Published online Jan 27, 2024. doi: 10.4240/wjgs.v16.i1.6
Peer-review started: November 18, 2023
First decision: December 6, 2023
Revised: December 11, 2023
Accepted: December 26, 2023
Article in press: December 26, 2023
Published online: January 27, 2024
Processing time: 67 Days and 23.8 Hours
Gastric cancer (GC) is a prevalent malignant tumor within the digestive system, with over 40% of new cases and deaths related to GC globally occurring in China. Despite advancements in treatment modalities, such as surgery supplemented by adjuvant radiotherapy or chemotherapeutic agents, the prognosis for GC remains poor. New targeted therapies and immunotherapies are currently under investigation, but no significant breakthroughs have been achieved. Studies have indicated that GC is a heterogeneous disease, encompassing multiple subtypes with distinct biological characteristics and roles. Consequently, personalized treatment based on clinical features, pathologic typing, and molecular typing is crucial for the diagnosis and management of precancerous lesions of gastric cancer (PLGC). Current research has categorized GC into four subtypes: Epstein-Barr virus-positive, microsatellite instability, genome stability, and chromosome instability (CIN). Technologies such as multi-omics analysis and gene sequencing are being employed to identify more suitable novel testing methods in these areas. Among these, ultrasensitive chromosomal aneuploidy detection (UCAD) can detect CIN at a genome-wide level in subjects using low-depth whole genome sequencing technology, in conjunction with bioinformatics analysis, to achieve qualitative and quantitative detection of chromosomal stability. This editorial reviews recent research advancements in UCAD technology for the diagnosis and management of PLGC.
Core Tip: The purpose of this editorial is to provide an overview of the current diagnostic and therapeutic guidelines for gastric precancerous lesions, and to explore the potential clinical application of ultrasensitive chromosomal aneuploidy detection in the field of gastric cancer prevention and control. By doing so, this article aims to advance future research in this area.
- Citation: Qian ST, Xie FF, Zhao HY, Liu QS, Cai DL. Prospects in the application of ultrasensitive chromosomal aneuploidy detection in precancerous lesions of gastric cancer. World J Gastrointest Surg 2024; 16(1): 6-12
- URL: https://www.wjgnet.com/1948-9366/full/v16/i1/6.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v16.i1.6
Gastric cancer (GC) is the third leading cause of cancer-related deaths globally, with the highest incidence particularly in East Asia, Central, and Eastern Europe[1]. The number of incidences of GC will likely increase in the future due to higher socioeconomic status and aging populations[2]. In recent times, the prevalence of pan-cancer screening and gastroscopy has led to early identification and treatment of a growing number of patients in the initial (gastric pre-cancer) phase. The use of endoscopic mucosal resection (EMR) and endoscopic submucosal dissection (ESD) has significantly enhanced patient survival. However, the majority of cases are still diagnosed at an advanced stage, resulting in the majority of patients succumbing to GC[3]. Despite the advancements in medical technology, a significant number of patients are reluctant to undergo endoscopy. Thus, researchers are still actively investigating the stages of precancerous lesions of gastric cancer (PLGC) and searching for more expedited and streamlined diagnostic approaches. These remain pressing issues in current PLGC research.
Due to the subtle symptoms of GC, the condition is typically discovered at an advanced stage in patients who seek medical attention, leading to higher mortality rates and a poorer prognosis. Advancements in next-generation sequencing and other genomic technologies have spurred new research into the molecular characteristics of GC[4]. Gene mutations, chromosomal aberrations, differential gene expression, and epigenetic alterations are some of the genetic and epigenetic factors that influence the pathogenesis of GC. Thus, The Cancer Genome Atlas (TCGA) network proposes a four-subtype classification scheme for GC based on the molecular biology of potential tumors for each subtype, including Epstein-Barr virus (EBV)-positive, microsatellite instability (MSI), genome stability (GS) type, and chromosome instability (CIN) type[5]. One of the most frequent genetic alterations is CIN, often referred to as chromosomal copy number aberration (CNA). The ultrasensitive chromosomal aneuploidy detection (UCAD) technology relies on low-coverage whole-genome sequencing (LC-WGS) to diagnose or predict a patient's tumor risk by identifying chromosomal CNA in a sample.
The purpose of this editorial is to offer an updated overview of the established diagnostic and treatment criteria for gastric precancerous lesions. It covers emerging diagnostic and treatment techniques, and explores the potential for deploying UCAD technology as an early diagnostic tool.
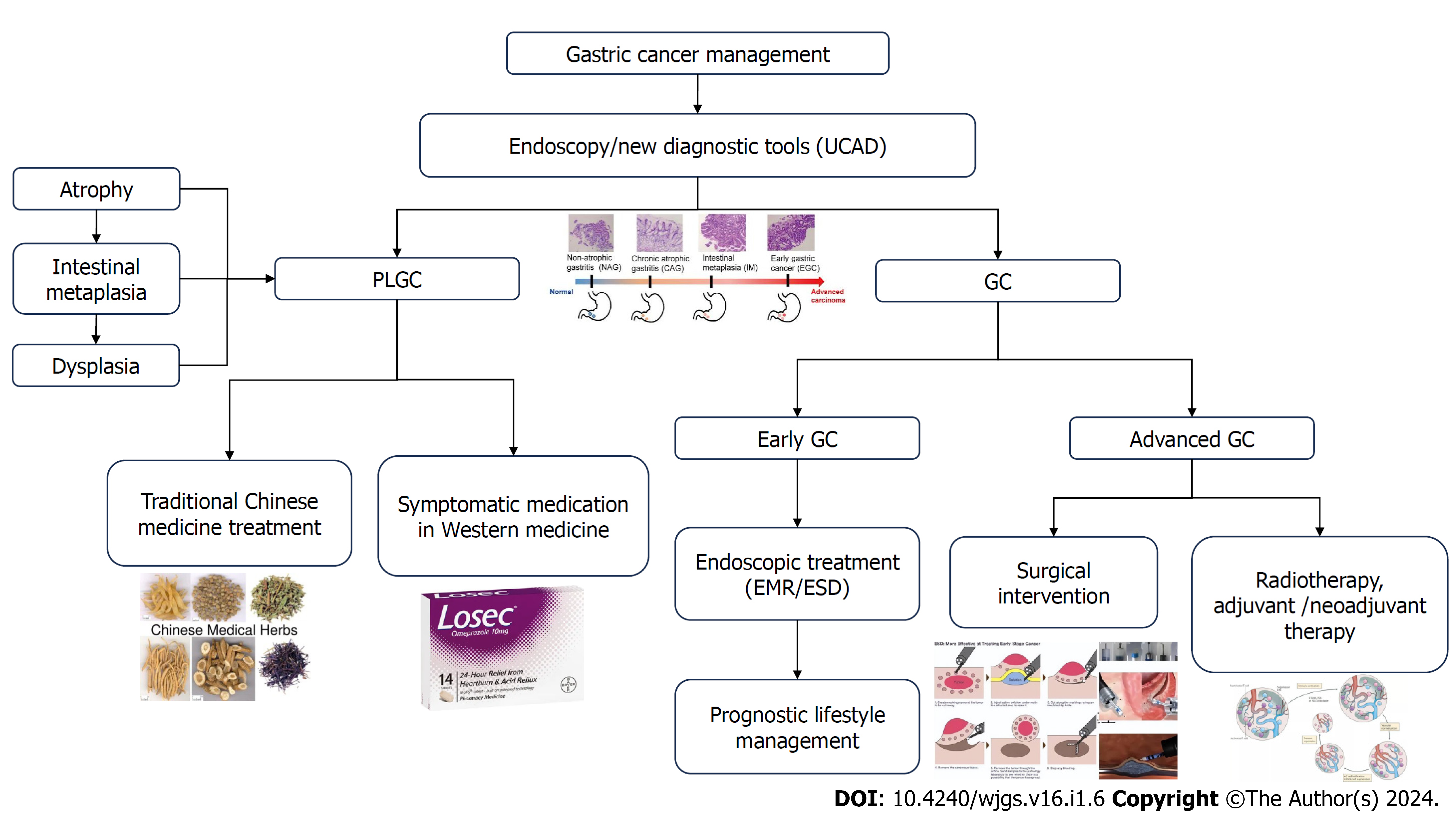
At this stage, the clinical consensus of PLGC can be categorized into atrophy (loss of gastric glands), intestinal metaplasia (replacement of gastric epithelium by intestinal epithelium), and dysplasia (intraepithelial neoplasia, including low-grade intraepithelial neoplasia and high-grade intraepithelial neoplasia)[6].
Helicobacter pylori (H. pylori) infection, the most common chronic bacterial infection in humans, is the strongest known risk factor for gastric carcinogenesis. Chronic and persistent inflammation resulting from H. pylori in the stomach lining may give rise to progressive atrophic gastritis and intestinal metaplasia[7].
H. pylori strains have a cytotoxin-associated gene A (CagA) that encodes a 120-140 kDa CagA protein, an oncoprotein that affects tumor cells. It also contains additional virulence factors, including vacuolating cytotoxin A, duodenal ulcer-promoting gene A protein, outer inflammatory protein A, and gamma-glutamyl transpeptidase. Most individuals infected with H. pylori are asymptomatic, yet have an increased risk of developing peptic ulcers or gastric adenocarcinoma[8,9].
In addition to H. pylori, age, tobacco and alcohol consumption, high salt intake and a diet low in fruits and vegetables, familial susceptibility, previous gastric surgery, and pernicious anemia are all relevant risk factors. It is clear that control of the above risk factors is an important basis for the prevention of PLGC[10].
Screening for cancer risk factors is a straightforward process; however, precisely identifying at-risk groups for early screening poses a challenge. To mitigate this, it is essential to implement effective strategies that optimize the selection of individuals who are most likely to benefit from early screening. The primary diagnostic method currently used is endoscopic biopsy, with the challenge being the identification of individuals at risk of developing PLGC in the absence of positive signs[11]. It is apparent that endoscopy, as an invasive medical examination, receives limited acceptance from certain demographics who may also be restricted by underlying diseases and age. The development and implementation of novel early cancer diagnostic tools must adhere to the reliability, reproducibility, and cost control standards set by the World Health Organization. Furthermore, they must prioritize ease of use and patient comfort[12].
Available studies have shown a significant level of heterogeneity in the histopathology and molecular biology of GC. Screening for hallmark molecules of PLGC involves identifying genetic or protein markers that can be used for early diagnosis and prognostic determination. Most of the screened molecules such as oncogenes, intercellular adhesion molecules, growth factors, and certain hormone receptors exhibit deficiencies in sensitivity, specificity, and reliability, with only a few being recognized.
A paper published in Nature a few years ago by TCGA proposed dividing GC into four subtypes based on molecular characteristics[13].
EBV-positive phenotype is frequently associated with PIK3CA mutations, DNA hypermethylation, and amplification of JAK2, CD274, and PDCD1LG2. MSI is a type characterized by a high mutation rate, including mutations in genes that activate oncogene signaling pathways. The GS type, which occurs most often in the histologically diffuse form, is caused by mutations in RHOA or fusions of GTPase-activating protein genes in the THO family. CIN type, which has the hallmark heterozygous chromosomes and in situ amplification of the receptor lysine kinase.
The conventional method of devising treatment plans based on tumor phenotypic characteristics will be replaced by a mode that considers gene alterations instead[14]. Utilizing single gene expression changes can direct targeted medication therapy, which is a more rational, efficient and personalized mode of treatment compared to chemotherapy for the same morphology type. Additionally, multi-gene detection will become an essential research area[15].
Early GC without lymph node metastasis may be treated through endoscopic or surgical means, depending on the extent of tumor invasion. Adjuvant radiotherapy or chemotherapy is not necessary after surgery.
Locally advanced GC or early GC with lymph node metastasis should be managed using comprehensive surgical treatment. The decision of direct radical surgery vs preoperative neoadjuvant chemotherapy, before radical surgery, depends on the depth of tumor invasion and whether or not lymph node metastasis is present. For patients with locally advanced GC who have undergone successful radical surgery, adjuvant treatment planning (including adjuvant chemotherapy and, if necessary, adjuvant chemoradiotherapy) should be based on the postoperative pathological staging.
Recurrent or metastatic GC requires comprehensive treatment consisting mainly of medication. Palliative surgery, radiotherapy, interventional therapy, and local therapies should also be administered as needed, along with optimal supportive therapies, including analgesia, stenting, and nutrition support. It is essential to give supportive therapies actively at the appropriate time[16].
EMR is the endoscopic removal of mucosal lesions, in whole or in part, for the diagnosis and treatment of superficial tumors of the gastrointestinal tract[17].
ESD is a new technology developed on the basis of EMR. According to the lesions with different parts, sizes and infiltration depths, special electrocautery knives such as IT knives, Dual knives, Hook knives, etc. are selected for endoscopic gradual separation of tissues between the mucous membrane layer and the intrinsic muscular layer, and finally the lesion mucous membrane and submucosal layer are completely removed[18].
Other endoscopic treatments include laser therapy, argon knife, microwave therapy, etc. They can only remove the tumor, but cannot obtain complete pathological specimens or be sure that the tumor is completely removed. Therefore, they are mostly used for the treatment of precancerous gastric lesions and require close follow-up after treatment, and are not recommended as the first choice of treatment for early GC[19,20].
PLGC surgery is divided into radical surgery and non-radical surgery. Radical surgery should completely remove the primary lesion and thoroughly clear the regional lymph nodes, mainly including standard surgery, modified surgery and extended surgery; non-radical surgery mainly includes palliative surgery and tumor reduction surgery[21,22].
Chemotherapy is divided into palliative chemotherapy, adjuvant chemotherapy, neoadjuvant chemotherapy, and conversion therapy.
Neoadjuvant chemotherapy is recommended for locally advanced GC without distant metastases (T3/4, N+). The regimen should consist of a two-drug combination of platinum and fluorouracil or a three-drug combination based on the two-drug regimen combined with paclitaxel. It should not be used as a single agent[11].
The GC management process is shown in Figure 1.
In the article "Hallmarks of cancer: The next generation" in Cell, the authors systematically summarized various hallmarks of cancer development. They highlighted genomic instability (or chromosomal instability) as the fundamental cause of cancer development, present in almost all malignant tumors. Thus, genomic instability is considered the root cause of cancer[23].
CIN is a feature of certain cells that results in the production of daughter cells that have an altered chromosome number or structure. This is a common feature in cancer cells. The fundamental aspects of chromosomal instability include changes in chromosome copy number, which leads to the emergence of cells with aneuploidy, as well as alterations in chromosome structure. These changes can include polyploidy, chromosomal translocations, genomic chaos, and non-clonal chromosomal aberrations[24].
The mechanism behind cancer cells is the irregular distribution of chromosomes during mitosis in zygote cells and the persistence of errors in chromosome segregation. This leads to modifications in chromosome copy number, as well as intrachromosomal segment amplification or deletion[25].
The substantial loss, gain, and rearrangement of DNA accumulate, resulting in chromosomal instability. This instability leads to extensive genomic complexity, which is a hallmark of cancer that is either present or expected. Chromosomal instability is strongly associated with tumor staging, metastasis, poor prognosis, and treatment resistance. This includes the loss or amplification of driver genes, focal rearrangements, extrachromosomal DNA, micronucleus formation, and the activation of innate immune signaling. These mechanisms lead to the diversification of tumor subclones in terms of space and time. This facilitates metastasis, accelerates the phenotypic adaptation of tumors, promotes cellular immortality, the escape from immune surveillance, and the development of resistance to drug treatments, among other processes[26].
CIN has been extensively studied as an innovative test. Using patient-derived tumor organoids to imitate colorectal cancer, researchers confirmed the existence of anaphase chromatin bridges in colorectal cancer cell lines. Causes consisted of insufficient disassembly of sister chromatids, telomere fusion producing bicentric chromosomes, and incomplete replication. These manifestations of CIN are prevalent in organs resembling colorectal cancer and have a significant impact on tumor evolution and therapeutic response. The levels of CIN and tolerance to mitotic errors play a crucial role in shaping the aneuploid landscapes and karyotypic heterogeneity[27].
Copy number and structural changes due to CIN are common features of multiple myeloma (MM). Primary and secondary genetic events caused by CIN lead to an increase in malignant plasma cell genomic instability by interfering with cell cycle checkpoints, thus accelerating proliferation. Thus, an assessment of CIN in MM and its precursor states may help to mitigate the progression of symptomatic disease and the risk of relapse[28].
CIN plays a role in the development of some subtypes of diffuse infiltrating gliomas. In isocitrate dehydrogenase-mutant astrocytomas, copy number variation (CNV) levels rise overall with tumor grade, duration, and proximity of infiltrating cells within the same tumor. The detection of CIN through CNV, DNA methylation, and/or gene expression profiling can effectively identify gliomas influenced by this molecular mechanism[29].
UCAD is a cutting-edge approach for identifying cellular chromosomal instability. This technique leverages body fluid free DNA or tissue genomic DNA as a template and utilizes LC-WGS, together with bioinformatics analysis, to identify chromosomal instability at the genome-wide scale. This technique has the potential to aid in the detection of tumors, monitor the effectiveness of treatment, and assess the likelihood of recurrence and metastasis. Samples including peripheral blood, exfoliated cells, menstrual blood, fresh and frozen tissues, formalin-fixed paraffin-embedded, urine, and others, can be analyzed for diagnostic purposes.
UCAD has been applied in clinical studies at this stage. Ye et al[30] collected samples of pancreatic cystic fluid from 102 patients with pancreatic cystic neoplasms (PCN) and employed UCAD to examine distinct CIN characteristics among different types of PCN. The deletion of chr3p and chr6p was used to define subtypes of serous cystadenoma. Meanwhile, the gain of chr1q and chr8q was associated with latent malignant PCN and facilitated the detection of high-risk intraductal papillary mucinous neoplasm.
Wang et al[31] collected plasma samples from 47 patients suspected of having lesions in the biliary tract. They utilized UCAD to analyze free DNA for CNV analysis through low-coverage whole genome sequencing. The results demonstrated the most frequent copy number gains in chr3q (7/29) and chr8q (6/29), with the most prevalent copy number losses noted in chr7p (6/29), chr17p (6/29), and chr19p (6/29). The sensitivity and specificity of the plasma CNV assay for the diagnosis of biliary tract cancer were 89.7% and 88.9%, respectively.
Feng et al[32] obtained 196 plasma samples from two groups of patients: a discovery cohort of individuals with PLC who were not eligible for surgery, and a validation cohort of patients who underwent pathologically confirmed hepatectomies. Of the 172 individuals, 22 (95.7%) surgically ineligible hepatocellular carcinomas were identified as having CNV in at least 1 of the 29 segments. 54 (69.4%) of the hepatocellular carcinomas eligible for surgery received positive screenings, and subsequently, confirmed to be cancerous by pathologic examination. Additionally, 26/27 non-cancers were identified with negative screenings.
Ye et al[33] gathered mucosal samples from 40 patients with GC from the hospital. 20 were microbiome-enriched, 5 tested positive for EBV DNA, and 15 had H. pylori DNA. Meanwhile, 20 of the samples were found to have CIN. UCAD can be used to identify 3 distinct subtypes of GC in the Chinese population, providing valuable guidance for future research on GC treatment and prevention. UCAD can identify three distinct subtypes of GC in the Chinese population. This finding could provide valuable guidance for further research on the prevention and treatment of GC.
In recent years, there has been an acceleration in precision treatment for PLGC, however, obstacles remain. The evolving endoscopic technology and surgical treatment have not sufficiently addressed the high recurrence rate after surgery. While there is a clinical benefit trend in anti-HER2 therapy and anti-CLDN18.2 monoclonal antibody therapy, additional research is necessary to optimize treatment and maximize patient benefit[34,35]. Early diagnostic treatment remains a pressing and essential need for improvement with a significant demand for less invasive or non-invasive testing and highly specific biomarkers[36]. Research efforts must focus on developing more convenient, comfortable, and highly accurate diagnostic tests, like tests on body fluids such as peripheral blood, urine or saliva, and gastric fluids, for early assessment of the likelihood of patient progression. Early treatment is critical for reducing the risk of developing GC.
In this context, UCAD has been researched and applied as a new test for early prevention in patients with positive early sequencing results or for gastroscopy and treatment to timely remove lesions at a stage when PLGC has not progressed. With the gradual decrease in the cost of second-generation sequencing, UCAD assays are expected to become more cost-effective in the near future. Meanwhile, the UCAD assay can capture both human and microbiome DNA while detecting specimens from mucosal, blood, and urine sources. This convenience, comfort, and information make it a superior technology for GC subtyping compared to other methods. Overall, this new approach may improve diagnostic and therapeutic strategies for PLGC in a cost-effective manner, achieving true precision.
We gratefully acknowledge the kind cooperation of all authors in the preparation of this paper.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kotelevets SM, Russia S-Editor: Zhang H L-Editor: A P-Editor: Zhang YL
| 1. | Röcken C. Predictive biomarkers in gastric cancer. J Cancer Res Clin Oncol. 2023;149:467-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 96] [Reference Citation Analysis (5)] |
| 2. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2847] [Article Influence: 569.4] [Reference Citation Analysis (5)] |
| 3. | Pimentel-Nunes P, Libânio D, Marcos-Pinto R, Areia M, Leja M, Esposito G, Garrido M, Kikuste I, Megraud F, Matysiak-Budnik T, Annibale B, Dumonceau JM, Barros R, Fléjou JF, Carneiro F, van Hooft JE, Kuipers EJ, Dinis-Ribeiro M. Management of epithelial precancerous conditions and lesions in the stomach (MAPS II): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG), European Society of Pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) guideline update 2019. Endoscopy. 2019;51:365-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 670] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 4. | Zeng Y, Jin RU. Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer. Semin Cancer Biol. 2022;86:566-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 103] [Article Influence: 34.3] [Reference Citation Analysis (1)] |
| 5. | Yuen ST, Leung SY. Genomics Study of Gastric Cancer and Its Molecular Subtypes. Adv Exp Med Biol. 2016;908:419-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Gullo I, Grillo F, Mastracci L, Vanoli A, Carneiro F, Saragoni L, Limarzi F, Ferro J, Parente P, Fassan M. Precancerous lesions of the stomach, gastric cancer and hereditary gastric cancer syndromes. Pathologica. 2020;112:166-185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (1)] |
| 7. | Toyoshima O, Nishizawa T, Koike K. Endoscopic Kyoto classification of Helicobacter pylori infection and gastric cancer risk diagnosis. World J Gastroenterol. 2020;26:466-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 101] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (2)] |
| 8. | de Brito BB, da Silva FAF, Soares AS, Pereira VA, Santos MLC, Sampaio MM, Neves PHM, de Melo FF. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J Gastroenterol. 2019;25:5578-5589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 198] [Cited by in RCA: 167] [Article Influence: 27.8] [Reference Citation Analysis (15)] |
| 9. | FitzGerald R, Smith SM. An Overview of Helicobacter pylori Infection. Methods Mol Biol. 2021;2283:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 10. | Eusebi LH, Telese A, Marasco G, Bazzoli F, Zagari RM. Gastric cancer prevention strategies: A global perspective. J Gastroenterol Hepatol. 2020;35:1495-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 11. | Hoshi H. Management of Gastric Adenocarcinoma for General Surgeons. Surg Clin North Am. 2020;100:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | Hu Y, Lv X, Wei W, Li X, Zhang K, Zhu L, Gan T, Zeng H, Yang J, Rao N. Quantitative Analysis on Molecular Characteristics Evolution of Gastric Cancer Progression and Prognosis. Adv Biol (Weinh). 2023;7:e2300129. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 13. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4848] [Article Influence: 440.7] [Reference Citation Analysis (2)] |
| 14. | Wadowska K, Bil-Lula I, Trembecki Ł, Śliwińska-Mossoń M. Genetic Markers in Lung Cancer Diagnosis: A Review. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (32)] |
| 15. | Bartlett J, Amemiya Y, Arts H, Bayani J, Eng B, Grafodatskaya D, Kamel Reid S, Lariviere M, Lo B, McClure R, Mittal V, Sadikovic B, Sadis S, Seth A, Smith J, Zhang X, Feilotter H. Multisite verification of the accuracy of a multi-gene next generation sequencing panel for detection of mutations and copy number alterations in solid tumours. PLoS One. 2021;16:e0258188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, Farjah F, Gerdes H, Gibson MK, Hochwald S, Hofstetter WL, Ilson DH, Keswani RN, Kim S, Kleinberg LR, Klempner SJ, Lacy J, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Outlaw D, Park H, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian N, Pluchino LA. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 957] [Article Influence: 319.0] [Reference Citation Analysis (0)] |
| 17. | Ahmed Y, Othman M. EMR/ESD: Techniques, Complications, and Evidence. Curr Gastroenterol Rep. 2020;22:39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 18. | Liu Q, Ding L, Qiu X, Meng F. Updated evaluation of endoscopic submucosal dissection versus surgery for early gastric cancer: A systematic review and meta-analysis. Int J Surg. 2020;73:28-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 19. | Niknam N, Obanor S, Lee LA. Endoscopic methods for the detection and treatment of gastric cancer. Curr Opin Gastroenterol. 2022;38:436-442. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Young E, Philpott H, Singh R. Endoscopic diagnosis and treatment of gastric dysplasia and early cancer: Current evidence and what the future may hold. World J Gastroenterol. 2021;27:5126-5151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 56] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (3)] |
| 21. | Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep. 2019;21:67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 316] [Article Influence: 52.7] [Reference Citation Analysis (1)] |
| 22. | Wang Y, Zhang L, Yang Y, Lu S, Chen H. Progress of Gastric Cancer Surgery in the era of Precision Medicine. Int J Biol Sci. 2021;17:1041-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 23. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47087] [Article Influence: 3363.4] [Reference Citation Analysis (5)] |
| 24. | Kuang X, Li J. Chromosome instability and aneuploidy as context-dependent activators or inhibitors of antitumor immunity. Front Immunol. 2022;13:895961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Bach DH, Zhang W, Sood AK. Chromosomal Instability in Tumor Initiation and Development. Cancer Res. 2019;79:3995-4002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 26. | Drews RM, Hernando B, Tarabichi M, Haase K, Lesluyes T, Smith PS, Morrill Gavarró L, Couturier DL, Liu L, Schneider M, Brenton JD, Van Loo P, Macintyre G, Markowetz F. A pan-cancer compendium of chromosomal instability. Nature. 2022;606:976-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 174] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 27. | Bolhaqueiro ACF, Ponsioen B, Bakker B, Klaasen SJ, Kucukkose E, van Jaarsveld RH, Vivié J, Verlaan-Klink I, Hami N, Spierings DCJ, Sasaki N, Dutta D, Boj SF, Vries RGJ, Lansdorp PM, van de Wetering M, van Oudenaarden A, Clevers H, Kranenburg O, Foijer F, Snippert HJG, Kops GJPL. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat Genet. 2019;51:824-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 157] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 28. | Neuse CJ, Lomas OC, Schliemann C, Shen YJ, Manier S, Bustoros M, Ghobrial IM. Genome instability in multiple myeloma. Leukemia. 2020;34:2887-2897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 29. | Richardson TE, Walker JM, Abdullah KG, McBrayer SK, Viapiano MS, Mussa ZM, Tsankova NM, Snuderl M, Hatanpaa KJ. Chromosomal instability in adult-type diffuse gliomas. Acta Neuropathol Commun. 2022;10:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Ye M, Zhang B, Han X, Wei X, Wang Y, Cao W, Wu J, Chen C, Sun X, Sun K, Li H, Zhang Q, Liang T. Low-Pass Genomic Sequencing Reveals Novel Subtypes of Pancreatic Cystic Neoplasms. Ann Surg Oncol. 2023;30:5804-5812. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 31. | Wang X, Fu XH, Qian ZL, Zhao T, Duan AQ, Ruan X, Zhu B, Yin L, Zhang YJ, Yu WL. Non-invasive detection of biliary tract cancer by low-coverage whole genome sequencing from plasma cell-free DNA: A prospective cohort study. Transl Oncol. 2021;14:100908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 32. | Feng S, Ding Z, Wang J, Qian Z, Li S, Zhang C, Xin H, Liu S, Ding G, Hu M, Meng Y, Li N. Investigation of Plasma cell-free cancer genome chromosomal instability as a tool for targeted minimally invasive biomarkers for primary liver cancer diagnoses. Cancer Med. 2020;9:5075-5085. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Ye LP, Mao XL, Zhou XB, Wang Y, Xu SW, He SQ, Qian ZL, Zhang XG, Zhai LJ, Peng JB, Gu BB, Jin XX, Song YQ, Li SW. Cost-effective low-coverage whole-genome sequencing assay for the risk stratification of gastric cancer. World J Gastrointest Oncol. 2022;14:690-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Reference Citation Analysis (0)] |
| 34. | Janjigian YY, Kawazoe A, Yañez P, Li N, Lonardi S, Kolesnik O, Barajas O, Bai Y, Shen L, Tang Y, Wyrwicz LS, Xu J, Shitara K, Qin S, Van Cutsem E, Tabernero J, Li L, Shah S, Bhagia P, Chung HC. The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer. Nature. 2021;600:727-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 477] [Article Influence: 119.3] [Reference Citation Analysis (1)] |
| 35. | Zhong W, Lu Y, Ma Z, He Y, Ding Y, Yao G, Zhou Z, Dong J, Fang Y, Jiang W, Wang W, Huang Y. Development of a Humanized VHH Based Recombinant Antibody Targeting Claudin 18.2 Positive Cancers. Front Immunol. 2022;13:885424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 36. | Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, Bleotu C, Diaconu CC, Chivu-Economescu M. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029-2044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 307] [Cited by in RCA: 298] [Article Influence: 49.7] [Reference Citation Analysis (3)] |