Published online Mar 15, 2024. doi: 10.4239/wjd.v15.i3.552
Peer-review started: September 25, 2023
First decision: December 6, 2023
Revised: December 13, 2023
Accepted: February 2, 2024
Article in press: February 2, 2024
Published online: March 15, 2024
Processing time: 172 Days and 2 Hours
The association of single nucleotide polymorphism of KCNQ1 gene rs2237895 with type 2 diabetes mellitus (T2DM) is currently controversial. It is unknown whether this association can be gene realized across different populations.
To determine the association of KCNQ1 rs2237895 with T2DM and provide reliable evidence for genetic susceptibility to T2DM.
We searched PubMed, Embase, Web of Science, Cochrane Library, Medline, Baidu Academic, China National Knowledge Infrastructure, China Biomedical Liter-ature Database, and Wanfang to investigate the association between KCNQ1 gene rs2237895 and the risk of T2DM up to January 12, 2022. Review Manager 5.4 was used to analyze the association of the KCNQ1 gene rs2237895 polymorphism with T2DM and to evaluate the publication bias of the selected literature.
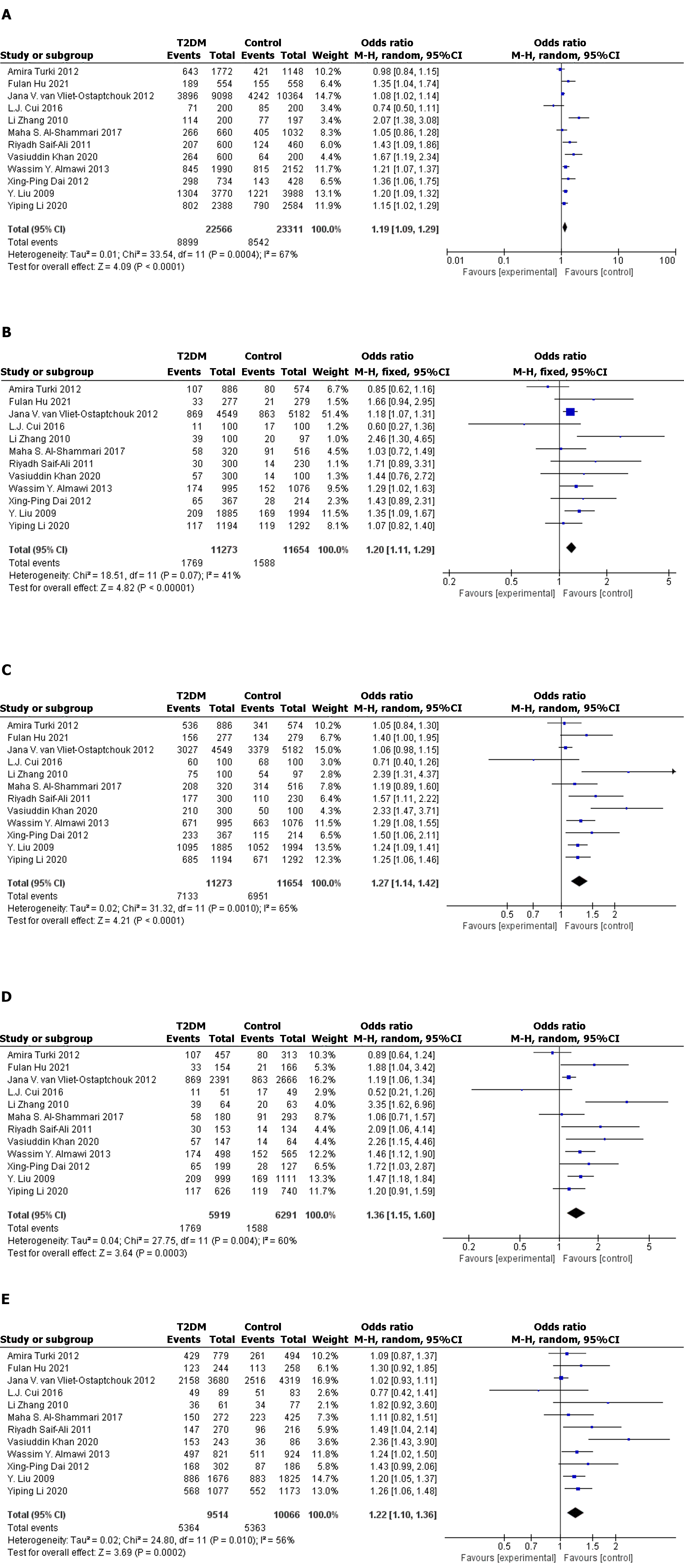
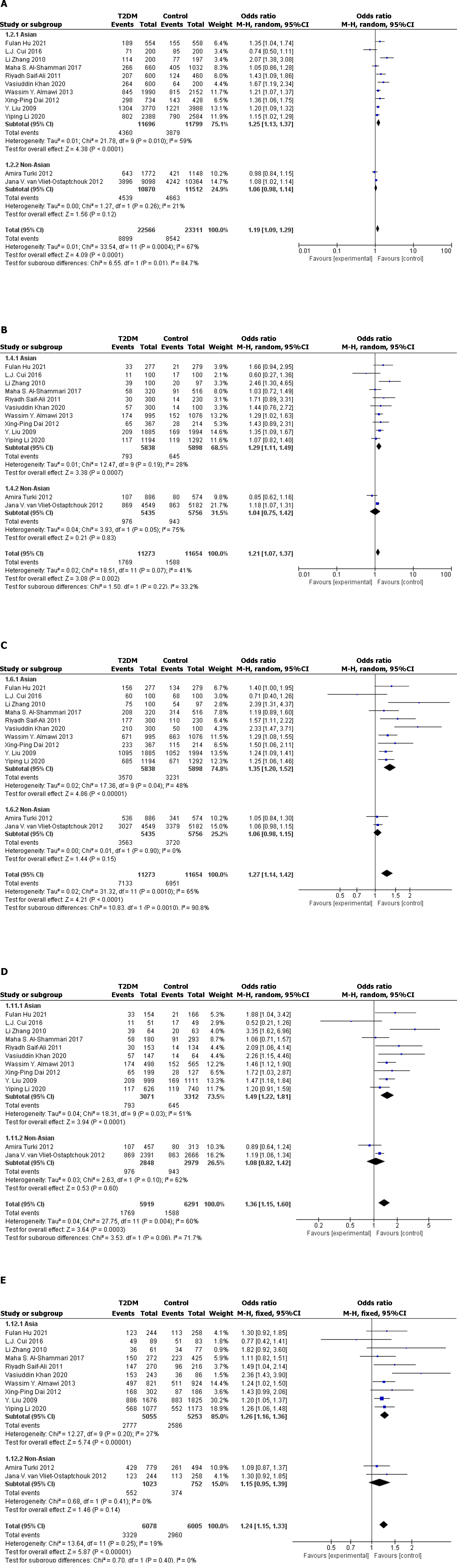
Twelve case–control studies (including 11273 cases and 11654 controls) met our inclusion criteria. In the full population, allelic model [odds ratio (OR): 1.19; 95% confidence interval (95%CI): 1.09–1.29; P < 0.0001], recessive model (OR: 1.20; 95%CI: 1.11–1.29; P < 0.0001), dominant model (OR: 1.27. 95%CI: 1.14–1.42; P < 0.0001), and codominant model (OR: 1.36; 95%CI: 1.15–1.60; P = 0.0003) (OR: 1.22; 95%CI: 1.10–1.36; P = 0.0002) indicated that the KCNQ1 gene rs2237895 polymorphism was significantly correlated with susceptibility to T2DM. In stratified analysis, this association was confirmed in Asian populations: allelic model (OR: 1.25; 95%CI: 1.13–1.37; P < 0.0001), recessive model (OR: 1.29; 95%CI: 1.11–1.49; P = 0.0007), dominant model (OR: 1.35; 95%CI: 1.20–1.52; P < 0.0001), codominant model (OR: 1.49; 95%CI: 1.22–1.81; P < 0.0001) (OR: 1.26; 95%CI: 1.16–1.36; P < 0.0001). In non-Asian populations, this association was not significant: Allelic model (OR: 1.06, 95%CI: 0.98–1.14; P = 0.12), recessive model (OR: 1.04; 95%CI: 0.75–1.42; P = 0.83), dominant model (OR: 1.06; 95%CI: 0.98–1.15; P = 0.15), codominant model (OR: 1.08; 95%CI: 0.82–1.42; P = 0.60. OR: 1.15; 95%CI: 0.95–1.39; P = 0.14).
KCNQ1 gene rs2237895 was significantly associated with susceptibility to T2DM in an Asian population. Carriers of the C allele had a higher risk of T2DM. This association was not significant in non-Asian populations.
Core Tip: In Asian populations, the rs2237895 polymorphism in the KCNQ1 gene was significantly associated with susceptibility to type 2 diabetes mellitus (T2DM), and C allele carriers had an increased risk of developing T2DM. The CC and AC genotypes of KCNQ1 rs2237895 significantly increased the susceptibility to T2DM. In non-Asian populations, this association was not significant.
- Citation: Li DX, Yin LP, Song YQ, Shao NN, Zhu H, He CS, Sun JJ. KCNQ1 rs2237895 gene polymorphism increases susceptibility to type 2 diabetes mellitus in Asian populations. World J Diabetes 2024; 15(3): 552-564
- URL: https://www.wjgnet.com/1948-9358/full/v15/i3/552.htm
- DOI: https://dx.doi.org/10.4239/wjd.v15.i3.552
Type 2 diabetes mellitus (T2DM) is a common multifactorial, metabolic disease whose pathogenesis is influenced by a combination of genetic and environmental factors. The rise and large-scale application of genome-wide association studies have contributed to the understanding of genetic factors related to T2DM. T2DM remains a health problem that plagues the world to this day. As of January 4, 2021, the number of people with diabetes worldwide had reached 537 million. Even more alarmingly, this number is expected to increase to 643 million by 2030. The various expenditures due to diabetes have exceeded $966 billion, and this figure has grown at an annual rate of 63% since 2006[1]. The etiology of T2DM is complex and has not yet been fully elucidated. T2DM is characterized by defective insulin secretion and reduced sensitivity, leading to chronic hyperglycemia and severe metabolic dysfunction in patients[2,3]. Hyperglycemia affects the physiological function of several tissues and organs in the body, among which the most common are neuropathy and vascular complications[1].
Studies have not provided an accurate description of the etiology of T2DM, and a genome-wide scan of Japanese by Nawata et al[4] showed that KCNQ1 is a susceptibility gene for T2DM in Japan. In addition, genes such as ADRA2A, KCNJ11 and CDKAL1 may be associated with the development of T2DM[4,5]. KCNQ1 is a potassium channel subunit that is mainly found in adipose and pancreatic tissues. It was found that KCNQ1 affects the process of islet β-cell depolarization by regulating potassium channel currents, thereby limiting insulin secretion from pancreatic β-cells and leading to the development of T2DM[6].
Previous studies have found that C allele carriers of the KCNQ1 gene rs2237895 may have an increased risk of developing T2DM[7]. rs2237895 is present in three genotypes in the population, AA, AC and CC. The A gene is wild type and the C gene is mutant, and their gene frequencies in the population are approximately 66% and 34%[8]. A study by Cui et al[7] in Kazakhs living in China showed that rs2237895 single nucleotide polymorphism (SNP) of KCNQ1 gene was not significantly associated with T2DM. A study by Afshardoost et al[9] on Iranians also showed no significant association between rs2237895 and T2DM; while in a study by Khan et al[10] on Indians, they confirmed a significant association between the SNP of rs2237895 and T2DM. Previously, a similar study has been conducted by Sun et al[11], but we consider that their inclusion criteria were more lenient and the strength of the proof may be weakened. Meanwhile, their work was > 10 years old and many new studies have been published during this period and that meta-analysis is in urgent need of updating. To address the above issues, we performed the present meta-analysis.
The following nine electronic databases were searched: PubMed, Embase, Web of Science, Cochrane Library, Medline, Baidu Academic, China National Knowledge Infrastructure (CNKI), China Biomedical Literature Database (CBM), and Wanfang Database, with the following search formulas: Subject (T2DM) and keywords (KCNQ1) and keywords (rs2237895). The last search date was January 12, 2022. Chinese and English literature on the association of the rs2237895 SNP in the KCNQ1 gene with T2DM was collected. The inclusion criteria for the articles were: (1) T2DM patients in the case group met the diagnostic criteria for diabetes published by WHO in 1999 or American Diabetes Association in 2010; (2) the type of experiment was a case–control study or a cohort study; (3) there was sufficient information in the text to describe the genotype and allele frequencies of the case and control groups; (4) the patients in the control group all met the Hardy–Weinberg genetic equilibrium model; (5) patients were randomly selected with no special restrictions on age, sex, or family history; and (6) for duplicate or data-identical literature, the one with the most complete information. Exclusion criteria were: (1) Incomplete study data; (2) literature reviews; (3) studies with gestational diabetes as an endpoint; and (4) exclusion of studies with familial diabetes as a basis.
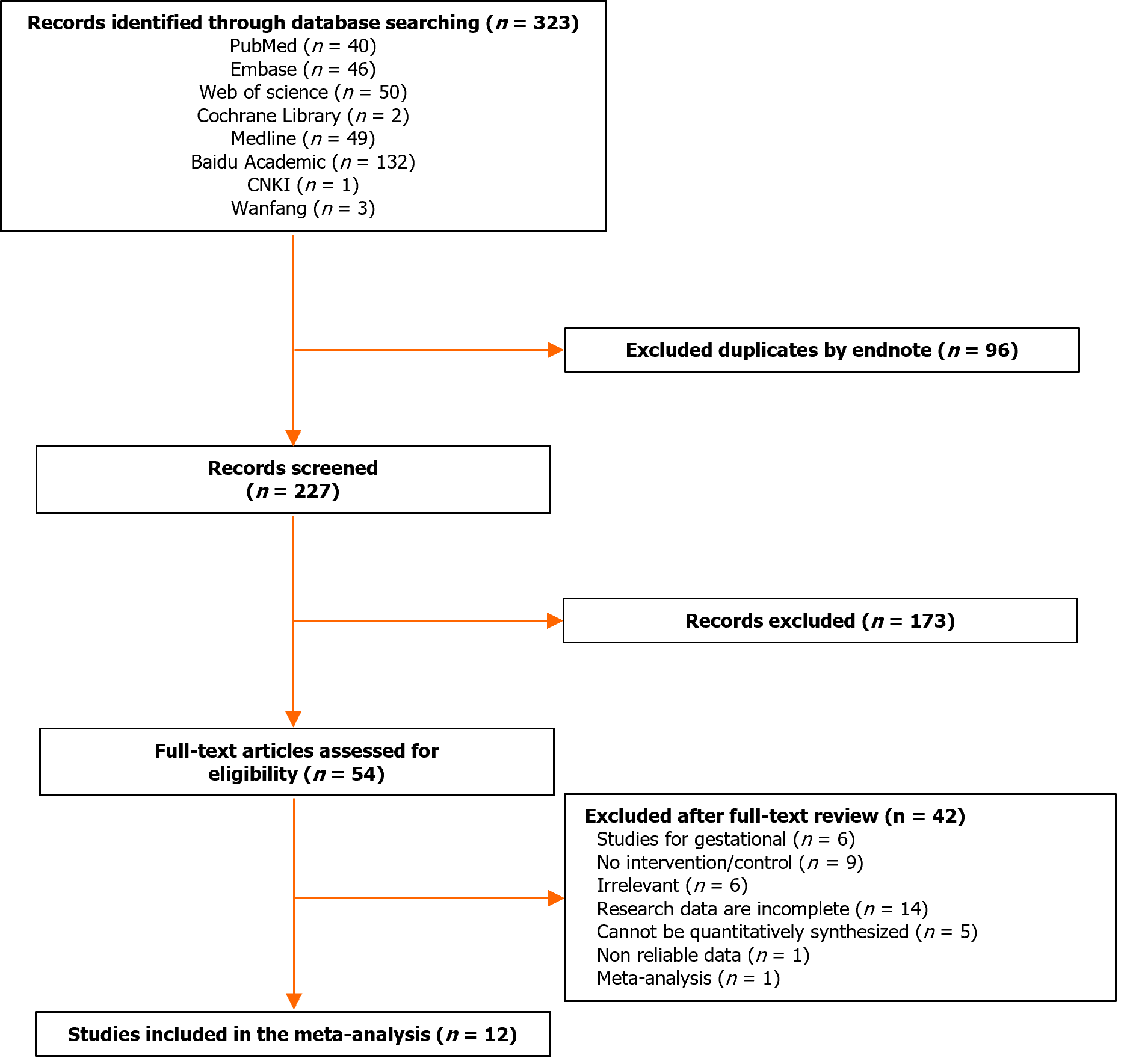
Two researchers independently performed literature screening and extraction of information based on the above criteria. A third researcher was required to discuss and agree on the results when difficult differences were encountered. For each article, we collected the basic information that needed to be used for Meta-analysis, and the literature screening process is shown in Figure 1.
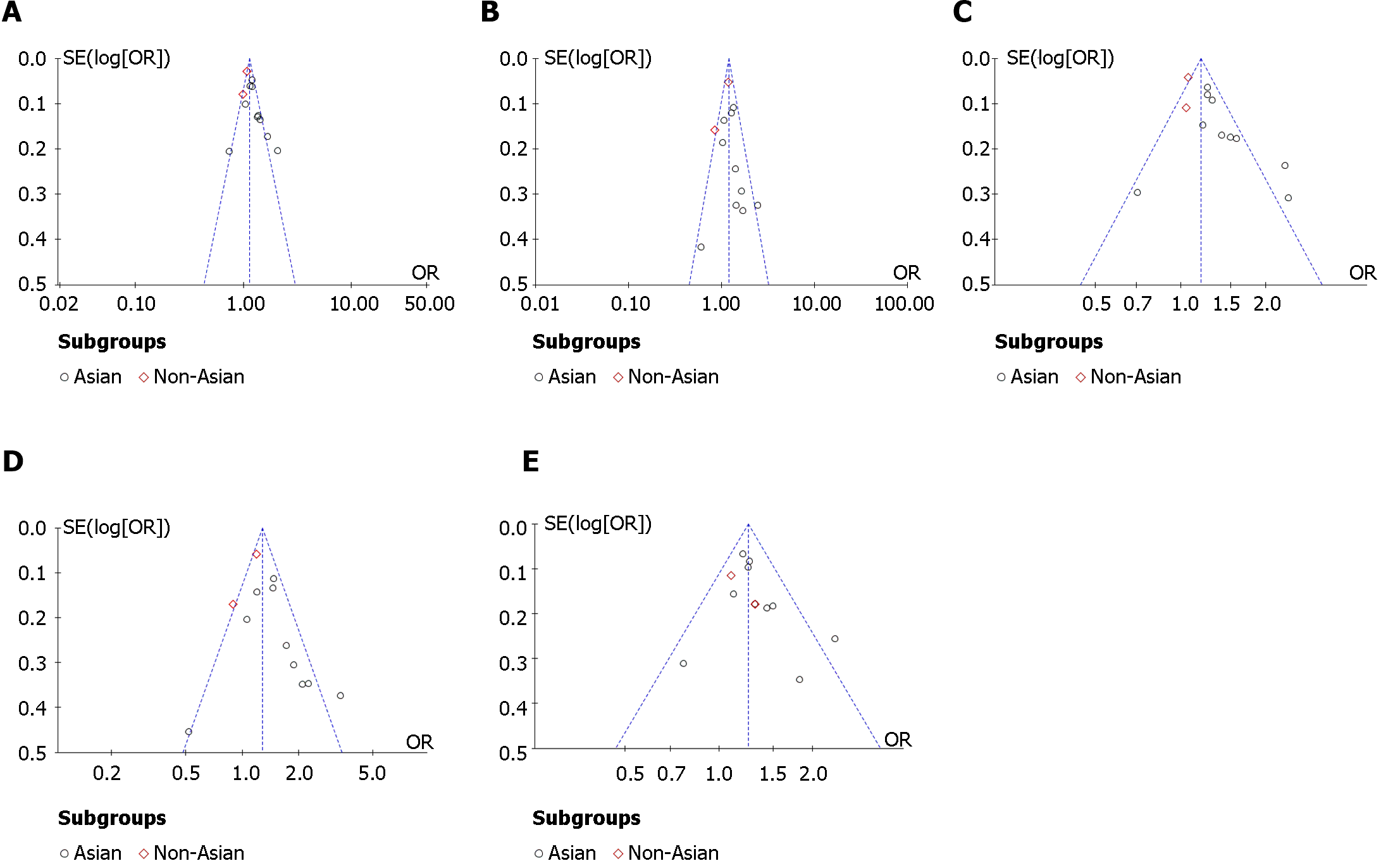
The data were processed using Review Manager 5.4. The strength of association between SNPs in the KCNQ1 gene rs2237895 and the risk of T2DM was assessed using the odds ratio (OR) and its corresponding 95% confidence interval (95%CI) as a criterion in the data statistics. The forest plots were used to show the OR and its 95%CI for each study. The pooled results were directly observed on the forest plots. The difference was considered significant when the 95%CI did not include 1. Allelic model (C vs A), recessive model (CC vs AA + AC), dominant model (CC + AC vs AA) and codominant model (CC vs AA and AC vs AA) were used to assess the genetic effects of the genes. The significance level was set at P < 0.05. The random-effect model was used to calculate the effect size when the heterogeneity was I2 > 50%, and the fixed-effect model was used when I2 was < 50%. Publication bias was assessed by Egger’s test and funnel plot. In the funnel plot, the dashed line perpendicular to the horizontal axis indicated the combined effect size. It suggested that the studies were without publication bias when the distribution of studies in the funnel plot was approximately symmetrical.
According to the research strategy, 323 relevant papers were retrieved from the databases. Some duplicates were found and we removed them by Endnote software. We also screened the citations of the paper to ensure the comprehensiveness of the search. After a stepwise screening process, 12 eligible papers were finally included for meta-analysis, which included 11273 patients with T2DM and 11654 controls. Five of the datasets were from China[12-16], five from the rest of Asia[7,10,17-19], one from Europe[20], and one from Africa[21]. The basic information of the studies is shown in Table 1.
| Ref. | Ethnicity | N | Age (yr) | Case | Control | ||||||
| Case | Control | Case | Control | AA | AC | CC | AA | AC | CC | ||
| Cui et al[7], 2016 | Kazakh | 100 | 100 | 51.21 ± 11.60 | 49.85 ± 12.41 | 40 | 49 | 11 | 32 | 51 | 17 |
| Khan et al[10], 2020 | Indian | 300 | 100 | 40.33 ± 9.76 | 35.29 ± 7.96 | 90 | 153 | 57 | 50 | 36 | 14 |
| Liu et al[12], 2009 | Chinese | 1885 | 1994 | 63.9 ± 9.50 | 58.10 ± 9.40 | 790 | 886 | 209 | 942 | 883 | 169 |
| Zhang[13], 2010 | Chinese | 100 | 97 | 63.90 ± 9.50 | 58.1 ± 9.40 | 25 | 36 | 39 | 43 | 34 | 20 |
| Dai et al[14], 2012 | Chinese | 367 | 214 | 49.13 ± 10.79 | 47.55 ± 10.93 | 134 | 168 | 65 | 99 | 87 | 28 |
| Li et al[15], 2020 | Chinese | 1194 | 1292 | 52.49 ± 12.10 | 52.70 ± 10.52 | 509 | 568 | 117 | 621 | 552 | 119 |
| Hu et al[16], 2021 | Chinese | 277 | 279 | 52.26 ± 9.49 | 52.26 ± 9.49 | 121 | 123 | 33 | 145 | 113 | 21 |
| Saif-Ali et al[17], 2011 | Malaysian | 300 | 230 | 49.80 ± 7.42 | 52.90 ± 9.15 | 123 | 147 | 30 | 120 | 96 | 14 |
| Almawi et al[18], 2013 | Arabs | 995 | 1076 | 58.6 ± 13.40 | 57.30 ± 10.40 | 324 | 497 | 174 | 413 | 511 | 152 |
| Al-Shammari et al[19], 2017 | Arabs | 320 | 516 | 51.50 ± 8.75 | 48.75 ± 6.85 | 122 | 150 | 58 | 202 | 223 | 91 |
| van Vliet-Ostaptchouk et al[20], 2012 | Dutch | 4549 | 5182 | 64.36 ± 10.6 | 51.16 ± 10.10 | 1522 | 2158 | 869 | 1803 | 2516 | 863 |
| Turki et al[21], 2012 | Arabs | 886 | 574 | 61.20 ± 9.70 | 52.00 ± 11.9 | 350 | 429 | 107 | 233 | 261 | 80 |
The 12 datasets that met the inclusion criteria were pooled for meta-analysis, and allelic, recessive, dominant, and codominant models were used to investigate the association of rs2237895 with T2DM. Since the study population was predominantly Asian, we performed stratified analysis of Asian and non-Asian populations (Figure 2 and Figure 3).
In the full population, allelic model (OR: 1.19; 95%CI: 1.09–1.29; P < 0.0001), recessive model (OR: 1.20; 95%CI: 1.11–1.29; P < 0.0001), dominant model (OR: 1.27. 95%CI: 1.14–1.42; P < 0.0001), and codominant model (OR: 1.36; 95%CI: 1.15–1.60; P = 0.0003. OR: 1.22; 95%CI: 1.10–1.36; P = 0.0002) all showed significant association between rs2237895 and T2DM. In the subgroup of the Asian population, allelic model (OR: 1.25; 95%CI: 1.13–1.37; P < 0.0001), recessive model (OR: 1.29; 95%CI: 1.11–1.49; P = 0.0007), dominant model (OR: 1.35; 95%CI: 1.20–1.52; P < 0.0001), and codominant model (OR: 1.49; 95%CI: 1.22–1.81; P < 0.0001. OR: 1.26; 95% CI: 1.16–1.36; P < 0.0001) also showed a significant association between rs2237895 and T2DM, which was consistent with the whole population. C allele carriers had an increased risk of developing T2DM. The CC and AC genotypes significantly increased the risk of T2DM compared to the AA genotype. However, in the non-Asian population subgroup, allelic model (OR: 1.06; 95%CI: 0.98–1.14; P = 0.12), recessive model (OR: 1.04; 95%CI: 0.75–1.42; P = 0.83), dominant model (OR: 1.06; 95%CI: 0.98–1.15; P = 0.15), and codominant model (OR: 1.08; 95%CI: 0.82–1.42; P = 0.60) (OR: 1.15; 95%CI: 0.95–1.39; P = 0.14) all showed no significant association between rs2237895 and T2DM.
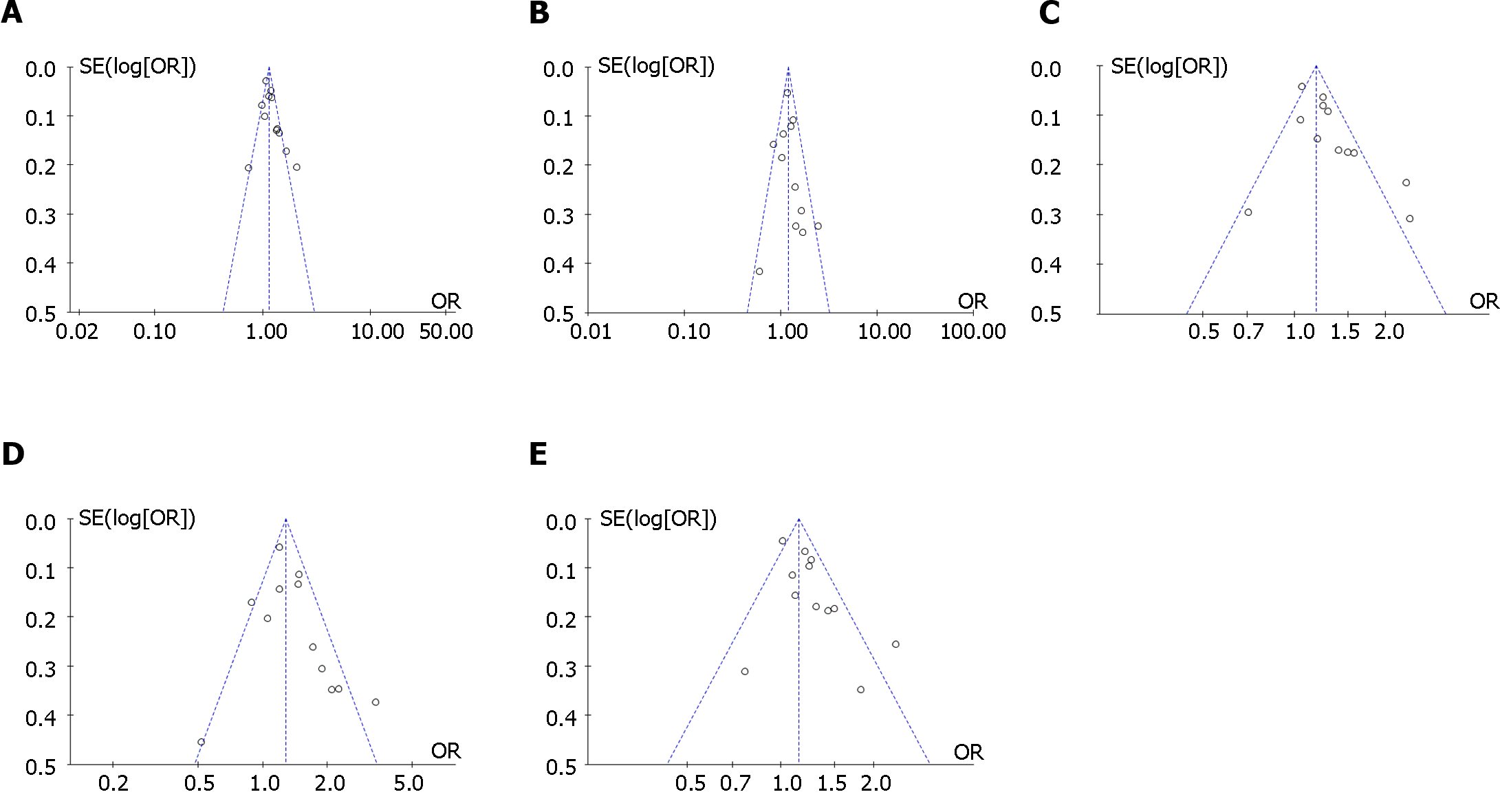
The funnel plots showed no significant publication bias was found in the meta-analysis (Figure 4 and Figure 5). Egger’s test showed no significant publication bias for the allelic model (t = 1.84, P = 0.095), recessive model (t = 0.48, P = 0.64), dominant model (t = 1.44, P = 0.18), and codominant model (t = 1.33, P = 0.21; t = 1.79, P = 0.10).
We performed a sensitivity analysis. After sequentially excluding one study in the allelic model, recessive model, dominant model, and codominant model, we calculated the pooled effect sizes for the remaining studies. By calculation, no qualitative change occurred between the pooled results of the remaining studies and the original results. Sensitivity analysis proved that the results of the meta-analysis were reliable.
Compared with previous studies, we increased the inclusion criteria of cases, excluded the interference of other factors (e.g., gestational diabetes), improved the strength of proof of the study, and made the results more reliable and stable. Our meta-analysis supported the findings of Khan et al[10], suggesting that the rs2237895 SNP in the KCNQ1 gene is significantly associated with the development of T2DM in Asian populations. In the study by Cui et al[7], the study population had an overall overweight problem, which increased the risk of T2DM prevalence and thus confounded the findings[22].
T2DM is a multifactorial, chronic, metabolic disease[23]. The idea that genetic factors have a significant role in the development of T2DM is now more widely accepted[23], although only a few genes have been confirmed as a risk for the development of T2DM. However, many genetic characteristics associated with T2DM, such as effect sizes and risk allele frequencies, need to be explored[24]. There is a need for researchers to identify risk genetic loci for T2DM and characterize the variation at the loci, thus providing a basis for elucidating the genetic pathogenesis of T2DM.
Previous studies have shown that the KCNQ1, miR-21, and Arg972 may be risk genes for T2DM[25,26]. KCNQ1 gene has now been shown to be located on chromosome 11p15.5, which is approximately 400 kb in length and consists of 17 exons ranging from 47 to 1122 bp in length[27]. KCNQ1 is associated with voltage-gated K+ channels, and mutations in the KCNQ1 gene lead to dysfunction of K+ channels, which would cause diseases such as QT syndrome and familial atrial fibrillation. KCNQ1 is expressed in many tissues[27,28], and the more studied about the KCNQ1 gene is expressed in cardiac and pancreatic tissues[29]. Current studies suggest that the main mechanisms of T2DM development are insulin resistance and islet β-cell dysfunction[2,23]. Variants in the KCNQ1 gene may lead to increased susceptibility to T2DM in the population by altering insulin secretion from pancreatic β-cells[30,31]. It was hypothesized that variants in the KCNQ1 gene would lead to increased expression of KCNQ1 protein on pancreatic β-cells, which in turn would alter the open state of voltage-gated potassium channels, decrease insulin secretion, and impair glucose storage and utilization[32].
This meta-analysis of the KCNQ1 gene rs2237895 SNP and T2DM association study involved 12 studies, including 11273 T2DM patients and 11654 controls. This analysis showed that the rs2237895 polymorphism was significantly associated with an elevated risk of developing T2DM in an Asian population, which is consistent with Khan et al’s[10] findings. In Asian populations, C allele carriers have an increased risk of developing T2DM. The risk of T2DM is also increased in people with the CC and AC genotypes compared to the AA genotype. This is consistent with the previous findings of Hu et al[16]. Also, their findings showed that rs2237895 was associated with hypertension, body mass index, and hypertriglyceridemia. In non-Asian populations, this association was not significant. A 2015 study by Ríos et al[33] in Europeans also showed that the KCNQ1 gene rs2237895 SNP was not significantly associated with T2DM, which is consistent with our findings. Our work provided strong evidence for the genetic pathogenesis of T2DM and helped to fully reveal the pathogenesis of T2DM.
This study showed that the rs2237895 SNP of the KCNQ1 gene was differentially associated with T2DM in different populations. The reasons for this variation may be mutations in the regulatory region of the KCNQ1 gene in particular populations[33], which interfere with the expression of the KCNQ1 gene; or it may be due to the existence of different genotypes and allele frequencies in populations with different clinical characteristics, geographical distribution and ethnic origin; or differences in the external influences associated with T2DM, such as lifestyle and behavior, in different populations[4,23-25]. The possibility of false-negative results in non-Asian populations with small study sample sizes cannot be excluded.
In the Asian population, there was a significant association between the KCNQ1 gene rs2237895 SNP and T2DM onset. C allele carriers were at increased risk of T2DM, and the CC and AC genotypes significantly increased the susceptibility to T2DM. However, in the non-Asian population, the association between rs2237895 and T2DM onset was not significant.
The association between the rs2237895 single nucleotide polymorphism (SNP) in the KCNQ1 gene and the prevalence of type 2 diabetes mellitus (T2DM) has been controversial in different studies.
The aim of this study was to investigate the association between the KCNQ1 gene rs2237895 and the prevalence of T2DM, and to provide help in establishing the pathogenesis of T2DM.
Demonstration of the association of the rs2237895 SNP in the KCNQ1 gene with the prevalence of T2DM. Also, to explore whether this relationship differs in different populations.
We searched nine databases. Two authors independently screened the literature according to the established inclusion and exclusion criteria. Finally, data extraction was performed and the data were meta-analyzed.
Twelve case–control studies met our inclusion criteria. After analysis, the rs2237895 SNP in the KCNQ1 gene was associated with T2DM prevalence in Asian populations. However, this association was not significant in non-Asian populations.
In Asian populations, carriers of the rs2237895 C allele of the KCNQ1 gene were highly susceptible to T2DM compared to those who did not carry the C allele. However, in non-Asian populations, the association between the rs2237895 SNP and T2DM was not significant.
We should continue to search for T2DM susceptibility genes through advanced technologies (e.g., genome-wide association strategy) and gradually elucidate the pathogenesis of T2DM.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Beg MMA, Kyrgyzstan; Horowitz M, Australia S-Editor: Lin C L-Editor: A P-Editor: Chen YX
| 2. | García-Chapa EG, Leal-Ugarte E, Peralta-Leal V, Durán-González J, Meza-Espinoza JP. Genetic Epidemiology of Type 2 Diabetes in Mexican Mestizos. Biomed Res Int. 2017;2017:3937893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 3. | Xu J, Zhang W, Song W, Cui J, Tian Y, Chen H, Huang P, Yang S, Wang L, He X, Shi B, Cui W. Relationship Between KCNQ1 Polymorphism and Type 2 Diabetes Risk in Northwestern China. Pharmgenomics Pers Med. 2021;14:1731-1751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 4. | Nawata H, Shirasawa S, Nakashima N, Araki E, Hashiguchi J, Miyake S, Yamauchi T, Hamaguchi K, Yoshimatsu H, Takeda H, Fukushima H, Sasahara T, Yamaguchi K, Sonoda N, Sonoda T, Matsumoto M, Tanaka Y, Sugimoto H, Tsubouchi H, Inoguchi T, Yanase T, Wake N, Narazaki K, Eto T, Umeda F, Nakazaki M, Ono J, Asano T, Ito Y, Akazawa S, Hazegawa I, Takasu N, Shinohara M, Nishikawa T, Nagafuchi S, Okeda T, Eguchi K, Iwase M, Ishikawa M, Aoki M, Keicho N, Kato N, Yasuda K, Yamamoto K, Sasazuki T. Genome-wide linkage analysis of type 2 diabetes mellitus reconfirms the susceptibility locus on 11p13-p12 in Japanese. J Hum Genet. 2004;49:629-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Xu N, Zhang TT, Han WJ, Yin LP, Ma NZ, Shi XY, Sun JJ. Association of CDKAL1 RS10946398 Gene Polymorphism with Susceptibility to Diabetes Mellitus Type 2: A Meta-Analysis. J Diabetes Res. 2021;2021:1254968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Rosengren AH, Braun M, Mahdi T, Andersson SA, Travers ME, Shigeto M, Zhang E, Almgren P, Ladenvall C, Axelsson AS, Edlund A, Pedersen MG, Jonsson A, Ramracheya R, Tang Y, Walker JN, Barrett A, Johnson PR, Lyssenko V, McCarthy MI, Groop L, Salehi A, Gloyn AL, Renström E, Rorsman P, Eliasson L. Reduced insulin exocytosis in human pancreatic β-cells with gene variants linked to type 2 diabetes. Diabetes. 2012;61:1726-1733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 7. | Cui LJ, Chang XY, Zhu LY, Feng G, Zhou T, Zhang CX, Chong KY, Sun K. Relationship between the polymorphisms in KCNQ1 and type 2 diabetes in Chinese Kazakh population. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jørgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 531] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 9. | Afshardoost S, Sarhangi N, Afshari M, Aghaei Meybodi HR, Hasanzad M. The influence of a genetic variant in the KCNQ1 gene on type 2 diabetes mellitus development. Gene Rep. 2019;17:100529. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Khan V, Verma AK, Bhatt D, Khan S, Hasan R, Goyal Y, Ramachandran S, Alsahli MA, Rahmani AH, Almatroudi A, Shareef MY, Meena B, Dev K. Association of Genetic Variants of KCNJ11 and KCNQ1 Genes with Risk of Type 2 Diabetes Mellitus (T2DM) in the Indian Population: A Case-Control Study. Int J Endocrinol. 2020;2020:5924756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Sun Q, Song K, Shen X, Cai Y. The association between KCNQ1 gene polymorphism and type 2 diabetes risk: a meta-analysis. PLoS One. 2012;7:e48578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, Zhang Z, Ning M, Hu X, Yang YF, Zhang ZF, Yu L, He L, Xu H. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia. 2009;52:1315-1321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Zhang L. The relationship of polymorphisms of rs2237895 in KCNQ1 gene with type 2 diabetes in Han population of Bengbu distric. M.Med. Thesis, BengBu Medical College. 2010. Available from: https://d.wanfangdata.com.cn/thesis/ChJUaGVzaXNOZXdTMjAyMzA5MDESB0Q2NjcxNTcaCGN5cnp4Y3I0. |
| 14. | Dai XP, Huang Q, Yin JY, Guo Y, Gong ZC, Lei MX, Jiang TJ, Zhou HH, Liu ZQ. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2012;39:462-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Li Y, Shen K, Li C, Yang Y, Yang M, Tao W, He S, Shi L, Yao Y. Identifying the association between single nucleotide polymorphisms in KCNQ1, ARAP1, and KCNJ11 and type 2 diabetes mellitus in a Chinese population. Int J Med Sci. 2020;17:2379-2386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Hu F, Zhang Y, Qin P, Zhao Y, Liu D, Zhou Q, Tian G, Li Q, Guo C, Wu X, Qie R, Huang S, Han M, Li Y, Zhang M, Hu D. Integrated analysis of probability of type 2 diabetes mellitus with polymorphisms and methylation of KCNQ1 gene: A nested case-control study. J Diabetes. 2021;13:975-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 17. | Saif-Ali R, Ismail IS, Al-Hamodi Z, Al-Mekhlafi HM, Siang LC, Alabsi AM, Muniandy S. KCNQ1 haplotypes associate with type 2 diabetes in Malaysian Chinese Subjects. Int J Mol Sci. 2011;12:5705-5718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Almawi WY, Nemr R, Keleshian SH, Echtay A, Saldanha FL, AlDoseri FA, Racoubian E. A replication study of 19 GWAS-validated type 2 diabetes at-risk variants in the Lebanese population. Diabetes Res Clin Pract. 2013;102:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Al-Shammari MS, Al-Ali R, Al-Balawi N, Al-Enazi MS, Al-Muraikhi AA, Busaleh FN, Al-Sahwan AS, Al-Elq A, Al-Nafaie AN, Borgio JF, AbdulAzeez S, Al-Ali A, Acharya S. Type 2 diabetes associated variants of KCNQ1 strongly confer the risk of cardiovascular disease among the Saudi Arabian population. Genet Mol Biol. 2017;40:586-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | van Vliet-Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, Bilo HJ, Klungel OH, de Boer A, van Diemen CC, Wijmenga C, Boezen HM, Dekker JM, van 't Riet E, Nijpels G, Welschen LM, Zavrelova H, Bruin EJ, Elbers CC, Bauer F, Onland-Moret NC, van der Schouw YT, Grobbee DE, Spijkerman AM, van der A DL, Simonis-Bik AM, Eekhoff EM, Diamant M, Kramer MH, Boomsma DI, de Geus EJ, Willemsen G, Slagboom PE, Hofker MH, 't Hart LM. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS One. 2012;7:e32148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Turki A, Mtiraoui N, Al-Busaidi AS, Khirallah M, Mahjoub T, Almawi WY. Lack of association between genetic polymorphisms within KCNQ1 locus and type 2 diabetes in Tunisian Arabs. Diabetes Res Clin Pract. 2012;98:452-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Naude CE, Brand A, Schoonees A, Nguyen KA, Chaplin M, Volmink J. Low-carbohydrate versus balanced-carbohydrate diets for reducing weight and cardiovascular risk. Cochrane Database Syst Rev. 2022;1:CD013334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1609] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 24. | Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Müller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stančáková A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutškov K, Langford C, Leander K, Lindholm E, Lobbens S, Männistö S, Mirza G, Mühleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurðsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvänen AC, Eriksson JG, Peltonen L, Nöthen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njølstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyövälti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jöckel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1477] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 25. | Li Y, Yang J, Tao W, Yang M, Wang X, Lu T, Li C, Yang Y, Yao Y. The Single Nucleotide Polymorphisms (rs1292037 and rs13137) in miR-21 Were Associated with T2DM in a Chinese Population. Diabetes Metab Syndr Obes. 2022;15:189-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Mannino GC, Andreozzi F, Sesti G. Pharmacogenetics of type 2 diabetes mellitus, the route toward tailored medicine. Diabetes Metab Res Rev. 2019;35:e3109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 27. | Han WJ, Deng JY, Jin H, Yin LP, Yang JX, Sun JJ. Association of KCNQ1rs2237892C. T Gene with Type 2 Diabetes Mellitus: A Meta-Analysis. J Diabetes Res. 2021;2021:6606830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 28. | Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Fauré S, Gary F, Coumel P, Petit C, Schwartz K, Guicheney P. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet. 1997;15:186-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 603] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 29. | Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 725] [Cited by in RCA: 724] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 30. | Ullrich S, Su J, Ranta F, Wittekindt OH, Ris F, Rösler M, Gerlach U, Heitzmann D, Warth R, Lang F. Effects of I(Ks) channel inhibitors in insulin-secreting INS-1 cells. Pflugers Arch. 2005;451:428-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 31. | Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, Nilsson P, Groop L, Lyssenko V. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes. 2009;58:2409-2413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 32. | Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5794] [Cited by in RCA: 5813] [Article Influence: 363.3] [Reference Citation Analysis (0)] |
| 33. | Ríos R, Lupiañez CB, Campa D, Martino A, Martínez-López J, Martínez-Bueno M, Varkonyi J, García-Sanz R, Jamroziak K, Dumontet C, Cayuela AJ, Wętek M, Landi S, Rossi AM, Lesueur F, Reis RM, Moreno V, Marques H, Jurczyszyn A, Andersen V, Vogel U, Buda G, Orciuolo E, Jacobsen SE, Petrini M, Vangsted AJ, Gemignani F, Canzian F, Jurado M, Sainz J. Type 2 diabetes-related variants influence the risk of developing multiple myeloma: results from the IMMEnSE consortium. Endocr Relat Cancer. 2015;22:545-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |