths[5]. Patients with CRC may present with clinical symptoms such as celialgia, abdominal mass, hemoproctia, abnormal weight loss, ab
GLI1 is a gene that mediates pathophysiological processes and changes in various tumors. It is involved in the proliferation, migration, invasiveness, growth, vascular invasion, and self-renewal of cancer stem cells in various malignancies[9,10]. According to Du et al[11], GLI1, as a component of the hedgehog (Hh)/GLI1 signaling pathway in CRC, can affect the inhibition of resveratrol on the growth and metastasis of HCT116 cells. Park et al[12] found that GLI1 influences the survival of patients with CRC by participating in hypoxic mechanisms and interacting with CCT2. PTTG1, a carcinogenic agent closely associated with tumor metastasis, participates in organ development, metabolism, DNA damage/repair, and cell cycle regulation. It is aberrantly overexpressed in various tumors, including those in the pituitary gland, thyroid, breast, ovary, uterus, and colon[13,14]. The promotion by PTTG1 of tumor metastasis and invasion has been linked to its direct regulation of basic fibroblast growth factor, epidermal growth factor, and β-catenin/transcription factor[15].
Research on the expression of GLI1 and PTTG1 in patients with CRC undergoing radical surgery and their correlation with lymph node metastasis (LNM) is limited. This study sought to bridge this gap to contribute to the improved clinical management of CRC.
We enrolled 103 patients who underwent radical surgery for CRC at Shanghai Zhangjiang Institute of Medical Innovation between April 2020 and April 2023. Samples of CRC and adjacent tissues were collected from all patients. Of the 103 patients, 53 were male and 50 were female. Furthermore, 42 were < 60 years old, and 61 were ≥ 60 years old. Tumors were located in the colon in 43 cases and in the rectum in 60 cases. In 23 cases, tumors were well differentiated, whereas in 80 cases, they were moderately and poorly differentiated. Furthermore, 48 cases were pathologically staged I–II, whereas 55 were staged III–IV. LNM was observed in 58 cases.
The inclusion criteria were as follows: Diagnosis of CRC by surgical pathology and colonoscopic biopsy; met corresponding indications for surgical treatment and underwent radical surgery, with complete cancer and para-cancer tissue specimens obtained; no mental and cognitive abnormalities; and intact clinical data.
The exclusion criteria were as follows: systemic inflammatory diseases; previous use of chemoradiotherapy or targeted therapy; presence of other malignant tumors; estimated survival < 3 months; inability to attend follow-up visits or loss to follow-up; lactating or pregnant women; infectious diseases or blood disorders.
Samples of CRC and adjacent tissues of about 2–3 g were collected from all patients during the surgery and subsequently processed into sections (5–10 μm thick) after preliminary treatment. The specimen sections were placed in an electrothermal constant-temperature air-blowing drying oven at 50°C–60°C for 3–4 h to make them adhere to the slide. After baking, disodium hydrogen phosphate–sodium dihydrogen phosphate buffer was used for antigen retrieval, and mouse antihuman GLI1 and PTTG1 monoclonal antibodies were added overnight. Next, each tissue sample was stained by immunohistochemistry following the SP kit instructions. Then, the positive rates of GLI1 and PTTG1 expression in each tissue sample were observed using an automatic immunohistochemical staining instrument.
The criteria for judging the positive expression rate were as follows: (1) According to the product of the staining intensity and the number of positive cells in the field of view, a percentage of positive cells < 5% was counted as 0 points, 5%–25% as 1 point, 26%–50% as 2 points, 51%–75% as 3 points, and > 75% as 4 points; and (2) Based on dyeing intensity, 0, 1, 2, and 3 points were given for colorless, light yellow, brownish yellow, and tan, respectively. Positive expression was determined if the sum of the two results was greater than 3 points. The immunohistochemical staining instrument was set to counting mode in counting the number of GLI1- and PTTG1-positive cells in the visual field. Finally, the expression levels of GLI1 and PTTG1 were calculated.
The positive expression rates and protein levels of GLI1 and PTTG1 in CRC and adjacent tissues were statistically compared. The correlations between GLI1 and PTTG1 expression and clinicopathological characteristics, such as sex, age, tumor location, degree of tumor differentiation, pathological stage, and LNM, were evaluated. The differences in GLI1 and PTTG1 expression levels between patients with and without LNM were examined.
All analyses were performed using SPSS 20.0 at a significance level of P < 0.05. Categorical variables were expressed as number (percentage) and comparatively analyzed using the χ2 test. Continuous variables were expressed as mean ± SE and analyzed using the independent sample t-test, respectively.
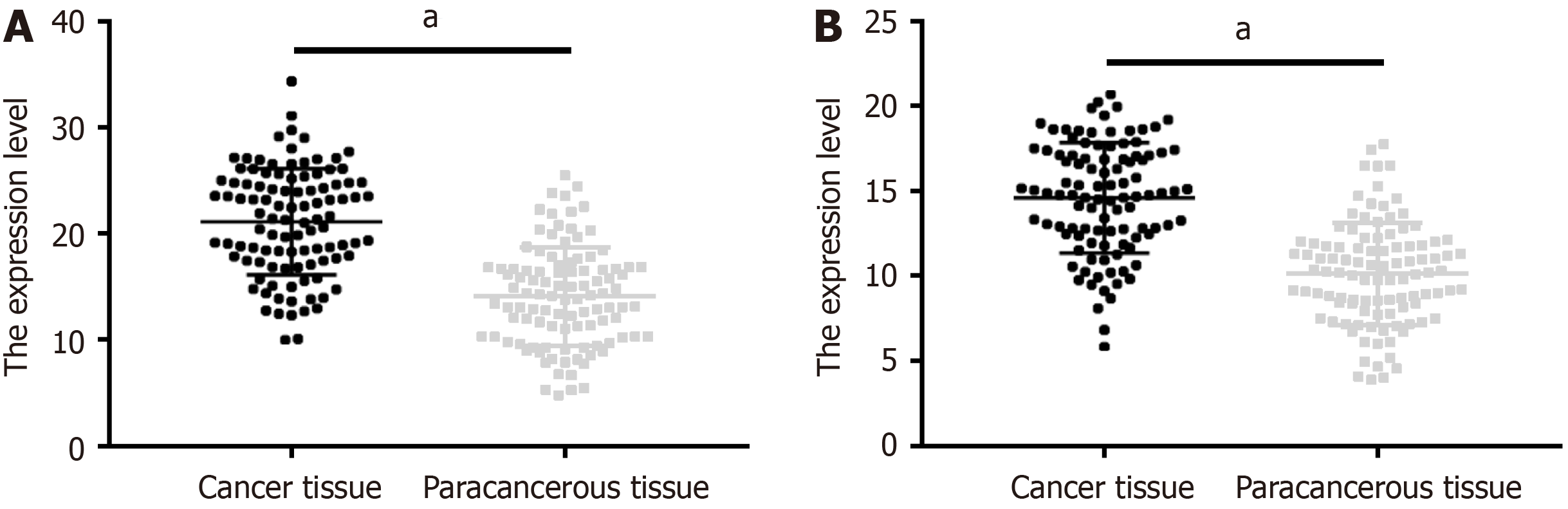
The positive expression rates of GLI1 in CRC and adjacent tissues were 63.11% and 12.62%, respectively, and the corresponding positive expression rates of PTTG1 were 56.31% and 5.83%, respectively. Significantly higher positive rates of GLI1 and PTTG1 were observed in cancer tissues than in adjacent tissues (P < 0.001; Table 1).
| Grouping | n | Cancerous tissue | Para-cancerous tissue | χ2 | P value |
| GLI1 positive | 103 | 65 (63.11) | 13 (12.62) | 55.792 | < 0.001 |
| PTTG1 positive | 103 | 58 (56.31) | 6 (5.83) | 61.292 | < 0.001 |
Significantly higher GLI1 and PTTG1 levels were observed in cancer tissue samples than in adjacent tissues (P < 0.05; Figure 1).
Neither GLI1 or PTTG1 expression were strongly associated with sex, age, tumor location, degree of tumor differentiation, or pathological stage (P > 0.05). However, the expression of both genes was significantly associated with LNM (P < 0.05; Tables 2 and 3).
| Grouping | n | GLI1 positive (n = 65), n (%) | GLI1 negative (n = 38), n (%) | χ2 | P value |
| Sex | 0.403 | 0.526 | |||
| Male | 53 | 35 (53.85) | 18 (47.37) | ||
| Female | 50 | 30 (46.15) | 20 (52.63) | ||
| Age | 3.504 | 0.061 | |||
| < 60 yr old | 42 | 22 (33.85) | 20 (53.63) | ||
| ≥ 60 yr old | 61 | 43 (66.15) | 18 (47.37) | ||
| Tumor location | 0.221 | 0.638 | |||
| Colon | 43 | 26 (40.00) | 17 (44.74) | ||
| Rectum | 60 | 39 (60.00) | 21 (55.26) | ||
| Differentiation degree | 0.552 | 0.458 | |||
| High differentiation | 23 | 13 (20.00) | 10 (26.32) | ||
| Medium–low differentiation | 80 | 52 (80.00) | 28 (73.68) | ||
| Pathological staging | 3.086 | 0.079 | |||
| I–II | 48 | 26 (40.00) | 22 (57.89) | ||
| III–IV | 55 | 39 (60.00) | 16 (42.11) | ||
| Lymph node metastasis | 5.233 | 0.022 | |||
| Positive | 58 | 44 (67.69) | 17 (44.74) | ||
| Negative | 45 | 21 (32.31) | 21 (55.26) |
| Grouping | n | PTTG1 positive (n = 58), n (%) | PTTG1 negative (n = 45), n (%) | χ2 | P value |
| Sex | 2.728 | 0.099 | |||
| Male | 53 | 34 (58.62) | 19 (42.22) | ||
| Female | 50 | 24 (41.38) | 26 (57.78) | ||
| Age | 3.534 | 0.060 | |||
| < 60 yr old | 42 | 19 (32.76) | 23 (51.11) | ||
| ≥ 60 yr old | 61 | 39 (67.24) | 22 (48.89) | ||
| Tumor location | 2.327 | 0.127 | |||
| Colon | 43 | 28 (48.28) | 15 (33.33) | ||
| Rectum | 60 | 30 (51.72) | 30 (66.67) | ||
| Differentiation degree | 0.867 | 0.352 | |||
| High differentiation | 23 | 11 (18.97) | 12 (26.67) | ||
| Low–moderate differentiation | 80 | 47 (81.03) | 33 (73.33) | ||
| Pathological staging | 0.616 | 0.433 | |||
| I–II | 48 | 29 (50.00) | 19 (42.22) | ||
| III–IV | 55 | 29 (50.00) | 26 (57.78) | ||
| Lymph node metastasis | 17.150 | < 0.001 | |||
| Positive | 58 | 43 (74.14) | 15 (33.33) | ||
| Negative | 45 | 15 (25.86) | 30 (66.67) |
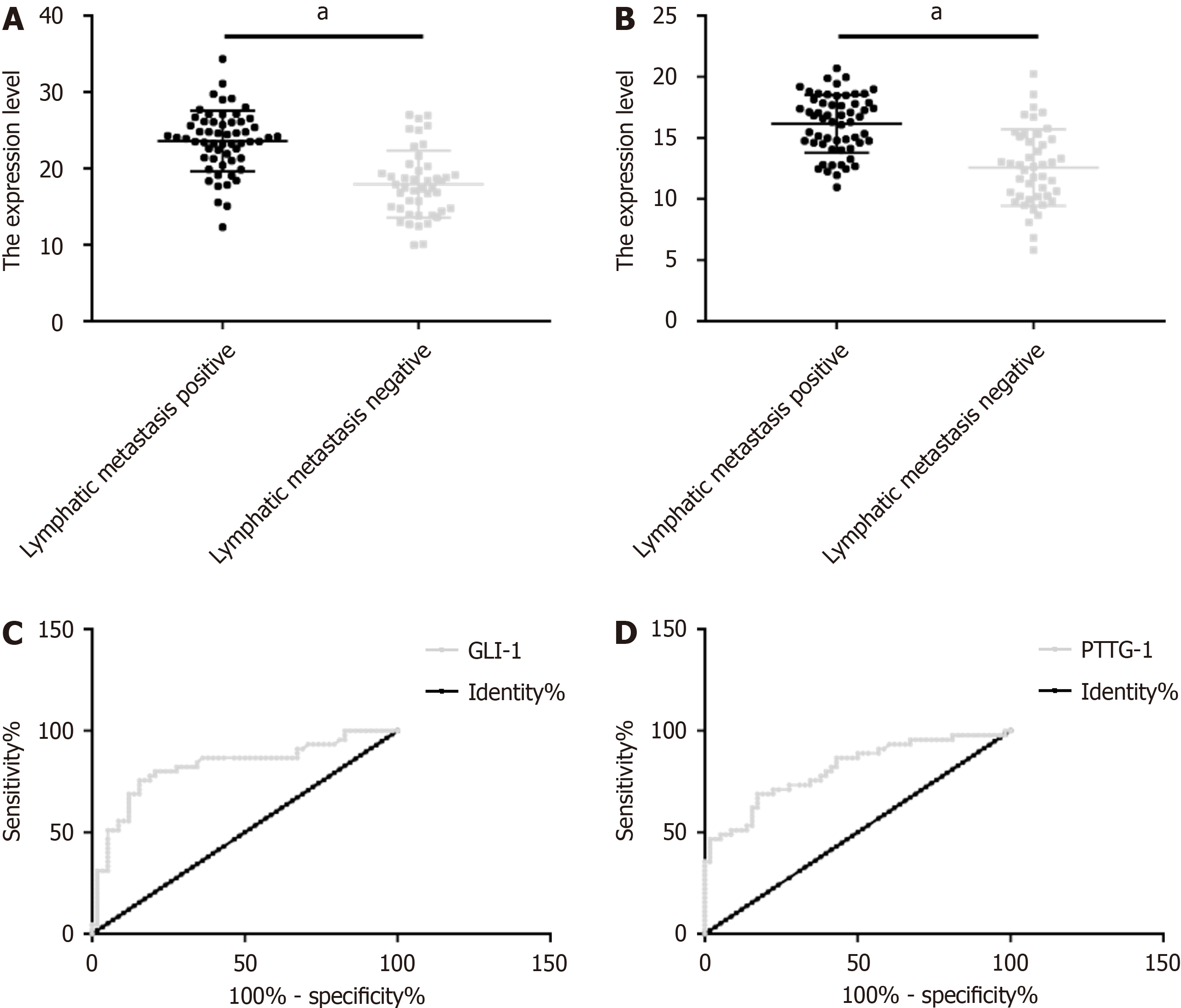
Patients with LNM showed obviously higher GLI1 and PTTG1 levels than those without. The area under the curve (AUC) of GLI1 for predicting LNM in patients with CRC undergoing radical surgery was 0.824 (sensitivity: 75.56%; specificity: 84.48%), and the optimal cut-off was 19.76. The AUC, sensitivity, specificity, and the optimal cut-off of PTTG1 for predicting LNM were 0.811, 68.89%, 82.76%, and 13.97, respectively (Figure 2 and Table 4).
| Grouping | AUC | 95%CI | SE | Cut-off | Sensitivity (%) | Specificity (%) |
| GLI1 | 0.824 | 0.739–0.910 | 0.044 | 19.76 | 75.56 | 84.48 |
| PTTG1 | 0.811 | 0.726–0.895 | 0.043 | 13.97 | 68.89 | 82.76 |
Screening for CRC in patients based on early symptoms is difficult; imaging and pathological examination are often required[16]. However, in patients with CRC who underwent radical resection of the primary tumor, the accuracy of the said methods of disease assessment will be greatly affected[17]. To maximize the curative effect of radical surgery for CRC and promote patients’ postoperative recovery, more accurate and effective means of predicting disease changes are needed.
This study included 103 patients with CRC who underwent radical surgery and analyzed samples of cancer and adjacent tissues. First, we observed a significantly higher positive rate and expression of GLI1 in cancer tissue samples versus adjacent tissues, as well as a markedly higher positive rate and expression of PTTG1, suggesting the overexpression of GLI1 and PTTG1 and the possible involvement of both genes in tumorigenesis in CRC. Wu et al[18] found high GLI1 expression in human small-cell lung cancer, which indicates that GLI1 may mediate the onset and progression of small-cell lung cancer by activating the target gene downstream of Hh signaling. Chen et al[19] also reported high GLI1 expression in hepatocellular carcinoma cells and found that downregulating its expression inhibited the adhesion, mo
Analysis of the clinicopathological data showed that GLI1 and PTTG1 had no significant relationship with sex, age, tumor location, degree of tumor differentiation, and pathological stage but were significantly related to LNM. That is, patients with LNM had significantly higher expression rates of GLI1 and PTTG1. Similarly, we found significantly ele
This study has several limitations. First, the single-center study design and small sample size would have affected the universality of the research results. Second, no basic investigation was conducted to explore the specific molecular mechanisms of GLI1, PTTG1, and LNM in CRC. Third, no analysis was performed to identify the risk factors that affect the efficacy of radical surgery or LNM in patients with CRC. Continuous optimization and supplementation should be implemented in future investigations to address the above deficiencies.
GLI1 and PTTG1 were aberrantly overexpressed in patients undergoing radical surgery for CRC. The expression of both genes was significantly correlated with LNM and can be used as predictors of LNM. Furthermore, GLI1 and PTTG1 expression has certain clinical implications on the evaluation of cases and the development of treatment plans.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report’s classification
Scientific Quality: Grade C
Novelty: Grade C
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Alonso C, Spain S-Editor: Lin C L-Editor: A P-Editor: Xu ZH
| 1. | Zhou E, Rifkin S. Colorectal Cancer and Diet: Risk Versus Prevention, Is Diet an Intervention? Gastroenterol Clin North Am. 2021;50:101-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 2. | Mattiuzzi C, Sanchis-Gomar F, Lippi G. Concise update on colorectal cancer epidemiology. Ann Transl Med. 2019;7:609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 203] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 3. | Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 444] [Cited by in RCA: 1060] [Article Influence: 176.7] [Reference Citation Analysis (1)] |
| 4. | Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 1566] [Article Influence: 261.0] [Reference Citation Analysis (2)] |
| 5. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 152] [Article Influence: 38.0] [Reference Citation Analysis (2)] |
| 6. | Sawicki T, Ruszkowska M, Danielewicz A, Niedźwiedzka E, Arłukowicz T, Przybyłowicz KE. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 453] [Article Influence: 113.3] [Reference Citation Analysis (0)] |
| 7. | Han CJ, Yang GS, Syrjala K. Symptom Experiences in Colorectal Cancer Survivors After Cancer Treatments: A Systematic Review and Meta-analysis. Cancer Nurs. 2020;43:E132-E158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 8. | Kim HJ, Choi GS. Clinical Implications of Lymph Node Metastasis in Colorectal Cancer: Current Status and Future Perspectives. Ann Coloproctol. 2019;35:109-117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 9. | Giannopoulou AI, Kanakoglou DS, Piperi C. Transcription Factors with Targeting Potential in Gliomas. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Chetty R. Gene of the month: GLI-1. J Clin Pathol. 2020;73:228-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Du Z, Zhou F, Jia Z, Zheng B, Han S, Cheng J, Zhu G, Huang P. The hedgehog/Gli-1 signaling pathways is involved in the inhibitory effect of resveratrol on human colorectal cancer HCT116 cells. Iran J Basic Med Sci. 2016;19:1171-1176. [PubMed] |
| 12. | Park SH, Jeong S, Kim BR, Jeong YA, Kim JL, Na YJ, Jo MJ, Yun HK, Kim DY, Kim BG, Lee DH, Oh SC. Activating CCT2 triggers Gli-1 activation during hypoxic condition in colorectal cancer. Oncogene. 2020;39:136-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 13. | Smith VE, Franklyn JA, McCabe CJ. Pituitary tumor-transforming gene and its binding factor in endocrine cancer. Expert Rev Mol Med. 2010;12:e38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Zhou C, Tong Y, Wawrowsky K, Melmed S. PTTG acts as a STAT3 target gene for colorectal cancer cell growth and motility. Oncogene. 2014;33:851-861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ, Yoo KC, Lee GH, Cui YH, Oh YS, Gye MC, Lee YY, Park IC, An S, Hwang SG, Park MJ, Suh Y, Lee SJ. PTTG1 oncogene promotes tumor malignancy via epithelial to mesenchymal transition and expansion of cancer stem cell population. J Biol Chem. 2012;287:19516-19527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 16. | Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 236] [Article Influence: 78.7] [Reference Citation Analysis (1)] |
| 17. | Gupta S. Screening for Colorectal Cancer. Hematol Oncol Clin North Am. 2022;36:393-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 18. | Wu J, Di D, Zhao C, Pan Q, Liu Y, Zhang X, Zhao X, Chen H. Clinical Significance of Gli-1 And Caveolin-1 Expression in the Human Small Cell Lung Cancer. Asian Pac J Cancer Prev. 2018;19:401-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 19. | Chen JS, Li HS, Huang JQ, Zhang LJ, Chen XL, Wang Q, Lei J, Feng JT, Liu Q, Huang XH. Down-regulation of Gli-1 inhibits hepatocellular carcinoma cell migration and invasion. Mol Cell Biochem. 2014;393:283-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Ding YL, Zhou Y, Xiang L, Ji ZP, Luo ZH. Expression of glioma-associated oncogene homolog 1 is associated with invasion and postoperative liver metastasis in colon cancer. Int J Med Sci. 2012;9:334-338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Ren Q, Jin B. The clinical value and biological function of PTTG1 in colorectal cancer. Biomed Pharmacother. 2017;89:108-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 22. | Zheng Y, Guo J, Zhou J, Lu J, Chen Q, Zhang C, Qing C, Koeffler HP, Tong Y. FoxM1 transactivates PTTG1 and promotes colorectal cancer cell migration and invasion. BMC Med Genomics. 2015;8:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Yang Z, Zhang C, Qi W, Cui Y, Xuan Y. GLI1 promotes cancer stemness through intracellular signaling pathway PI3K/Akt/NFκB in colorectal adenocarcinoma. Exp Cell Res. 2018;373:145-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 24. | Papadopoulos V, Tsapakidis K, Riobo Del Galdo NA, Papandreou CN, Del Galdo F, Anthoney A, Sakellaridis N, Dimas K, Kamposioras K. The Prognostic Significance of the Hedgehog Signaling Pathway in Colorectal Cancer. Clin Colorectal Cancer. 2016;15:116-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Wang X, Yao Y, Zhu X. The influence of aberrant expression of GLI1/p-S6K on colorectal cancer. Biochem Biophys Res Commun. 2018;503:3198-3204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Feng ZZ, Chen JW, Yang ZR, Lu GZ, Cai ZG. Expression of PTTG1 and PTEN in endometrial carcinoma: correlation with tumorigenesis and progression. Med Oncol. 2012;29:304-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |