Published online Feb 15, 2024. doi: 10.4239/wjd.v15.i2.260
Peer-review started: September 24, 2023
First decision: December 6, 2023
Revised: December 13, 2023
Accepted: January 15, 2024
Article in press: January 15, 2024
Published online: February 15, 2024
Processing time: 132 Days and 18.3 Hours
Podocyte apoptosis plays a vital role in proteinuria pathogenesis in diabetic nephropathy (DN). The regulatory relationship between long noncoding RNAs (lncRNAs) and podocyte apoptosis has recently become another research hot spot in the DN field.
To investigate whether lncRNA protein-disulfide isomerase-associated 3 (Pdia3) could regulate podocyte apoptosis through miR-139-3p and revealed the under
Using normal glucose or high glucose (HG)-cultured podocytes, the cellular functions and exact mechanisms underlying the regulatory effects of lncRNA Pdia3 on podocyte apoptosis and endoplasmic reticulum stress (ERS) were explored. LncRNA Pdia3 and miR-139-3p expression were measured through quantitative real-time polymerase chain reaction. Relative cell viability was de
The expression of lncRNA Pdia3 was significantly downregulated in HG-cultured podocytes. Next, lncRNA Pdia3 was involved in HG-induced podocyte apoptosis. Furthermore, the dual luciferase reporter assay confirmed the direct interaction between lncRNA Pdia3 and miR-139-3p. LncRNA Pdia3 overexpression attenuated podocyte apoptosis and ERS through miR-139-3p in HG-cultured podocytes.
Taken together, this study demonstrated that lncRNA Pdia3 overexpression could attenuate HG-induced podocyte apoptosis and ERS by acting as a competing endogenous RNA of miR-139-3p, which might provide a potential therapeutic target for DN.
Core Tip: The expression of long noncoding RNA (lncRNA) protein-disulfide isomerase-associated 3 (Pdia3) was significantly downregulated in high glucose (HG)-cultured podocytes. LncRNA Pdia3 was involved in HG-induced podocyte apoptosis. LncRNA Pdia3 overexpression attenuated HG-induced podocyte apoptosis and endoplasmic reticulum stress by acting as a competing endogenous RNA of miR-139-3p.
- Citation: He YX, Wang T, Li WX, Chen YX. Long noncoding RNA protein-disulfide isomerase-associated 3 regulated high glucose-induced podocyte apoptosis in diabetic nephropathy through targeting miR-139-3p. World J Diabetes 2024; 15(2): 260-274
- URL: https://www.wjgnet.com/1948-9358/full/v15/i2/260.htm
- DOI: https://dx.doi.org/10.4239/wjd.v15.i2.260
Diabetic nephropathy (DN) is one of the prominent and serious complications of diabetes mellitus. It is caused by changes in kidney structure and function, frequently resulting in end-stage renal disease and death[1]. Similar to many renal diseases[2,3], DN is characterized by progressive proteinuria, followed by a decline in glomerular filtration along with glomerulosclerosis, ultimately causing renal failure. Albuminuria results from renal glomerular filtration barrier disruption, which increases barrier permeability and protein leakage into the urine. Podocytes, also known as glomerular epithelial cells, are a crucial component of this barrier. Podocytes, as terminal differentiation cells, cannot regenerate when injured. Podocyte depletion and structural changes could destroy the glomerular filtration membrane and induce albuminuria[4]. Earlier studies have indicated that podocyte loss is among the main reasons for diabetes-induced proteinuria and the hallmark events of DN[5-7]. Podocyte apoptosis is an inciting event in DN development and correlates with DN progression.
Accumulating evidence has indicated that endoplasmic reticulum stress (ERS) plays a crucial role in DN development and progression[8], including podocyte injury[9,10]. Under normal physiological conditions, newly synthesized polypeptides translocate into the ER lumen to undergo proper folding, so that they meet the cellular quality control criteria for exit from the ER. Disrupted homeostasis, such as oxidative stress or high glucose (HG), causes an imbalance between the protein loading and folding capacity of the ER, resulting in unfolded and/or misfolded protein accumulation and ER dilatation. This process is known as ERS which consequently triggers an unfolded protein response (UPR)[11]. Numerous studies have indicated ERS-induced apoptosis as a critical mechanism mediating podocyte injury in
Long noncoding RNAs (lncRNAs) belong to a class of noncoding RNAs of > 200 nucleotides in length lacking protein-coding potential. Several lncRNAs are involved in many biological processes, such as regulating transcription, translation, RNA modification, protein modification, and epigenetic modification of chromatin structures[16,17]. More importantly, lncRNAs are associated with the progression and occurrence of metabolic diseases, including diabetes and diabetic complications[18]. In particular, lncRNA TCF7 silencing attenuated HG-induced podocyte damage. Therefore, lncRNAs are a potential therapeutic target for alleviating DN development to search for novel lncRNAs and alter the expression of specific lncRNAs.
We here investigated lncRNA expression profiles and the associated competing endogenous RNA (ceRNA) network using high-throughput RNA-sequencing (RNA-seq) technologies in normal glucose (5.5 mmol/L, NG group) and HG (25 mmol/L, HG group) cultured mouse podocytes. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to determine the function of differentially expressed lncRNAs. The present study investigated the function and underlying molecular mechanism of novel lncRNAs in ERS-podocyte apoptosis, providing the hope of developing a new and effective therapeutic strategy against DN.
Conditionally immortalized mouse podocytes were cultured with Dulbecco’s modified eagle medium containing 10% fetal bovine serum and antibiotics (100 U/mL of penicillin and 0.1 mg/mL of streptomycin) at 37 °C under humidified conditions of 95% air and 5% CO2. The differentiated podocytes were used for the subsequent experiments at 80%-90% podocyte confluence. The podocytes were incubated in a medium containing 25 or 5.5 mmol/L of glucose to induce a hyperglycemic or normal condition, respectively.
TRIzol® Reagent (Life Technologies) was used to extract the total RNA from the podocytes of the NG and HG, according to the manufacturer’s instructions. The RNA concentration was measured using the Qubit® RNA Assay kit in Qubit®2.0 Fluorometer (Life Technologies, CA, United States). The total RNA was purified by depleting rRNA using the Ribo-off rRNA Depletion kit (Vazyme Biotech Co., Ltd, Nanjing, China). A cDNA library was then constructed using these samples and the VAHTS™ Stranded mRNA-seq Library Prep kit for Illumina® (Vazyme Biotech Co., Ltd, Nanjing, China). Sangon Biotech (Shanghai, China) used an Illumina Novaseq6000 sequencer (Illumina Inc., San Diego, CA, United States) to sequence the libraries. Differentially expressed lncRNAs with statistical significance between the NG and HG groups were determined through P value/false discovery rate (FDR) filtering. A volcano plot filtering approach [|log2 (fold change)| ≥ 1.0; q value ≤ 0.05] was used to identify significantly and differentially expressed lncRNAs between the two groups.
The cDNA fragments were cloned into the pcDNA 3.1 plasmid vector to construct lncRNA protein-disulfide isomerase-associated 3 (Pdia3) overexpressing plasmids (pcDNA3.1-lncRNA Pdia3) to overexpress ENSMUST00000153378 (lncRNA protein-disulfide isomerase-associated 3, lncRNA Pdia3 for short). The empty vector served as a control. The podocytes were transfected with small interfering RNA (siRNA) against lncRNA Pdia3 (siRNA-lncRNA Pdia3) to inhibit lncRNA Pdia3 expression. The corresponding scrambled RNA served as a negative control. Additionally, miR-139-3p mimics or inhibitors were used to increase or decrease miR-139-3p expression, respectively. The scrambled oligonucleotides (NC mimics or NC inhibitors) served as controls. The podocytes from each group were seeded into six-well plates and incubated at 37 °C for 24 h for transfection. The cells were transfected or co-transfected with the relevant plasmids using opti-MEM and Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol after attaining 80% podocyte confluence.
Cell counting kit-8 (CCK-8) was used to measure cell viability, as described by the manufacturer. The differentiated podocytes were seeded into 96-well plates and incubated at 37 °C overnight. CCK-8 solution of 10 μL was then added to each well. The cells were incubated at 37 °C for 2 h in the dark. An automatic microplate reader was used to measure the light absorbance value of each well at 450 nm of wavelength.
The podocyte apoptosis rate in the different groups was determined through flow cytometry using an Annexin V-FITC and propidium iodide (PI) double staining kit (MultiSciences Biotechnology Corporate Limited, China) after transfection for 48 h, following the manufacturer’s instructions. The cells from each group were collected and resuspended in the binding buffer to form single-cell suspensions (1 × 106 cells/mL) for staining. The podocytes were then dual stained with 10 μL annexin-V FITC and 5 μL PI at 37 °C for 5 min to avoid light exposure. Finally, flow cytometry detected the percentage of apoptotic podocytes.
The podocytes were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde at room temperature for 30 min after transfection for 48 h. Next, the cells were permeabilized with 0.5% Triton X-100 for 5 min. The cells were blocked in 5% bovine serum albumin (BSA) for 1 h at room temperature. Subsequently, the cells were incubated with primary antibodies (podocin, 1:100, Abcam, United States; nephrin, 1:100, Abcam, United States) at 4 °C overnight. The cells were washed with PBS and incubated with fluorescence-conjugated secondary antibodies for 1 h at room temperature. The podocytes were tinted with 6-diamidino-2-phenylindole (DAPI) for 5 min and photographed under a fluorescence microscope (Olympus FV10-ASW, Tokyo, Japan).
Following 48 h of transfection, the total RNA was extracted and purified from podocytes using Trizol reagents according to the manufacturer’s instructions. A NanoDrop spectrophotometer detected the concentration and purity of total RNA. Subsequently, a reverse transcription kit (QIAGEN, Valencia, CA, United States) was used to reverse transcribe total RNA into cDNA. The expressions of RNA were then quantified by quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR®Premix Ex Taq™ (Takara, Dalian, China). The GAPDH or U6 expressions were used as the endogenous control for lncRNA Pdia3 or miR-139-3p, respectively. The relative expression of RNA was analyzed using the 2-ΔΔCt method. Primers were displayed as follows: LncRNA Pdia3 (forward primers 5’-ATGCGCTTCAGCTGCCTA-3’, reverse primers 5’-CGTCAGTTCCAACACATCG-3’); miR-139-3p (forward primers 5’-TCACAGAGGTTGTCCCGGC-3’, reverse primers 5’-TATGGTTGTTCACGACTCCTTCAC-3’); GAPDH (forward primers 5’-GCAAGTTCAACGGCACAG-3’, reverse primers 5’-CTCGCTCCTGGAAGATGG-3’); U6 (forward primers 5’-CTCGCTTCGGCAGCACA-3’, reverse primers 5’-AACGCTTCACGAATTTGCGT-3’).
Fluorescence in situ hybridization (FISH), which was performed using the FISH kit (Boster Biological Technology Co. Ltd, Wuhan, China) following the manufacturer’s protocol, was used to analyze the subcellular localization of lncRNA Pdia3. The lncRNA Pdia3 FISH probe was designed and synthesized by Servicebio Technology (Wuhan, China). After 48 h of transfection, the podocytes were fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 5 min. The podocytes were incubated with the fluorescence probe at 37 °C overnight after blocking the permeabilized podocytes in 5% BSA. The podocytes were stained with DAPI for 5 min after hybridization. Finally, the images were observed under the fluorescence microscope (Olympus FV10-ASW, Japan).
The dual luciferase reporter assay was used to assess the direct interaction between lncRNA Pdia3 and miR-139-3p. A pmirGLO luciferase expression vector (Cosmo Bio, Tianjin, China) was used to construct the reporter plasmid. The predicted lncRNA Pdia3 3’-UTR sequence that interacts with miR-139-3p and artificially mutated sequences within the predicted target sites were synthesized and cloned into the pmirGLO luciferase vector, respectively. The wide-type (wt) or mutated (mut) luciferase reporter plasmid was then transfected with miR-139-3p mimics or NC mimics into podocytes using Lipofectamine 2000 reagents following the manufacturer’s instructions. A non-related miRNA was used as NC mimics. The luciferase assay kit (Promega, Madison, WI, United States) was used to measure the luciferase activity after transfection for 48 h. The relative luciferase activity was normalized to Renilla luciferase activity.
After transfection for 48 h, the podocytes from each group were lysed by whole-cell lysate for 10 min on ice. The radioimmunoprecipitation assay lysis buffer was used to extract total proteins from the cultured podocytes, and a bicinchoninic acid kit was utilized to determine their concentration. The total proteins were then isolated through sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After blocking in 5% skim milk at room temperature for 1 h, the membranes were probed with primary antibodies [glucose-regulated protein 78 (GRP78), 1:1000, Abcam, United States; C/EBP homologous protein (CHOP), 1:500, Abcam, United States; caspase-12, 1:1000, Abcam, United States] overnight at 4 °C and then incubated with the secondary antibody for 1 h. The membranes were washed three times with PBST for 10 min, incubated in Western LightningTM Chemiluminescence Reagent (PerkinElmer, United States) for 5 min, and visualized by the LabWorksTM imaging system. β-Tubulin (1:1000, Abcam, United States) was used as an internal control. ImageJ software (National Institutes of Health, Bethesda, MD, United States) was used to analyze the gray value of the target band.
Statistical Package for the Social Sciences version 20.0 was used for data analyses. All data were presented as mean ± SD. Unpaired Student’s t-tests were used to analyze differences between the two groups. One-way analysis of variance with Student-Newman-Keuls or Dunnett’s test was used to assess differences among multiple groups. The Benjamini-Hochberg method controlled the FDR using sequential modified Bonferroni correction for multiple hypothesis testing. P values of < 0.05 were considered statistically significant.
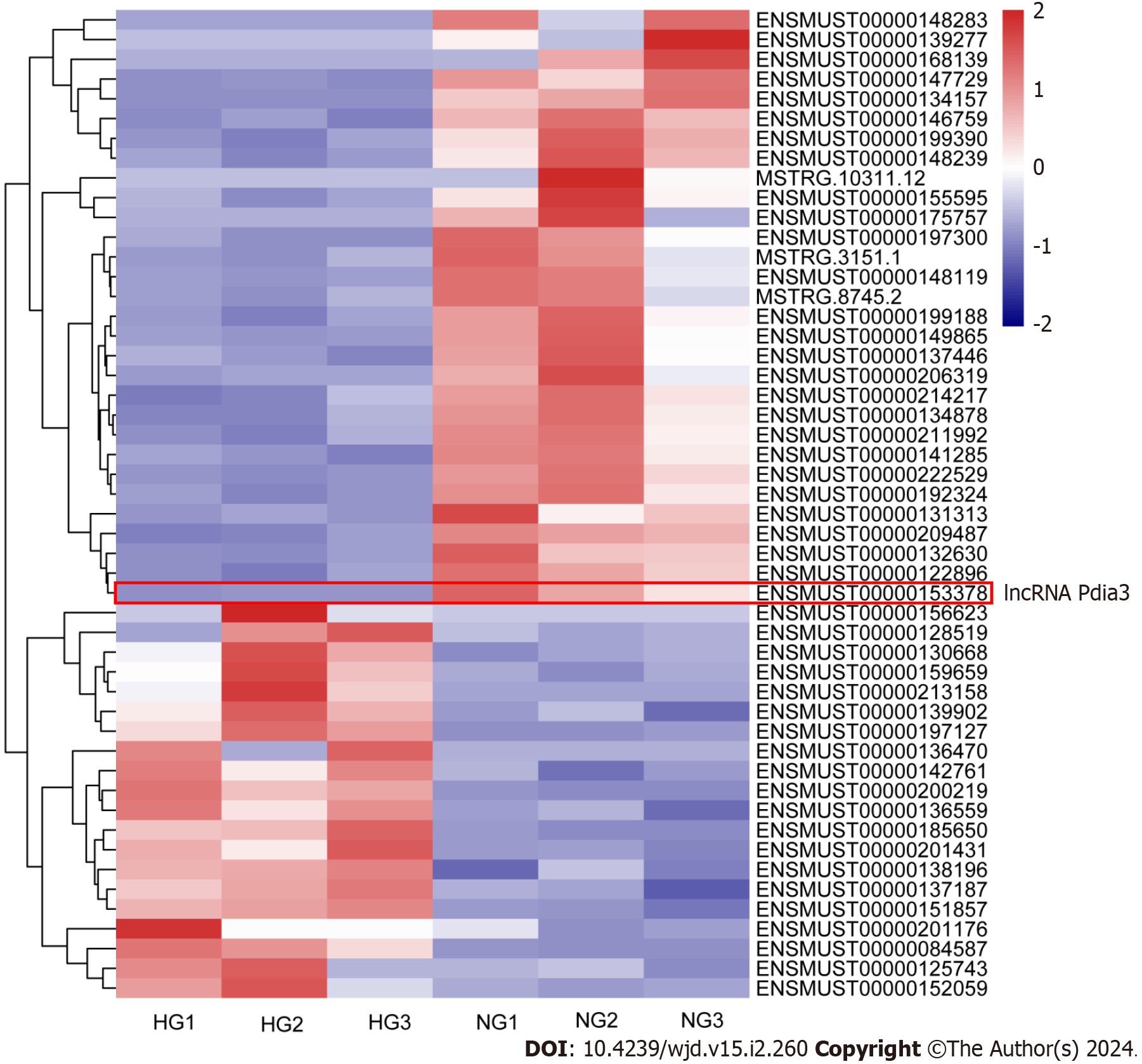
Mouse podocytes were cultured under NG or HG concentrations for 48 h. RNA-seq analysis was then performed to identify differentially expressed lncRNAs between NG and HG cultured podocytes. RNA-seq revealed that 51 lncRNAs were differentially expressed between the NG and HG groups using the following criteria: P value of < 0.001, q-value of < 0.01 and |log2 (fold change)| > 1, including 20 upregulated and 31 downregulated genes (Figure 1). Among them, lncRNA Pdia3 expression was markedly lower in the HG group than in the NG group. The GO and KEGG pathway enrichment analyses were conducted to determine the biological role of lncRNAs. Bioinformatic analysis revealed an association between lncRNA Pdia3 and ERS (Supplementary material). LncRNA Pdia3 was focused on to further study its potential function and action mechanism.
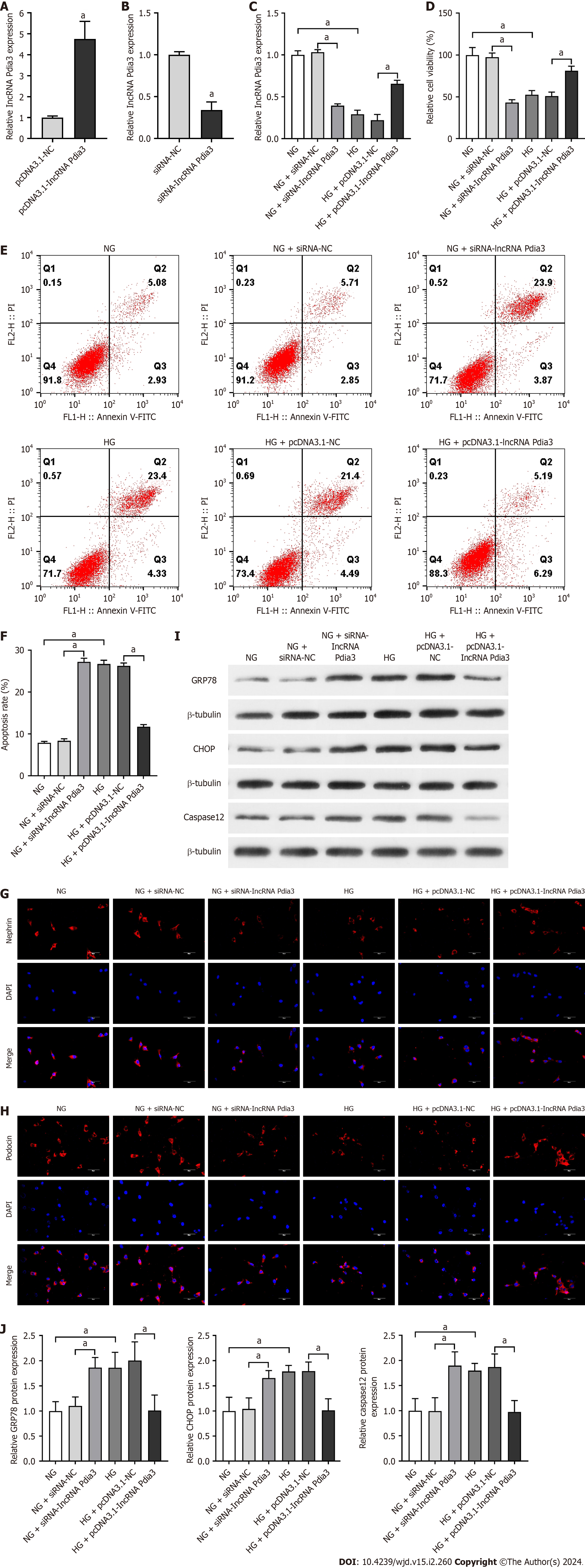
First, whether or not lncRNA Pdia3 overexpression attenuated the apoptosis of HG-cultured podocytes was evaluated. HG could reduce lncRNA Pdia3 expression according to qRT-PCR data, which was consistent with the result of RNA-seq. LncRNA Pdia3 expression significantly increased after pcDNA3.1-lncRNA Pdia3 treatment, which indicated successful transfection (Figure 2A). Following siRNA-lncRNA Pdia3 treatment, lncRNA Pdia3 expression was significantly reduced (Figure 2B), indicating the successful silencing efficiency. Compared with the NG + siRNA-NC group, qRT-PCR data indicated that the NG + siRNA-lncRNA Pdia3 group had decreased lncRNA Pdia3 expression (Figure 2C). LncRNA Pdia3 expression was significantly increased in the HG + pcDNA3.1-lncRNA Pdia3 group compared with the HG + pcDNA3.1-NC group (Figure 2C). Afterward, a CCK-8 assay revealed that siRNA-lncRNA Pdia3 transfection in NG-cultured podocytes caused a decline in cell viability. By contrast, cell viability was significantly enhanced in the HG + pcDNA3.1-lncRNA Pdia3 group compared with the HG + pcDNA3.1-NC group (Figure 2D). Furthermore, flow cytometry indicated that lncRNA Pdia3 silencing transfected by siRNA-lncRNA Pdia3 significantly increased cell apoptotic rate in NG-cultured podocytes. LncRNA Pdia3 overexpression transfected by pcDNA3.1-lncRNA Pdia3 obviously reduced cell apoptotic rate in HG-cultured podocytes (Figure 2E and F). Additionally, immunofluorescence revealed that podocin and nephrin expression were significantly decreased in the NG + siRNA-lncRNA Pdia3 group compared with the NG + siRNA-NC group. By contrast, the HG + pcDNA3.1-lncRNA Pdia3 group demonstrated greatly increased podocin and nephrin expression compared with the HG + pcDNA3.1-NC group (Figure 2G and H). These results indicated that lncRNA Pdia3 overexpression could attenuate podocyte apoptosis in HG-cultured podocytes.
We further investigated the mechanism underlying the regulatory role of lncRNA Pdia3 in podocyte apoptosis. We assessed whether lncRNA Pdia3 modulated ERS in the context of HG-induced podocyte apoptosis. Compared with the NG + siRNA-NC group, GRP78, CHOP, and caspase-12 levels significantly increased in the NG + siRNA-lncRNA Pdia3 group. After transfecting the podocytes with pcDNA3.1-lncRNA Pdia3 under the HG condition, lncRNA Pdia3 overexpression significantly reduced GRP78, CHOP, and caspase-12 levels (Figure 2I and J). These data indicated that lncRNA Pdia3 overexpression might ameliorate HG-induced ERS in podocytes.
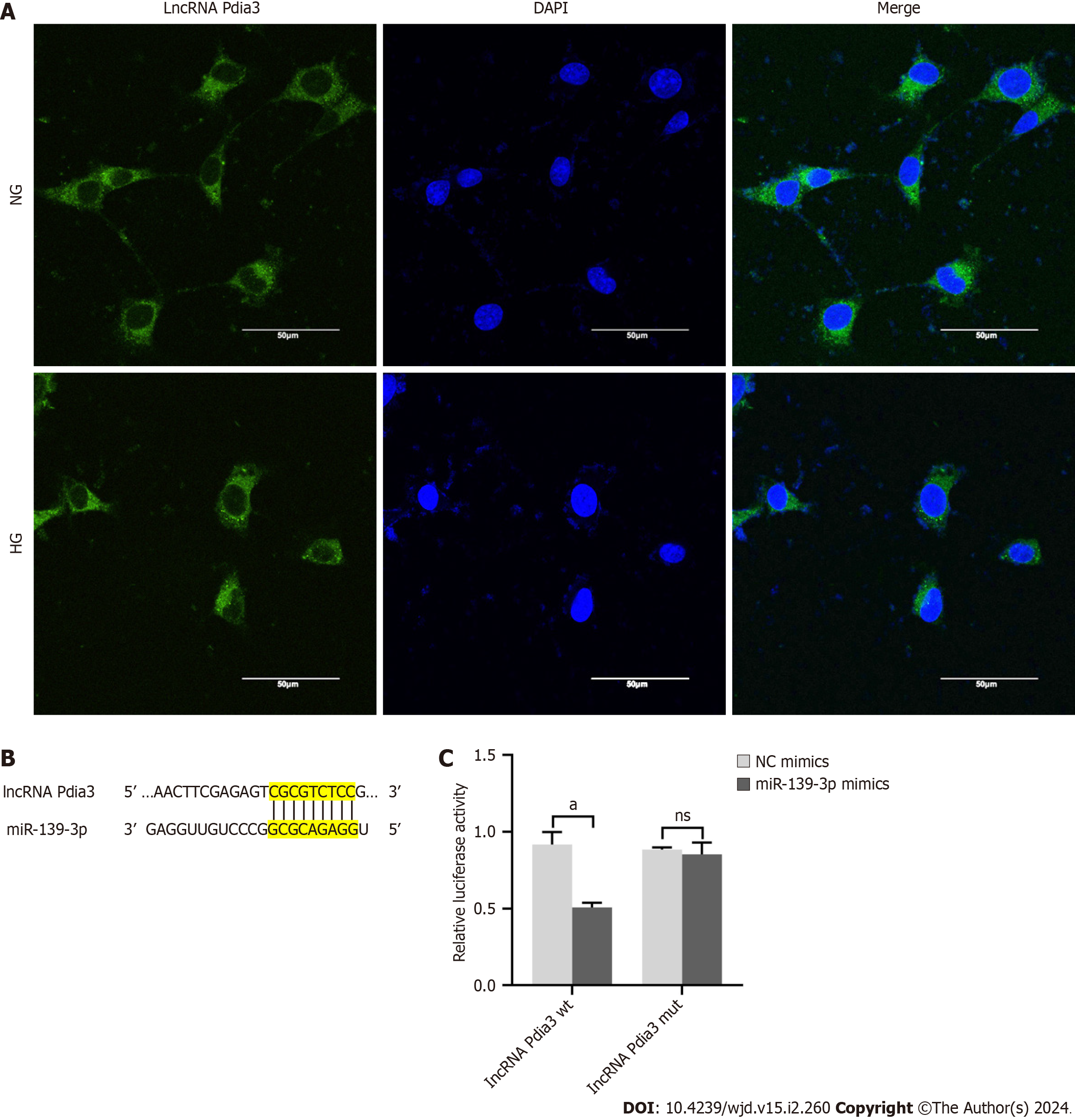
Based on the aforementioned results, we investigated how lncRNA Pdia3 regulated podocyte apoptosis. The subcellular localization of lncRNA Pdia3 was assessed using FISH under the assumption of the dependence of one lncRNA’s function on its subcellular distribution. As suggested by the subcellular fractionation results presented in Figure 3A, lncRNA Pdia3 was primarily expressed in the cytoplasm. Thus, we speculated that lncRNA alleviated HG-induced podocyte apoptosis maybe by serving as a ceRNA. Bioinformatics analysis revealed that miR-139-3p may be a possible target of lncRNA Pdia3. Figure 3B illustrated the binding sequence prediction of lncRNA Pdia3 and miR-139-3p. Moreover, the luciferase reporter assay demonstrated that lncRNA Pdia3-wt and miR-139-3p mimics co-transfection significantly decreased luciferase activity compared to lncRNA Pdia3-wt and NC mimics co-transfection. By contrast, no significant difference was observed when lncRNA Pdia3-mut was co-transfected with miR-139-3p mimics or NC mimics group (Figure 3C). Altogether, the dual luciferase reporter assay indicated that confirmed the in silico prediction of interaction between lncRNA Pdia3 and miR-139-3p.
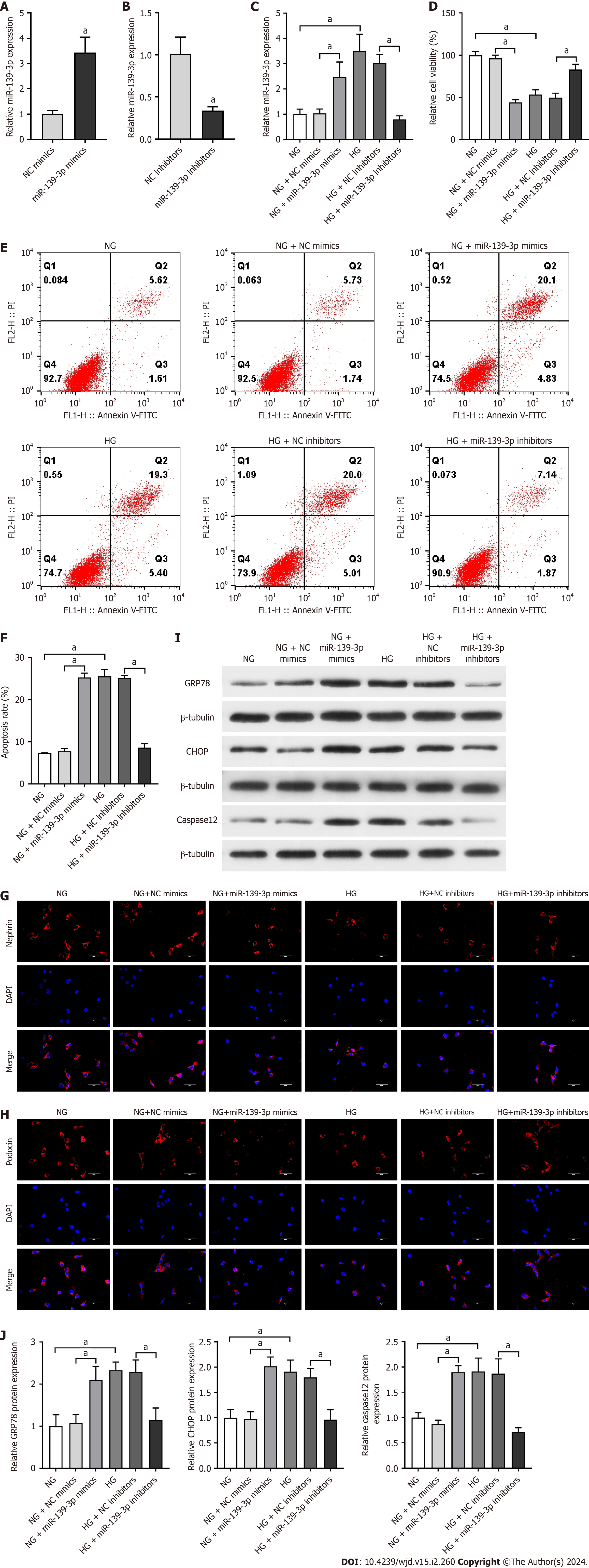
Subsequently, we investigated whether miR-139-3p participated in HG-induced podocyte apoptosis. According to the qRT-PCR data, miR-139-3p expression was significantly increased in the HG group compared with that in the NG group, which was consistent with the result of RNA-seq. The qRT-PCR results revealed the high transfection efficiency of miR-139-3p mimics (Figure 4A). After transfection with miR-139-3p inhibitors, miR-139-3p expression was significantly reduced (Figure 4B), indicating the successful silencing efficiency. The qRT-PCR data depicted that miR-139-3p expression was increased in the NG + miR-139-3p mimics group compared with the NG + NC mimics group. The miR-139-3p expression was significantly decreased by miR-139-3p inhibitors transfection in HG-cultured podocytes (Figure 4C). CCK-8 then revealed that miR-139-3p overexpression by miR-139-3p mimics significantly decreased the cell viability of the NG-cultured podocytes. Further, miR-139-3p inhibition transfected by miR-139-3p inhibitors significantly increased the cell viability of the HG-cultured podocytes (Figure 4D). Furthermore, flow cytometry indicated that miR-139-3p overexpression significantly increased the podocyte apoptosis in NG-cultured podocytes. Inhibiting miR-139-3p reduced the podocyte apoptosis in HG-cultured podocytes (Figure 4E and F). Additionalonly, immunofluorescence revealed that miR-139-3p overexpression significantly decreased podocin and nephrin expression in NG-cultured podocytes. By contrast, miR-139-3p inhibition greatly increased podocin and nephrin expression in HG-cultured podocytes (Figure 4G and H). These aforementioned results revealed the potential involvement of miR-139-3p in HG-induced podocyte apoptosis.
Furthermore, whether or not miR-139-3p regulated HG-induced ERS was determined. MiR-139-3p overexpression significantly increased GRP78, CHOP, and caspase-12 levels in NG-cultured podocytes. After transfecting the podocytes with miR-139-3p inhibitors under the HG condition, miR-139-3p inhibition significantly reduced GRP78, CHOP, and caspase-12 levels (Figure 4I and J). These aforementioned data confirmed that miR-139-3p inhibition ameliorated HG-induced ERS in podocytes.
Here, we focused on the function and underlying molecular mechanism of lncRNA Pdia3, which is a previously unidentified lncRNA, in hyperglycemia-induced podocyte apoptosis. Next, the interplay between lncRNA Pdia3 and miR-139-3p in podocytes during DN was investigated. LncRNA Pdia3 was significantly downregulated in the HG-cultured podocytes, where in HG simulated a DN microenvironment, compared with the NG-cultured podocytes. More im
LncRNA Pdia3 (Ensembl ID: ENSMUST00000153378), a 400 bp lncRNA, is located in chromosome 2 (chromosome 2: 121,244,364-121,255,082). In this study, lncRNA Pdia3 was first found to be involved in HG-induced podocyte apoptosis. In the HG-cultured podocytes, lncRNA Pdia3 expression was dramatically downregulated, which is relevant to podocyte apoptosis. Moreover, lncRNA Pdia3 overexpression with pcDNA3.1-lncRNA Pdia3 transfection significantly alleviated podocyte apoptosis in the HG-cultured podocytes. The crucial role of lncRNAs in regulating the pathological processes of podocyte apoptosis in DN, such as PVT1[19], lncRNA SPAG5 antisense RNA1 (SPAG5-AS1)[20], and lncRNA MIAT[21], has been confirmed. In our study, lncRNA Pdia3 was first proved to exert a protective effect against podocyte apoptosis.
Notably, lncRNA Pdia3 may be associated with HG-induced ERS. The ER is a key intracellular organelle with multiple functions, which is responsible for protein production, folding, processing, and secretion[22], as well as intracellular calcium storage and lipid production[23]. Newly synthesized proteins are properly folded and structurally corrected in the ER, and then transported to the Golgi apparatus. These folded proteins functioned as secretory or membrane proteins. Thus, maintaining ER homeostasis is crucial for cell survival, differentiation, development, and proliferation[24]. Acute and chronic hyperglycemia disrupts ER homeostasis, causing unfolded protein accumulation in the ER, which is known as ERS[24]. The UPR prevents misfolded protein overloading and restores ER homeostasis. ERS can result in cell death or apoptosis if the UPR system fails to restore the ER balance[25]. Emerging evidence has revealed the crucial role of ERS in regulating DN-related pathological processes[10]. The ER chaperone protein, GRP78, assists with the proper folding and assembly of proteins as a master modulator for UPR. Under ERS, GRP78 preferentially binds to misfolded or unfolded proteins and targets misfolded proteins for degradation[26]. Prolonged or intense stress has induced cell apoptosis by activating various apoptotic pathways, such as caspase-12 and CHOP. LncRNA Pdia3 silencing through siRNA-lncRNA Pdia3 transfection dramatically aggravated ERS in the NG-cultured podocytes in our study, including increasing caspase-12 and CHOP expression. In contrast, lncRNA Pdia3 overexpression through pcDNA3.1-lncRNA Pdia3 transfection dramatically alleviated ERS in the HG-cultured podocytes, including decreasing CHOP and caspase-12 expression. The results revealed that lncRNA Pdia3 overexpression ameliorated podocyte apoptosis by alleviating ERS. Several studies have extensively evaluated the relationship between lncRNAs and podocyte apoptosis, such as lncRNA 1500026
The target-mimetic, sponge/decoy function of lncRNAs on miRNAs recently gained widespread research atten
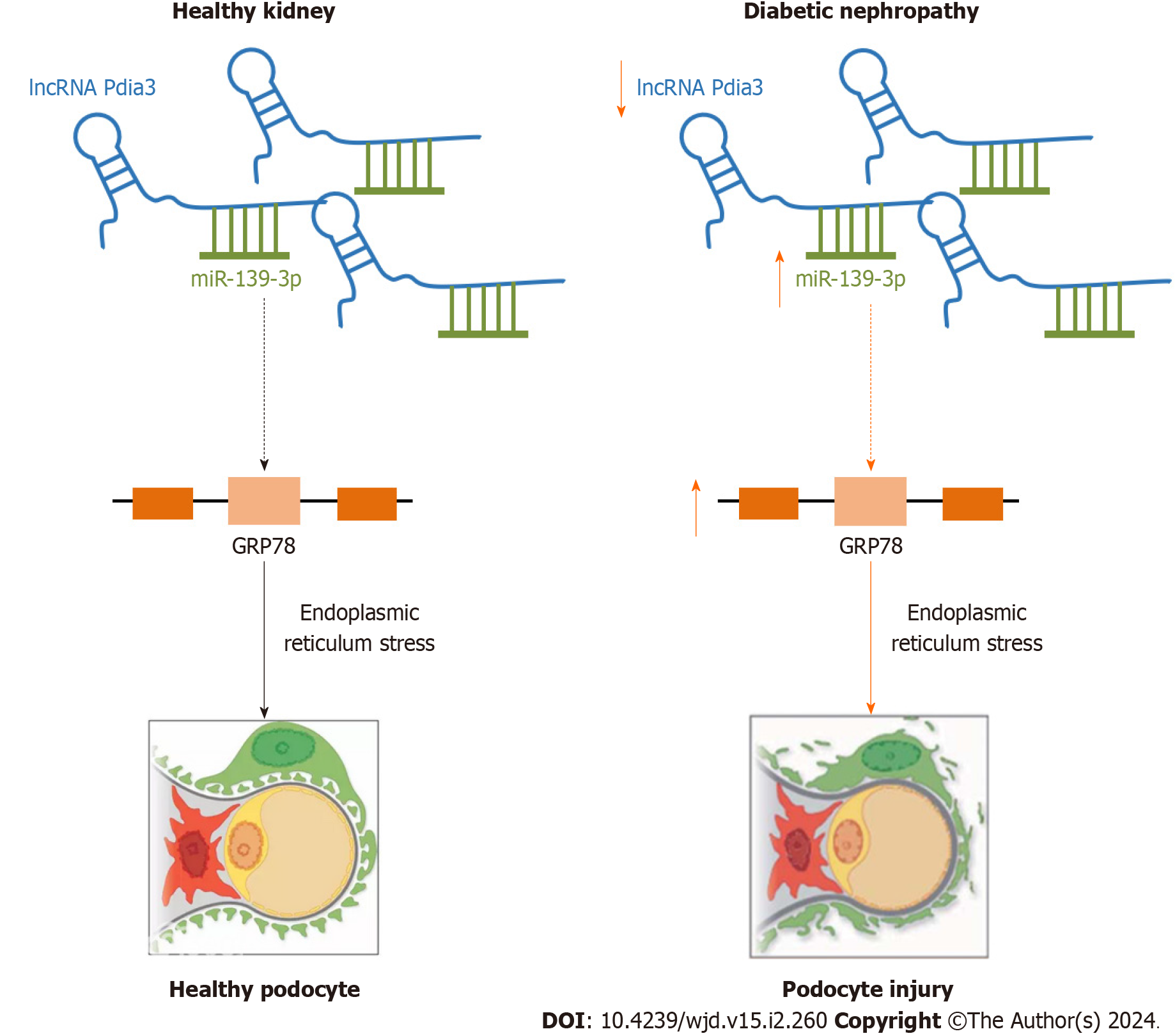
In conclusion, our study provided evidence that lncRNA Pdia3 downregulation is a significant contributing factor for podocyte apoptosis in DN. LncRNA Pdia3 downregulation could induce ERS and podocyte injury by serving as a ceRNA of miR-139-3p, thereby leading to DN progression (Figure 5). Thus, lncRNA Pdia3 played a substantial role in podocyte apoptosis in DN. So, it might act as a potential therapeutic target and offer an alternative therapy for DN. However, the current study had some limitations. Our study used glucose concentrations (25 mmol/L) to mimic hyperglycemic conditions in podocytes, which might not completely reflect the complex situation in DN patients. In addition, our experiment was conducted by using only mouse podocytes, which might restrict the generalization of the study results. According to their unique genetics, different cell types might respond differently to the same treatment. Whether the findings obtained in vitro can be applied to in vivo DN needs to be further investigated. We intend to detect the expression and underlying molecular mechanism of lncRNA Pdia3 in DN patients. Our study found that inhibition of miR-139-3p significantly reduced ERS. How does miR-139-3p act on ERS needs to be further investigated. The goal of our research is to produce knowledge that can be applied as widely as possible.
Podocyte apoptosis plays a vital role in proteinuria pathogenesis in diabetic nephropathy (DN). The regulatory relationship between long noncoding RNAs (lncRNAs) and podocyte apoptosis has recently become another research hot spot in the DN field. LncRNAs are a potential therapeutic target for alleviating DN development to search for novel lncRNAs and alter the expression of specific lncRNAs.
We here investigated lncRNA expression profiles and the associated competing endogenous RNA (ceRNA) network using high-throughput RNA-sequencing (RNA-seq) technologies in normal glucose (5.5 mmol/L, NG group) and high glucose (25 mmol/L, HG group) cultured mouse podocytes. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to determine the function of differentially expressed lncRNAs.
The present study investigated the function and underlying molecular mechanism of novel lncRNAs in endoplasmic reticulum stress (ERS)-podocyte apoptosis, providing the hope of developing a new and effective therapeutic strategy against DN.
Using NG or HG-cultured podocytes, the cellular functions and exact mechanisms underlying the regulatory effects of lncRNA protein-disulfide isomerase-associated 3 (Pdia3) on podocyte apoptosis and ERS were explored. LncRNA Pdia3 and miR-139-3p expression were measured through quantitative real-time polymerase chain reaction. Relative cell viability was detected through the cell counting kit-8 colorimetric assay. The podocyte apoptosis rate in each group was measured through flow cytometry. The interaction between lncRNA Pdia3 and miR-139-3p was examined through the dual luciferase reporter assay. Finally, western blotting was performed to detect the effect of lncRNA Pdia3 on podocyte apoptosis and ERS via miR-139-3p.
LncRNA Pdia3 was down-expressed in HG-cultured podocytes. LncRNA Pdia3 overexpression attenuated podocyte apoptosis and ERS in HG-cultured podocytes. LncRNA Pdia3 regulated podocyte apoptosis by serving as a ceRNA of miR-139-3p. Inhibition of miR-139-3p attenuated podocyte apoptosis and ERS in HG-cultured podocytes.
This study provided evidence that lncRNA Pdia3 downregulation is a significant contributing factor for podocyte apoptosis in DN. LncRNA Pdia3 downregulation could induce ERS and podocyte injury by serving as a ceRNA of miR-139-3p, thereby leading to DN progression.
In the future, whether the findings obtained in vitro can be applied to in vivo DN needs to be investigated. We intend to detect the expression and underlying molecular mechanism of lncRNA Pdia3 in DN patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sanyal D, India; Selamoglu Z, Turkey S-Editor: Wang JJ L-Editor: A P-Editor: Zheng XM
| 1. | Gnudi L, Coward RJM, Long DA. Diabetic Nephropathy: Perspective on Novel Molecular Mechanisms. Trends Endocrinol Metab. 2016;27:820-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 241] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 2. | Talas ZS, Ozdemir I, Yilmaz I, Gok Y. Antioxidative effects of novel synthetic organoselenium compound in rat lung and kidney. Ecotoxicol Environ Saf. 2009;72:916-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Talas ZS, Ozdemir I, Ciftci O, Cakir O, Gulhan MF, Pasaoglu OM. Role of propolis on biochemical parameters in kidney and heart tissues against L-NAME induced oxidative injury in rats. Clin Exp Hypertens. 2014;36:492-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Dai H, Liu Q, Liu B. Research Progress on Mechanism of Podocyte Depletion in Diabetic Nephropathy. J Diabetes Res. 2017;2017:2615286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 194] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 5. | Tagawa A, Yasuda M, Kume S, Yamahara K, Nakazawa J, Chin-Kanasaki M, Araki H, Araki S, Koya D, Asanuma K, Kim EH, Haneda M, Kajiwara N, Hayashi K, Ohashi H, Ugi S, Maegawa H, Uzu T. Impaired Podocyte Autophagy Exacerbates Proteinuria in Diabetic Nephropathy. Diabetes. 2016;65:755-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 257] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 6. | Lenoir O, Jasiek M, Hénique C, Guyonnet L, Hartleben B, Bork T, Chipont A, Flosseau K, Bensaada I, Schmitt A, Massé JM, Souyri M, Huber TB, Tharaux PL. Endothelial cell and podocyte autophagy synergistically protect from diabetes-induced glomerulosclerosis. Autophagy. 2015;11:1130-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 241] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 7. | Podgórski P, Konieczny A, Lis Ł, Witkiewicz W, Hruby Z. Glomerular podocytes in diabetic renal disease. Adv Clin Exp Med. 2019;28:1711-1715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 8. | Sankrityayan H, Oza MJ, Kulkarni YA, Mulay SR, Gaikwad AB. ER stress response mediates diabetic microvascular complications. Drug Discov Today. 2019;24:2247-2257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 9. | Lei J, Zhao L, Zhang Y, Wu Y, Liu Y. High Glucose-Induced Podocyte Injury Involves Activation of Mammalian Target of Rapamycin (mTOR)-Induced Endoplasmic Reticulum (ER) Stress. Cell Physiol Biochem. 2018;45:2431-2443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Shen H, Ming Y, Xu C, Xu Y, Zhao S, Zhang Q. Deregulation of long noncoding RNA (TUG1) contributes to excessive podocytes apoptosis by activating endoplasmic reticulum stress in the development of diabetic nephropathy. J Cell Physiol. 2019;234:15123-15133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 11. | Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21:421-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 1616] [Article Influence: 323.2] [Reference Citation Analysis (0)] |
| 12. | Cao Y, Hao Y, Li H, Liu Q, Gao F, Liu W, Duan H. Role of endoplasmic reticulum stress in apoptosis of differentiated mouse podocytes induced by high glucose. Int J Mol Med. 2014;33:809-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 13. | Li M, Ni W, Zhang M, Liu S, Chen M, Hong X, Ma Y, Yu X, Wang W, Yang M, Hua F. MicroRNA-30/Cx43 axis contributes to podocyte injury by regulating ER stress in diabetic nephropathy. Ann Transl Med. 2020;8:1674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Fan Y, Zhang J, Xiao W, Lee K, Li Z, Wen J, He L, Gui D, Xue R, Jian G, Sheng X, He JC, Wang N. Rtn1a-Mediated Endoplasmic Reticulum Stress in Podocyte Injury and Diabetic Nephropathy. Sci Rep. 2017;7:323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 15. | Zhang Y, Gao X, Chen S, Zhao M, Chen J, Liu R, Cheng S, Qi M, Wang S, Liu W. Cyclin-dependent kinase 5 contributes to endoplasmic reticulum stress induced podocyte apoptosis via promoting MEKK1 phosphorylation at Ser280 in diabetic nephropathy. Cell Signal. 2017;31:31-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Kunz M, Wolf B, Fuchs M, Christoph J, Xiao K, Thum T, Atlan D, Prokosch HU, Dandekar T. A comprehensive method protocol for annotation and integrated functional understanding of lncRNAs. Brief Bioinform. 2020;21:1391-1396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | He Y, Chen Y. The potential role of lncRNAs in osteoporosis. J Bone Miner Metab. 2021;39:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Chen Y, He Y, Zhou H. The potential role of lncRNAs in diabetes and diabetic microvascular complications. Endocr J. 2020;67:659-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Liu DW, Zhang JH, Liu FX, Wang XT, Pan SK, Jiang DK, Zhao ZH, Liu ZS. Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp Mol Med. 2019;51:1-15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 20. | Xu J, Deng Y, Wang Y, Sun X, Chen S, Fu G. SPAG5-AS1 inhibited autophagy and aggravated apoptosis of podocytes via SPAG5/AKT/mTOR pathway. Cell Prolif. 2020;53:e12738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Zhang M, Zhao S, Xu C, Shen Y, Huang J, Shen S, Li Y, Chen X. Ablation of lncRNA MIAT mitigates high glucose-stimulated inflammation and apoptosis of podocyte via miR-130a-3p/TLR4 signaling axis. Biochem Biophys Res Commun. 2020;533:429-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73:79-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 949] [Cited by in RCA: 1073] [Article Influence: 119.2] [Reference Citation Analysis (0)] |
| 23. | Sozen E, Ozer NK. Impact of high cholesterol and endoplasmic reticulum stress on metabolic diseases: An updated mini-review. Redox Biol. 2017;12:456-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 24. | Mustapha S, Mohammed M, Azemi AK, Jatau AI, Shehu A, Mustapha L, Aliyu IM, Danraka RN, Amin A, Bala AA, Ahmad WANW, Rasool AHG, Mustafa MR, Mokhtar SS. Current Status of Endoplasmic Reticulum Stress in Type II Diabetes. Molecules. 2021;26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 847] [Cited by in RCA: 1190] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 26. | Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 27. | Xia J, Sun W, Dun J. LncRNA 1500026H17Rik knockdown ameliorates high glucose-induced mouse podocyte injuries through the miR-205-5p/EGR1 pathway. Int Urol Nephrol. 2023;55:1045-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 28. | Fei B, Zhou H, He Z, Wang S. KCNQ1OT1 inhibition alleviates high glucose-induced podocyte injury by adsorbing miR-23b-3p and regulating Sema3A. Clin Exp Nephrol. 2022;26:385-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 29. | Xiao M, Bai S, Chen J, Li Y, Zhang S, Hu Z. CDKN2B-AS1 participates in high glucose-induced apoptosis and fibrosis via NOTCH2 through functioning as a miR-98-5p decoy in human podocytes and renal tubular cells. Diabetol Metab Syndr. 2021;13:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Jin J, Gong J, Zhao L, Li Y, He Q. LncRNA Hoxb3os protects podocytes from high glucose-induced cell injury through autophagy dependent on the Akt-mTOR signaling pathway. Acta Biochim Pol. 2021;68:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Long B, Wan Y, Zhang S, Lv L. LncRNA XIST protects podocyte from high glucose-induced cell injury in diabetic nephropathy by sponging miR-30 and regulating AVEN expression. Arch Physiol Biochem. 2023;129:610-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Liu H, Sun HL. LncRNA TCF7 triggered endoplasmic reticulum stress through a sponge action with miR-200c in patients with diabetic nephropathy. Eur Rev Med Pharmacol Sci. 2019;23:5912-5922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 33. | Bai X, Geng J, Li X, Wan J, Liu J, Zhou Z, Liu X. Long Noncoding RNA LINC01619 Regulates MicroRNA-27a/Forkhead Box Protein O1 and Endoplasmic Reticulum Stress-Mediated Podocyte Injury in Diabetic Nephropathy. Antioxid Redox Signal. 2018;29:355-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 34. | Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 674] [Article Influence: 74.9] [Reference Citation Analysis (0)] |
| 35. | Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 4121] [Article Influence: 374.6] [Reference Citation Analysis (1)] |
| 36. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1590] [Article Influence: 176.7] [Reference Citation Analysis (0)] |
| 37. | Zhang J, Song L, Ma Y, Yin Y, Liu X, Luo X, Sun J, Wang L. lncRNA MEG8 Upregulates miR-770-5p Through Methylation and Promotes Cell Apoptosis in Diabetic Nephropathy. Diabetes Metab Syndr Obes. 2020;13:2477-2483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 38. | Li F, Dai B, Ni X. Long non-coding RNA cancer susceptibility candidate 2 (CASC2) alleviates the high glucose-induced injury of CIHP-1 cells via regulating miR-9-5p/PPARγ axis in diabetes nephropathy. Diabetol Metab Syndr. 2020;12:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |