Published online Jun 15, 2023. doi: 10.4239/wjd.v14.i6.820
Peer-review started: December 10, 2022
First decision: December 26, 2022
Revised: December 27, 2022
Accepted: April 4, 2023
Article in press: April 4, 2023
Published online: June 15, 2023
Processing time: 186 Days and 22.5 Hours
The endoplasmic reticulum (ER) is closely related to a wide range of cellular functions and is a key component to maintain and restore metabolic health. Type 2 diabetes mellitus (T2DM) is a serious threat to human health, but the ER stress (ERS)-related mechanisms in T2DM have not been fully elucidated.
To identify potential ERS-related mechanisms and crucial biomarkers in T2DM.
We conducted gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) in myoblast and myotube form GSE166502, and obtained the differentially expressed genes (DEGs). After intersecting with ERS-related genes, we obtained ERS-related DEGs. Finally, functional analyses, immune infiltration, and several networks were established.
Through GSEA and GSVA, we identified several metabolic and immune-related pathways. We obtained 227 ERS-related DEGs and constructed several important networks that help to understand the mechanisms and treatment of T2DM. Finally, memory CD4+ T cells accounted for the largest proportion of immune cells.
This study revealed ERS-related mechanisms in T2DM, which might contribute to new ideas and insights into the mechanisms and treatment of T2DM.
Core Tip: This study revealed endoplasmic reticulum stress-related mechanisms in type 2 diabetes mellitus (T2DM), which might contribute to new ideas and insights for the mechanisms and treatment of T2DM.
- Citation: Liang B, Chen SW, Li YY, Zhang SX, Zhang Y. Comprehensive analysis of endoplasmic reticulum stress-related mechanisms in type 2 diabetes mellitus. World J Diabetes 2023; 14(6): 820-845
- URL: https://www.wjgnet.com/1948-9358/full/v14/i6/820.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i6.820
Diabetes is a chronic disease that occurs either when the pancreas does not produce enough insulin or when the body cannot effectively use the insulin it produces[1,2]. Hyperglycemia is a common effect of uncontrolled diabetes and over time leads to serious damage to many of the body’s systems, especially the heart, blood vessels, eyes, kidneys, and nerves[3,4]. Recently, the estimated prevalence of diabetes among children, adolescents, and adults has increased[5,6]. The majority of people with diabetes have type 2 diabetes mellitus (T2DM)[7]. Simple lifestyle measures have been shown to be effective in preventing or delaying the onset of T2DM[8]. Recently, with the in-depth understanding of the mechanisms of T2DM, many new drugs, such as sodium-glucose cotransporter-2 inhibitors, glucagon-like peptide-1 analogs, and dipeptidyl peptidase-4 inhibitors, have been gradually applied to clinical practice and achieved good results[9-11]. However, the residual risk of these populations remains high, especially when combined with other diseases[12].
The endoplasmic reticulum (ER) is closely related to a wide range of cellular functions and is a key component to maintain and restore metabolic health[13]. Protein handling, modification, and folding in the ER are tightly regulated processes that determine cell function, fate, and survival[14]. Many genetic and environmental damages hinder the ability of cells to correctly fold and post-translationally modify secreted and transmembrane proteins in the ER, resulting in the accumulation of misfolded proteins in this organelle, which is called ER stress (ERS)[15]. Chronic ERS is becoming a key factor in more human diseases, including T2DM[16,17]. Recently, the biological mechanisms of ERS in T2DM have been gradually explored. YIPF5 mutations can disrupt the ER-to-Golgi trafficking, thereby resulting in T2DM[16]. Inositol-requiring enzyme 1alpha upregulates miR-200a degradation and stimulates TXINP/NLRP3-pathway-mediated pyroptosis and renal damage in T2DM[18]. Mfn2 plays an important role in ERS, and Mfn2 silencing prevents mitochondrial Ca2+ overload-mediated mitochondrial dysfunction[19]. ATF5 is a regulator of ERS and β-cell apoptosis in different models of diabetes mellitus[20]. Lactogens modulate the ERS pathway, causing enhanced β-cell survival and reduced T2DM incidence[21]. The development of ERS for the treatment of T2DM has also emerged in clinical trials. A randomized placebo-controlled crossover trial indicated that decreased ERS may lead to improvement of insulin sensitivity mediated by hyperbaric oxygen[22]. Nevertheless, the role of ERS in T2DM, especially the related markers and mechanisms, is still lacking.
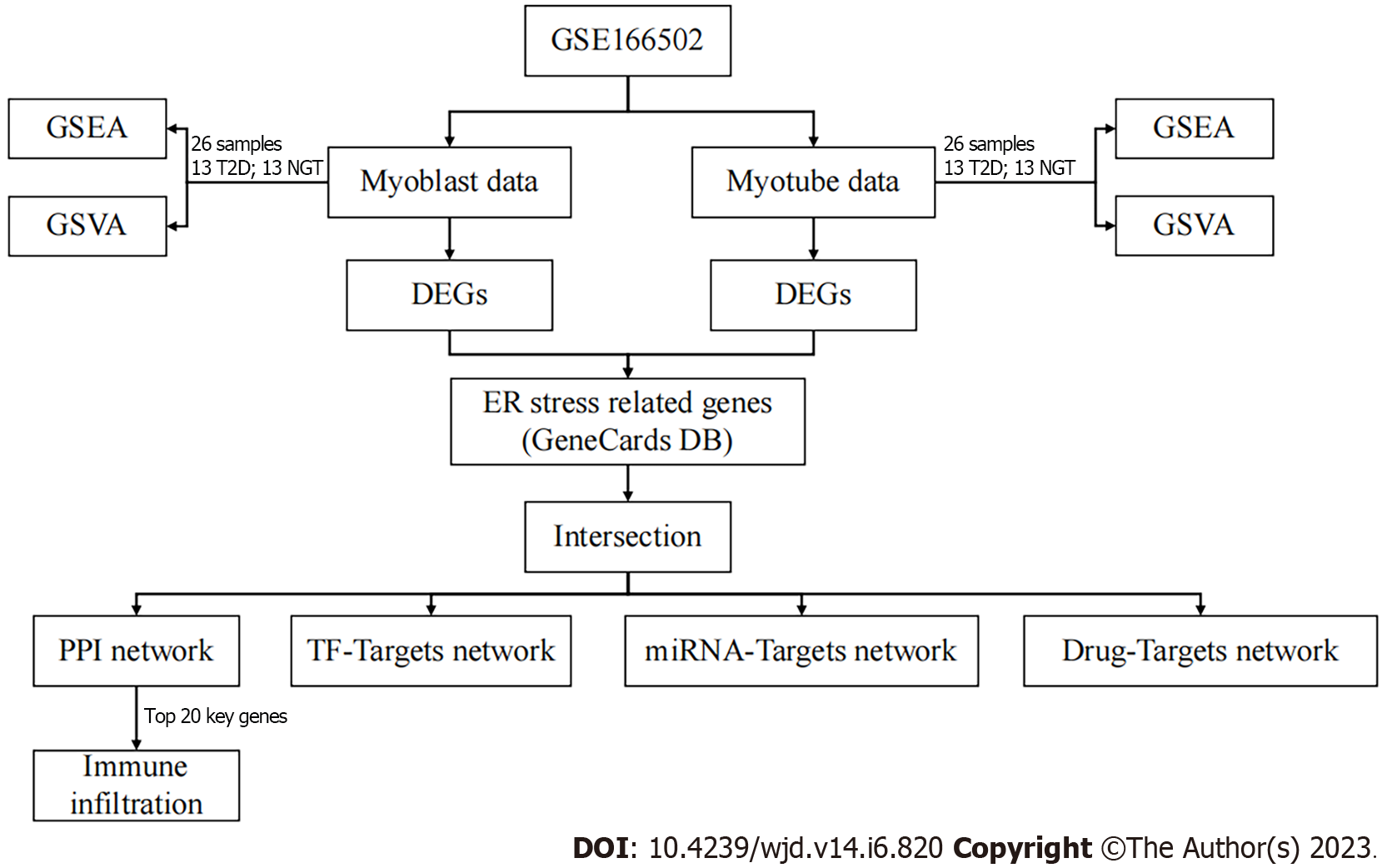
Here, we conducted gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) in both proliferating myoblasts and differentiated myotubes, which are important in T2DM. Then, the differentially expressed genes (DEGs) and ERS-related DEGs between T2DM patients and healthy populations were investigated, sequentially. Furthermore, functional enrichment analysis [Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG], immune infiltration analysis, and three networks [transcription factor (TF)–mRNA, miRNA–mRNA, and drug–mRNA] were detected to explore the mechanisms and potential therapeutic agents of ERS in T2DM. The flow chart is shown in Figure 1.
The raw data of the microarray expression dataset GSE166502[23] and its annotation file GPL10558 (Illumina HumanHT-12 V4.0 Expression BeadChip) were obtained from Gene Expression Omnibus[24]. GSE166502 holds the mRNA expression in proliferating myoblasts and differentiated myotubes in patients with T2DM (n = 13) or controls (n = 13).
We selected and downloaded c2.cp.v7.2.symbols.gmt gene set data through the GSEA database[25], and conducted GSEA on the proliferating myoblasts and differentiated myotubes through the clusterProfiler package (version 3.14.3)[26]. The statistical process of GSEA was to calculate the enrichment score, estimate the significance, and correct the multiple hypothesis tests. We also selected the same data from GSEA and conducted GSVA. The different pathways were obtained through the limma package (version 3.42.2)[27].
After the processing of raw data, we analyzed the data using the limma package with a fold change and P for DEGs. The threshold of DEGs was |log2fold change| > 0.263 and P < 0.05 as described previously, and the results were visualized as a heat map and volcano map using the pheatmap package (version 1.0.12).
GeneCards provides annotated and predicted human gene information, which integrates gene data from about 150 network sources, including genomics, transcriptomics, proteomics, genetics, and clinical and functional information[28]. In this study, ERS-related genes were downloaded through GeneCards with “endoplasmic reticulum stress” as the search keyword. Taking the intersection of DEGs and ERS-elated genes, we got the ERS-related DEGs and the Venn diagram was drawn through the Venndiagram package (version 1.6.20).
GO and KEGG pathway analysis can contribute to the interpretation of system-level data and enable discoveries[29,30]. In this work, GO terms and KEGG analysis of ERS-related DEGs and potential molecular complex were carried out using the clusterProfiler package with P < 0.05, and then visualized by the ggplot2 package (version 3.3.3), as described previously[31].
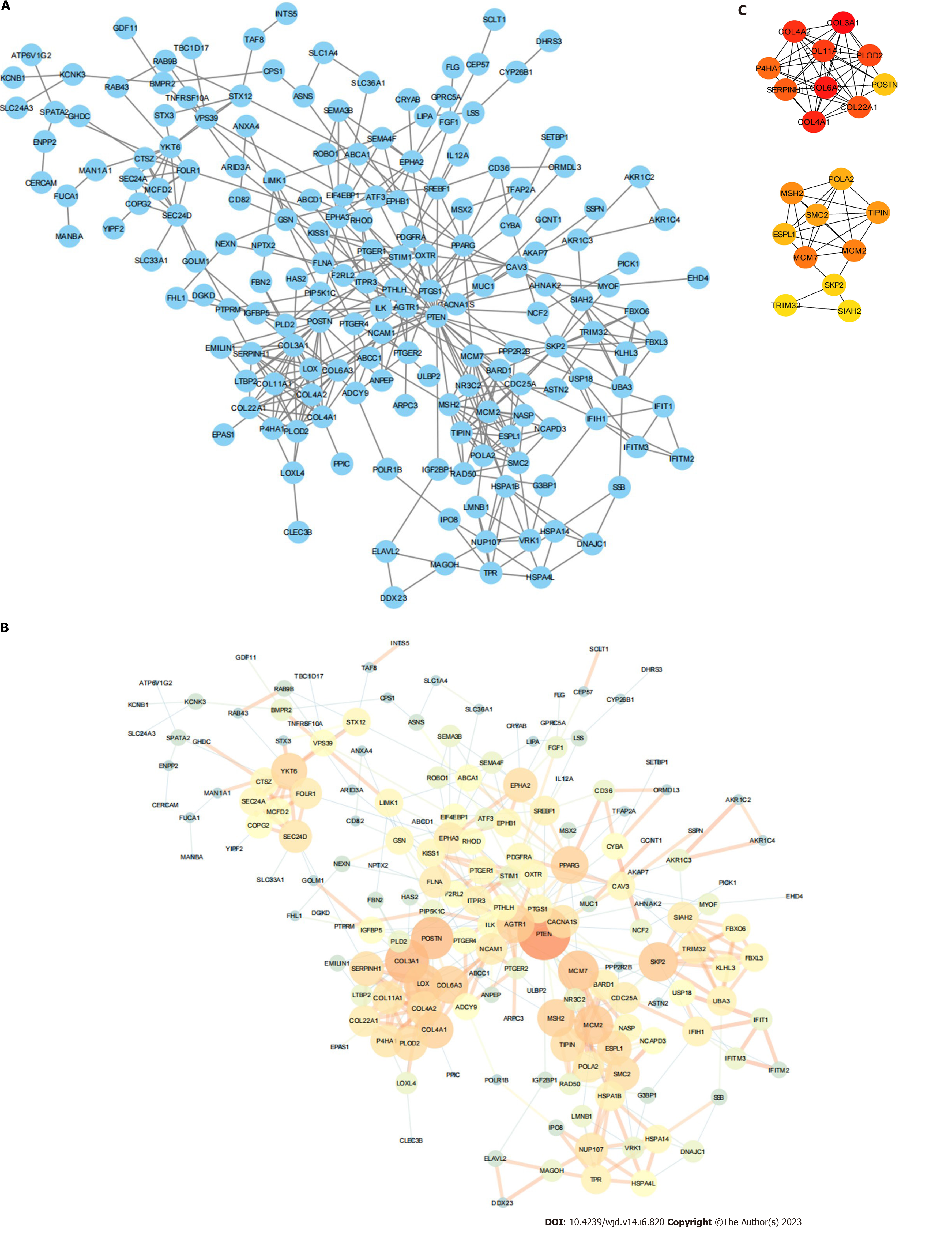
Protein–protein interaction (PPI) is one of the cores of cellular processing. The analysis of PPI makes the relationships among proteins clear and helps the function explanation of potential protein complexes or functional modules. In this work, PPI information was surveyed using the String database (version 11.0)[32]. The PPI network of ERS-related DEGs was uploaded to Cytoscape (version 3.8.2)[33] and the NetworkAnalyzer plugin was used to further processing and analysis. The cytoHubba plugin was used to select the top 20 key genes[34].
Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST, version 2) manually curated database of human and mouse transcriptional regulatory networks[35]. Current TRRUST contains 8444 and 6552 TF-target regulatory relationships of 800 human and 828 mouse TFs. TRRUST database also provides information on the mode of regulation (activation or repression). miRWalk (version 3.0) stores predicted data obtained with a machine-learning algorithm including experimentally verified miRNA–target interactions[36]. The drug–gene interaction database (DGIdb, version 4.2.0) builds drug–gene interactions mined from DrugBank, PharmGKB, Chembl, Drug Target Commons, Therapeutic Target Database, and others[37]. DGIdb contains > 40000 genes and > 10000 drugs involved in > 100000 drug–gene interactions or belonging to one of 42 potentially druggable gene categories. We obtained the TFs, miRNAs, and drugs of ERS-related DGEs, respectively, and then constructed the regulation relationship networks through Cytoscape.
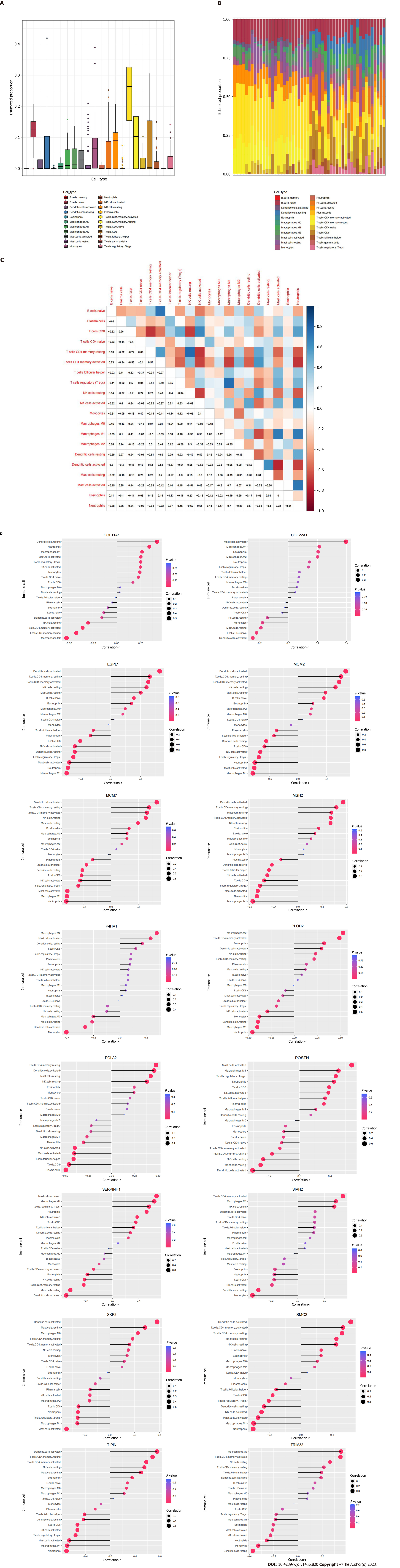
CIBERSORT (version 1.03) calculates the proportion of different types of cells according to LM22[38]. The proportion of different cell types can be calculated after the nonnegative matrix decomposition of the expression matrix. In this study, the immune infiltration of GSE166502 was analyzed by CIBERSORT, and the infiltration of 22 kinds of immune cells in the sample was analyzed. Finally, we analyzed the correlation between the expression of the top 20 key genes in the PPI network and the immune infiltration.
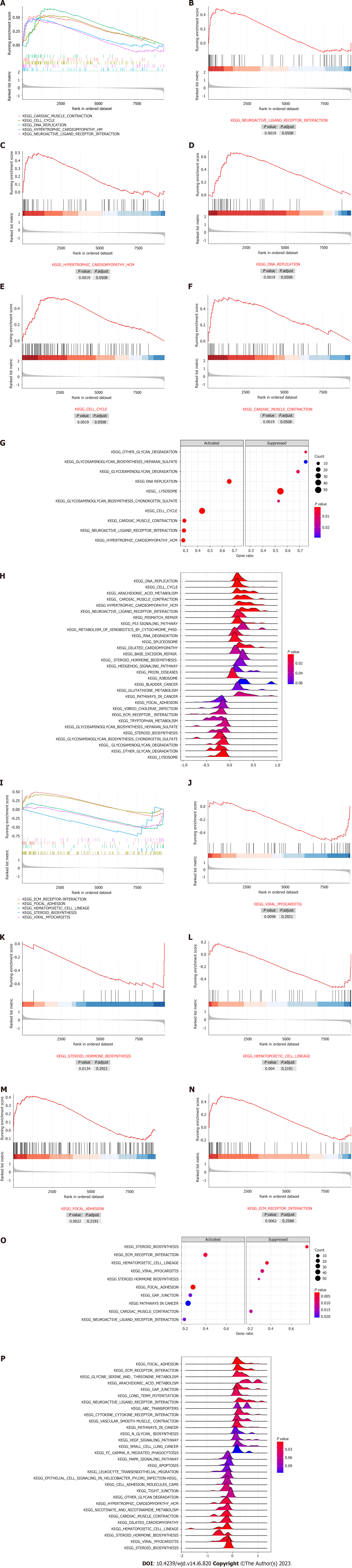
Through GSEA, we found that neuroactive ligand–receptor interaction, hypertrophic cardiomyopathy, DNA replication, cell cycle, and cardiac muscle contraction were the top five pathways in proliferating myoblasts (Figure 2A–F). DNA replication, cell cycle, cardiac muscle contraction, neuroactive ligand–receptor interaction, and hypertrophic cardiomyopathy were activated, whereas glycosaminoglycan biosynthesis heparan sulfate, glycosaminoglycan biosynthesis chondroitin sulfate, glycosaminoglycan degradation, other glycan degradation, and lysosome were suppressed (Figure 2G). Other pathways, such as arachidonic acid metabolism, mismatch repair, P53 signaling pathway, metabolism of xenobiotics by cytochrome P450, and prion diseases, were also enriched (Figure 2H). Similarly, we found that viral myocarditis, steroid hormone biosynthesis, hematopoietic cell lineage, focal adhesion, and extracellular matrix (ECM) receptor interaction were the top five pathways in differentiated myotubes (Figure 2I–N). Neuroactive ligand–receptor interaction, gap junction, pathways in cancer, focal adhesion, and ECM receptor interaction were activated, whereas steroid hormone biosynthesis, cardiac muscle contraction, viral myocarditis, hematopoietic cell lineage, and steroid biosynthesis were suppressed (Figure 2O). Vascular endothelial growth factor (VEGF) signaling pathway, cell adhesion molecules cams, mitogen-activated protein kinase (MAPK) signaling pathway, and apoptosis were also enriched (Figure 2P).
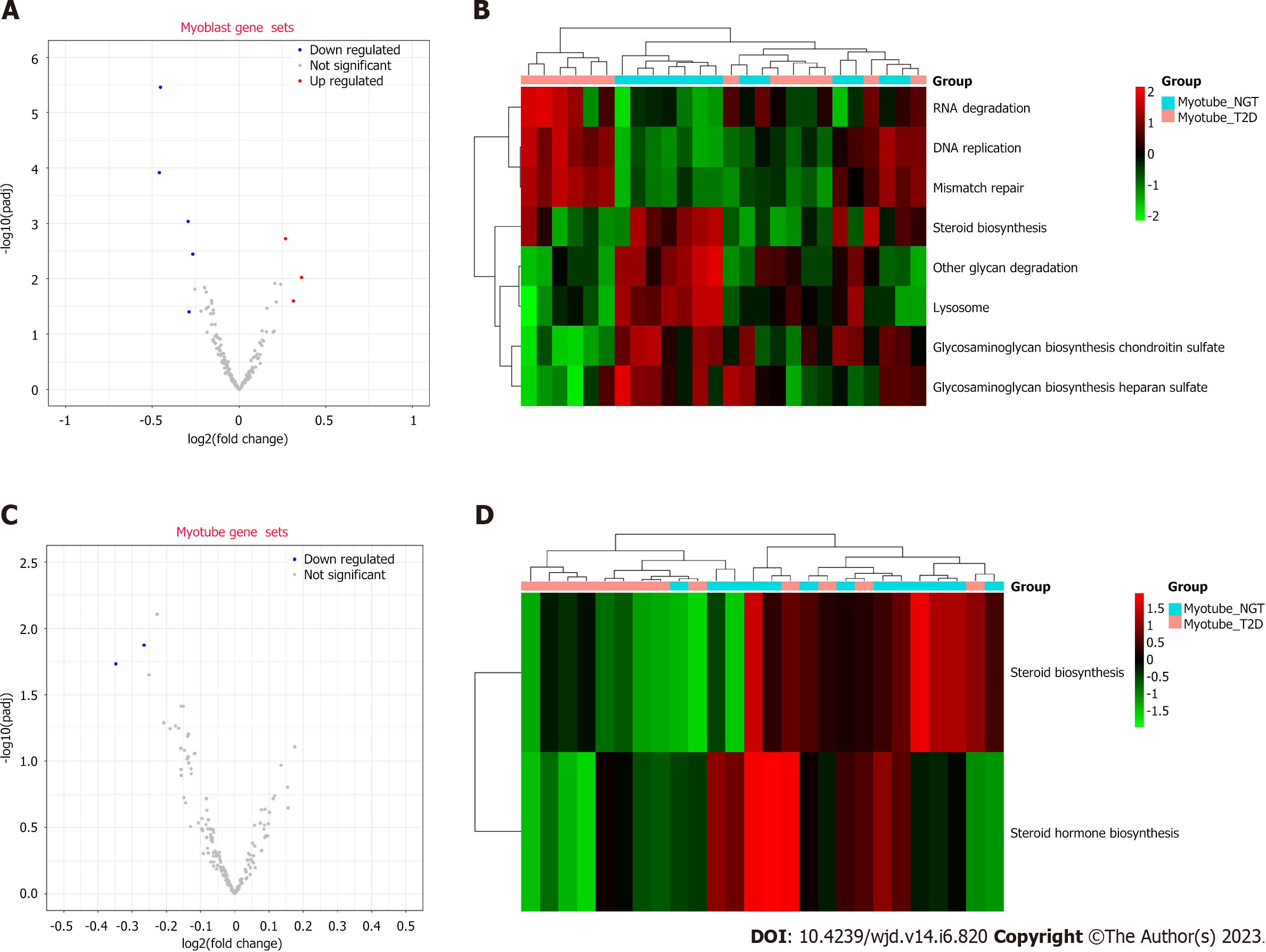
Through GSVA, eight pathways were enriched in proliferating myoblasts (Figure 3A and B). RNA degradation, DNA replication, and mismatch repair were upregulated, and glycosaminoglycan biosynthesis chondroitin sulfate, other glycan degradation, lysosome, glycosaminoglycan biosynthesis heparan sulfate, and steroid biosynthesis were downregulated. Two pathways (steroid hormone biosynthesis and steroid biosynthesis) were enriched in differentiated myotubes (Figure 3C and D).
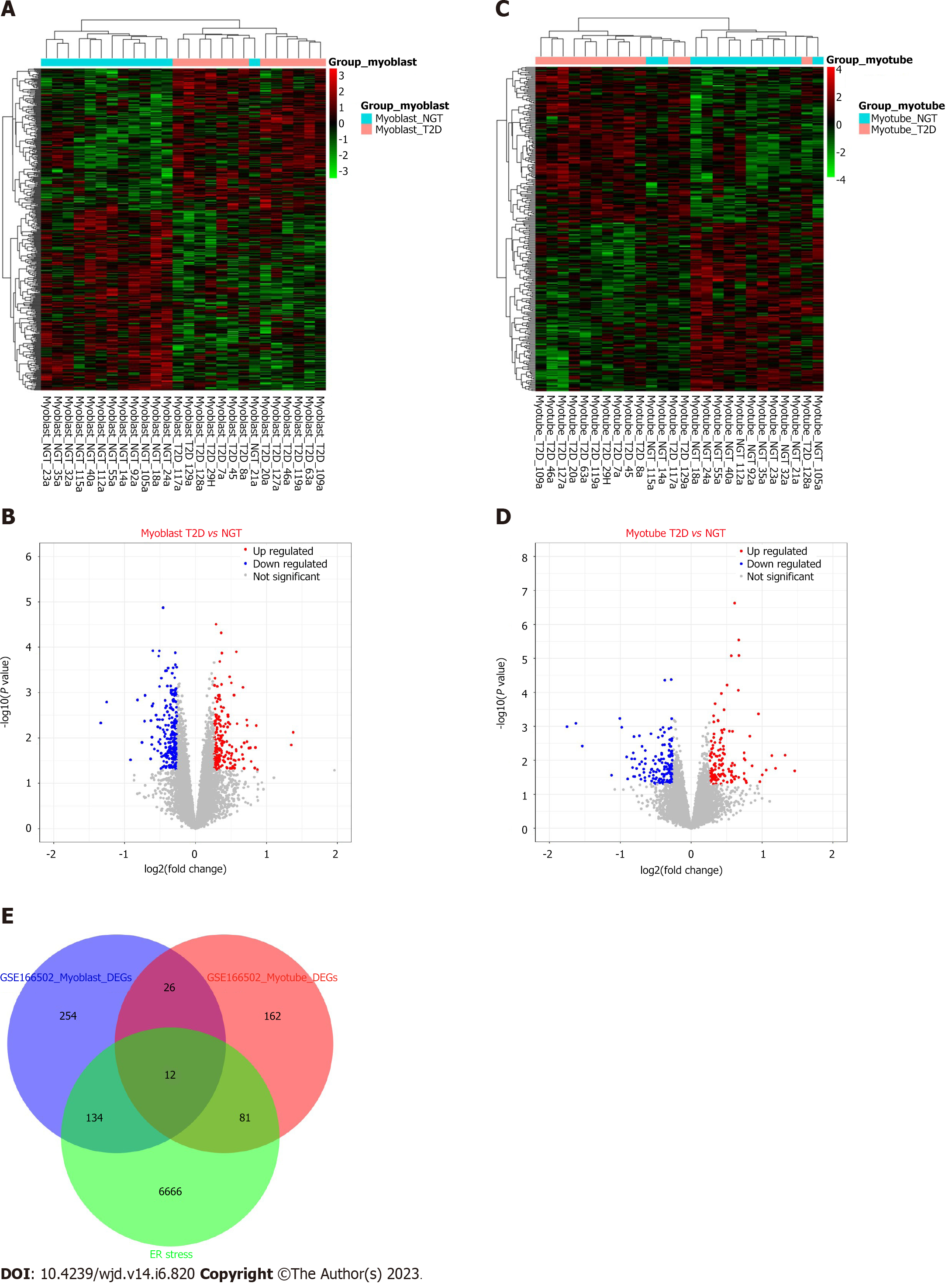
We performed DEG analysis on proliferating myoblasts and differentiated myotubes. We obtained 426 DEGs (188 upregulated and 238 downregulated, Figure 4A and B) and 281 DEGs (135 upregulated and 146 downregulated, Figure 4C and D) from proliferating myoblasts and differentiated myotubes, respectively. Through intersecting with 6893 ERS-related genes, we obtained 227 ERS-related DEGs (Figure 4E).
GO terms include biological processes, molecular functions, and cellular components. There were 227 ERS-related DEGs enriched in 875 biological process terms, 103 molecular function terms, and 81 cellular component terms. The results indicated that numerous biological processes were involved in extracellular structure organization, collagen fibril organization, ECM organization, cellular response to external stimulus, response to ketone, cellular response to fatty acid, cellular response to prostaglandin stimulus, response to fatty acid, response to mechanical stimulus, respiratory tube development, cellular response to extracellular stimulus, response to alcohol, and cell–substrate adhesion (Figure 5A). The results indicated that numerous cellular components were involved in the collagen-containing ECM, ECM component, collagen trimer, ER lumen, ER–Golgi intermediate compartment, Golgi-associated vesicle membrane, the complex of collagen trimers, membrane raft, membrane microdomain, membrane region, phagocytic vesicle, neuronal cell body, Golgi-associated vesicle, focal adhesion, cell–substrate adherens junction, cell–substrate junction, postsynaptic endosome, transport vesicle, COPII-coated ER to Golgi transport vesicle, and ER to Golgi transport vesicle membrane (Figure 5B). The results indicated that numerous molecular functions were involved in prostaglandin receptor activity, ECM structural constituent, prostanoid receptor activity, icosanoid receptor activity, growth factor binding, ECM structural constituent conferring tensile strength, platelet-derived growth factor binding, transmembrane receptor protein kinase activity, heat shock protein binding, sulfur compound transmembrane transporter activity, transmembrane receptor protein tyrosine kinase activity, transmembrane-ephrin receptor activity, oxidoreductase activity, ephrin receptor activity, virus receptor activity, and hijacked molecular function (Figure 5C). Through KEGG function enrichment analysis, 26 pathways were significant, such as axon guidance, protein digestion and absorption, focal adhesion, protein processing in the ER, cortisol synthesis and secretion, Fc gamma R-mediated phagocytosis, renin secretion, glutathione metabolism, AMPK signaling pathway, ECM–receptor interaction, DNA replication, calcium signaling pathway, thyroid cancer, aldosterone synthesis, and secretion, lipid and atherosclerosis, P53 signaling pathway, other glycan degradation, biosynthesis of amino acids, ABC transporters, phospholipase D signaling pathway, and steroid biosynthesis (Figure 5D).
The network consisted of 227 nodes and 416 edges (Figure 6A). We used the NetworkAnalyzer plugin to calculate the degree and combine the score (Figure 6B). We obtained 20 key genes (2 modules with 67 interactions) via the cytoHubba plugin (Table 1 and Figure 6C).
| Gene | Description | MCC-score | Degree | Closeness | Betweenness |
| COL3A1 | Collagen Type III Alpha 1 Chain | 11179 | 16 | 64.5 | 1189.37723 |
| COL6A3 | Collagen Type VI Alpha 3 Chain | 11168 | 12 | 62.03333 | 567.89507 |
| COL4A1 | Collagen Type IV Alpha 1 Chain | 11167 | 12 | 59.91667 | 480.86034 |
| COL4A2 | Collagen Type IV Alpha 2 Chain | 10806 | 10 | 56.23333 | 84.68666 |
| COL11A1 | Collagen Type XI Alpha 1 Chain | 10800 | 9 | 54.5 | 18.34225 |
| PLOD2 | Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 2 | 10326 | 10 | 56.88333 | 112.4573 |
| COL22A1 | Collagen Type XXII Alpha 1 Chain | 10080 | 8 | 51.80952 | 4.88333 |
| SERPINH1 | Serpin Family H Member 1 | 6001 | 10 | 60.3 | 1308.02191 |
| P4HA1 | Prolyl 4-Hydroxylase Subunit Alpha 1 | 5166 | 9 | 55.07857 | 68.74063 |
| MCM7 | Minichromosome Maintenance Complex Component 7 | 1804 | 14 | 68.24286 | 1418.45764 |
| MCM2 | Minichromosome Maintenance Complex Component 2 | 1801 | 13 | 63.94524 | 784.74991 |
| MSH2 | MutS Homolog 2 | 1645 | 12 | 62.22619 | 646.66407 |
| TIPIN | TIMELESS Interacting Protein | 1596 | 10 | 54.1631 | 37.22401 |
| SMC2 | Structural Maintenance of Chromosomes 2 | 1567 | 10 | 53.77976 | 230.10198 |
| POLA2 | DNA Polymerase Alpha 2, Accessory Subunit | 1560 | 8 | 52.6131 | 4.22778 |
| ESPL1 | Extra Spindle Pole Bodies Like 1, Separase | 962 | 10 | 56.52976 | 455.86734 |
| POSTN | Periostin | 860 | 15 | 66.51667 | 1995.77049 |
| SKP2 | S-Phase Kinase Associated Protein 2 | 771 | 13 | 64.5619 | 1861.813 |
| TRIM32 | Tripartite Motif Containing 32 | 722 | 8 | 54.52976 | 282.93712 |
| SIAH2 | Siah E3 Ubiquitin Protein Ligase 2 | 722 | 8 | 57.06905 | 982.96735 |
We obtained 27 TFs and 49 target genes from TRRUST to build the TF–mRNA network (Figure 7A). We obtained 51 miRNAs and 25 target genes from miRWalk to build the miRNA–mRNA network (Figure 7B). We also obtained 59 drugs and 22 target genes from DGIdb to build the drug–mRNA network (Figure 7C).
We demonstrated that memory CD4+ T cells accounted for the largest proportion of 22 immune cell types (Figure 8A). Figure 8B showed the distribution of different immune cells in each sample. Moreover, we evaluated the correlation between immune infiltration and each sample (Figure 8C). In 20 key genes, the enrichment degree of each immune cell was different (Figure 8D).
T2DM is a complex metabolic disease driven by interactions among diverse environmental and genetic susceptibilities[39]. Although environmental and epigenetic factors clearly play a contributory role in the pathogenesis of T2DM, genetic factors appear to be the primary contributors to the recent rise in T2DM prevalence[40]. More studies have shown that ERS is involved in T2DM[41]. In the present study, we first explored the potential pathways in proliferating myoblasts and differentiated myotubes, and obtained 227 ERS-related DEGs in T2DM, which may contribute to the occurrence and development of T2DM. Later enrichment analysis, immune infiltration, TF–mRNA network, and miRNA–mRNA network revealed the mechanisms of T2DM, which provided a way for clinical treatment of T2DM. In particular, the drug–mRNA network provided new insights and perspectives into the therapeutic reagents.
In GSEA and GSVA, we confirmed that DNA replication, cell cycle, neuroactive ligand–receptor interaction, glycosaminoglycan biosynthesis heparan sulfate, glycosaminoglycan biosynthesis chondroitin sulfate, glycosaminoglycan degradation, other glycan degradation, lysosome, arachidonic acid metabolism, mismatch repair, metabolism of xenobiotics by cytochrome P450, steroid hormone biosynthesis, focal adhesion, and ECM–receptor interaction, neuroactive ligand–receptor interaction, gap junction, steroid biosynthesis, and cell adhesion molecules were enriched. Moreover, the P53 signaling pathway, VEGF signaling pathway, MAPK signaling pathway, and apoptosis may contribute to T2DM. Previous studies have indicated that these biological processes are related to T2DM[42-44]. SRT2104 enhanced renal SIRT1 expression and activity, deacetylated P53, and activated NRF2 antioxidant signaling, providing remarkable protection against T2DM[45]. The p-ERK/p-JNK/VEGF/PKC signaling pathway may play an important role in pathological T2DM conditions[46]. TREM-2 negatively regulates p38 MAPK-mediated inflammatory response in T2DM[47]. These previous findings are consistent with our findings in this study.
We identified 227 ERS-related DEGs and later function enrichment analysis demonstrated that the enriched biological processes and pathways are highly consistent with the previous GSEA and GSVA results. The immune infiltration analysis revealed that memory CD4+ T cells accounted for the largest proportion of 22 immune cell types. T2DM patients are present with self-reactive T cells with a memory phenotype[48]. The memory CD4+ T cells develop directly from effector cells and thereby preserve features of their effector precursors are reserved[49]. Depending on the immune context, memory CD4+ T cells can contribute to immune protection, pathology, or tissue remodeling[50]. The memory CD4+ T cells could act as immunological markers for predicting change in β-cell function in T2DM[51]. TFs recognize specific DNA sequences to control chromatin and transcription, forming a complex system that guides the expression of the genome[52]. Here we obtained 27 TFs, which may contribute to T2DM. MiRNA is a class of endogenous noncoding RNA encoding 19–25 nucleotides, which is involved in the post-transcriptional regulation of genes[53]. Most of them have high sequence conservation, expression timing, and tissue specificity[54]. Recent studies have shown that miRNA is involved in a variety of regulatory pathways, we here identified 51 miRNAs to further explain the mechanisms of T2DM. Importantly, we also established a drug–mRNA network map to provide new ideas and directions for the treatment of T2DM. Immune infiltration plays an important role in the occurrence and development of T2DM[55,56]. The memory CD4+ T cells play central roles in immunity in health and disease[57]. We also explored the relationship between immune infiltration and T2DM, and we found that memory CD4+ T cells were the most numerous types of immune cells in T2DM. Previous studies indicated that CD4+ T cells contribute to the destruction of insulin-producing β-cells in type 1 diabetes mellitus[58,59], which confirmed our results.
This study has some limitations. First, all the results of the analysis were derived from previous data. Despite the efforts we have made in the present, our results still need verification experimentally and clinically. Moreover, the TF–mRNA, miRNA–mRNA, and drug–mRNA networks we built in this study provided some new ideas and insights for the mechanisms and treatment of T2DM. However, this is only the beginning, and more work is still needed in the follow-up.
This study revealed ERS-related mechanisms in T2DM, which might contribute to new ideas and insights for the mechanisms and treatment of T2DM.
The endoplasmic reticulum (ER) is closely related to a wide range of cellular functions and is a key component to maintain and restore metabolic health.
Type 2 diabetes mellitus (T2DM) is a serious threat to human health, but knowledge of the ER stress (ERS)-related mechanisms in T2DM is lacking.
Here, we conducted a bioinformatics analysis to identify potential ERS-related mechanisms and crucial biomarkers in T2DM.
We conducted gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) in myoblast and myotube form GSE166502, and obtained the differentially expressed genes (DEGs). After intersecting with ERS-related genes, we obtained ERS-related DEGs. Finally, functional analyses, immune infiltration, and several networks were established.
Through GSEA and GSVA, we identified several metabolic and immune-related pathways. We obtained 227 ERS-related DEGs and constructed several important networks that help to understand the mechanisms and treatment of T2DM. Finally, memory CD4+ T cells accounted for the largest proportion of immune cells.
This study revealed ERS-related mechanisms in T2DM.
Our study might contribute to new ideas and insights for the mechanisms and treatment of T2DM.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Adela R, India; Hassan FE, Egypt; Islam S, South Africa S-Editor: Fan JR L-Editor: A P-Editor: Chen YX
| 1. | Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1609] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 2. | O'Brien BJ, Faraoni EY, Strickland LN, Ma Z, Mota V, Mota S, Chen X, Mills T, Eltzschig HK, DelGiorno KE, Bailey-Lundberg JM. CD73-generated extracellular adenosine promotes resolution of neutrophil-mediated tissue injury and restrains metaplasia in pancreatitis. FASEB J. 2023;37:e22684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 3. | International Hypoglycaemia Study Group. Hypoglycaemia, cardiovascular disease, and mortality in diabetes: epidemiology, pathogenesis, and management. Lancet Diabetes Endocrinol. 2019;7:385-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 332] [Article Influence: 55.3] [Reference Citation Analysis (1)] |
| 4. | Hahr AJ, Molitch ME. Management of Diabetes Mellitus in Patients With CKD: Core Curriculum 2022. Am J Kidney Dis. 2022;79:728-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 5. | Wang L, Peng W, Zhao Z, Zhang M, Shi Z, Song Z, Zhang X, Li C, Huang Z, Sun X, Wang L, Zhou M, Wu J, Wang Y. Prevalence and Treatment of Diabetes in China, 2013-2018. JAMA. 2021;326:2498-2506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 558] [Article Influence: 139.5] [Reference Citation Analysis (0)] |
| 6. | Lawrence JM, Divers J, Isom S, Saydah S, Imperatore G, Pihoker C, Marcovina SM, Mayer-Davis EJ, Hamman RF, Dolan L, Dabelea D, Pettitt DJ, Liese AD; SEARCH for Diabetes in Youth Study Group. Trends in Prevalence of Type 1 and Type 2 Diabetes in Children and Adolescents in the US, 2001-2017. JAMA. 2021;326:717-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 370] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 7. | Stefan N, Cusi K. A global view of the interplay between non-alcoholic fatty liver disease and diabetes. Lancet Diabetes Endocrinol. 2022;10:284-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 349] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 8. | Einarson TR, Acs A, Ludwig C, Panton UH. Prevalence of cardiovascular disease in type 2 diabetes: a systematic literature review of scientific evidence from across the world in 2007-2017. Cardiovasc Diabetol. 2018;17:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1295] [Cited by in RCA: 1339] [Article Influence: 191.3] [Reference Citation Analysis (2)] |
| 9. | Liang B, Zhao YX, Zhang XX, Liao HL, Gu N. Reappraisal on pharmacological and mechanical treatments of heart failure. Cardiovasc Diabetol. 2020;19:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Wu CY, Shapiro L, Ouk M, MacIntosh BJ, Black SE, Shah BR, Swardfager W. Glucose-lowering drugs, cognition, and dementia: The clinical evidence. Neurosci Biobehav Rev. 2022;137:104654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 11. | Søndergaard CS, Esquivel PN, Dalamaga M, Magkos F. Use of Antihyperglycemic Drugs and Risk of Cancer in Patients with Diabetes. Curr Oncol Rep. 2023;25:29-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 12. | Ling J, Ng JKC, Chan JCN, Chow E. Use of Continuous Glucose Monitoring in the Assessment and Management of Patients With Diabetes and Chronic Kidney Disease. Front Endocrinol (Lausanne). 2022;13:869899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 13. | Lemmer IL, Willemsen N, Hilal N, Bartelt A. A guide to understanding endoplasmic reticulum stress in metabolic disorders. Mol Metab. 2021;47:101169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 192] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 14. | Chen X, Cubillos-Ruiz JR. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat Rev Cancer. 2021;21:71-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 798] [Article Influence: 199.5] [Reference Citation Analysis (0)] |
| 15. | Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015;10:173-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 1146] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 16. | De Franco E, Lytrivi M, Ibrahim H, Montaser H, Wakeling MN, Fantuzzi F, Patel K, Demarez C, Cai Y, Igoillo-Esteve M, Cosentino C, Lithovius V, Vihinen H, Jokitalo E, Laver TW, Johnson MB, Sawatani T, Shakeri H, Pachera N, Haliloglu B, Ozbek MN, Unal E, Yıldırım R, Godbole T, Yildiz M, Aydin B, Bilheu A, Suzuki I, Flanagan SE, Vanderhaeghen P, Senée V, Julier C, Marchetti P, Eizirik DL, Ellard S, Saarimäki-Vire J, Otonkoski T, Cnop M, Hattersley AT. YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J Clin Invest. 2020;130:6338-6353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 17. | Sahin E, Saglam N, Erdem S, Alvuroglu E, Abidin I, Yulug E, Alver A. 7,8-Dihydroxyflavone alleviates Endoplasmic Reticulum Stress in cafeteria diet-induced metabolic syndrome. Life Sci. 2022;306:120781. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 18. | Ke R, Wang Y, Hong S, Xiao L. Endoplasmic reticulum stress related factor IRE1α regulates TXNIP/NLRP3-mediated pyroptosis in diabetic nephropathy. Exp Cell Res. 2020;396:112293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 19. | Yuan M, Gong M, Zhang Z, Meng L, Tse G, Zhao Y, Bao Q, Zhang Y, Yuan M, Liu X, Li G, Liu T. Hyperglycemia Induces Endoplasmic Reticulum Stress in Atrial Cardiomyocytes, and Mitofusin-2 Downregulation Prevents Mitochondrial Dysfunction and Subsequent Cell Death. Oxid Med Cell Longev. 2020;2020:6569728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 20. | Ma J, Liu Y, Valladolid-Acebes I, Recio-López P, Peng G, Li J, Berggren PO, Juntti-Berggren L, Tong N. ATF5 is a regulator of ER stress and β-cell apoptosis in different mouse models of genetic- and diet-induced obesity and diabetes mellitus. Cell Signal. 2023;102:110535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 21. | Li R, Kondegowda NG, Filipowska J, Hampton RF, Leblanc S, Garcia-Ocana A, Vasavada RC. Lactogens Reduce Endoplasmic Reticulum Stress-Induced Rodent and Human β-Cell Death and Diabetes Incidence in Akita Mice. Diabetes. 2020;69:1463-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Sarabhai T, Mastrototaro L, Kahl S, Bönhof GJ, Jonuscheit M, Bobrov P, Katsuyama H, Guthoff R, Wolkersdorfer M, Herder C, Meuth SG, Dreyer S, Roden M. Hyperbaric oxygen rapidly improves tissue-specific insulin sensitivity and mitochondrial capacity in humans with type 2 diabetes: a randomised placebo-controlled crossover trial. Diabetologia. 2023;66:57-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 23. | Davegårdh C, Säll J, Benrick A, Broholm C, Volkov P, Perfilyev A, Henriksen TI, Wu Y, Hjort L, Brøns C, Hansson O, Pedersen M, Würthner JU, Pfeffer K, Nilsson E, Vaag A, Stener-Victorin E, Pircs K, Scheele C, Ling C. VPS39-deficiency observed in type 2 diabetes impairs muscle stem cell differentiation via altered autophagy and epigenetics. Nat Commun. 2021;12:2431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 24. | Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8769] [Cited by in RCA: 9703] [Article Influence: 421.9] [Reference Citation Analysis (0)] |
| 25. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37415] [Article Influence: 1870.8] [Reference Citation Analysis (0)] |
| 26. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22157] [Article Influence: 1704.4] [Reference Citation Analysis (0)] |
| 27. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16184] [Cited by in RCA: 25645] [Article Influence: 2564.5] [Reference Citation Analysis (0)] |
| 28. | Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1-1.30.33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1289] [Cited by in RCA: 2887] [Article Influence: 320.8] [Reference Citation Analysis (0)] |
| 29. | The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330-D338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2608] [Cited by in RCA: 3028] [Article Influence: 504.7] [Reference Citation Analysis (0)] |
| 30. | Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18868] [Cited by in RCA: 24696] [Article Influence: 987.8] [Reference Citation Analysis (0)] |
| 31. | Zhou JG, Liang B, Jin SH, Liao HL, Du GB, Cheng L, Ma H, Gaipl US. Development and Validation of an RNA-Seq-Based Prognostic Signature in Neuroblastoma. Front Oncol. 2019;9:1361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11733] [Article Influence: 1955.5] [Reference Citation Analysis (1)] |
| 33. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33512] [Article Influence: 1595.8] [Reference Citation Analysis (0)] |
| 34. | Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1658] [Cited by in RCA: 3751] [Article Influence: 341.0] [Reference Citation Analysis (0)] |
| 35. | Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380-D386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1155] [Cited by in RCA: 1295] [Article Influence: 185.0] [Reference Citation Analysis (1)] |
| 36. | Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;13:e0206239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 597] [Cited by in RCA: 1187] [Article Influence: 169.6] [Reference Citation Analysis (0)] |
| 37. | Freshour SL, Kiwala S, Cotto KC, Coffman AC, McMichael JF, Song JJ, Griffith M, Griffith OL, Wagner AH. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144-D1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 436] [Cited by in RCA: 617] [Article Influence: 154.3] [Reference Citation Analysis (0)] |
| 38. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8891] [Article Influence: 889.1] [Reference Citation Analysis (0)] |
| 39. | Smushkin G, Vella A. Genetics of type 2 diabetes. Curr Opin Clin Nutr Metab Care. 2010;13:471-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 40. | Franks PW, McCarthy MI. Exposing the exposures responsible for type 2 diabetes and obesity. Science. 2016;354:69-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 41. | Li W, Li W, Leng Y, Xiong Y, Xia Z. Ferroptosis Is Involved in Diabetes Myocardial Ischemia/Reperfusion Injury Through Endoplasmic Reticulum Stress. DNA Cell Biol. 2020;39:210-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 283] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 42. | Koh JH, Johnson ML, Dasari S, LeBrasseur NK, Vuckovic I, Henderson GC, Cooper SA, Manjunatha S, Ruegsegger GN, Shulman GI, Lanza IR, Nair KS. TFAM Enhances Fat Oxidation and Attenuates High-Fat Diet-Induced Insulin Resistance in Skeletal Muscle. Diabetes. 2019;68:1552-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 43. | Kehm R, Jähnert M, Deubel S, Flore T, König J, Jung T, Stadion M, Jonas W, Schürmann A, Grune T, Höhn A. Redox homeostasis and cell cycle activation mediate beta-cell mass expansion in aged, diabetes-prone mice under metabolic stress conditions: Role of thioredoxin-interacting protein (TXNIP). Redox Biol. 2020;37:101748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 44. | Liu P, Pu J, Zhang J, Chen Z, Wei K, Shi L. Bioinformatic analysis of miR-4792 regulates Radix Tetrastigma hemsleyani flavone to inhibit proliferation, invasion, and induce apoptosis of A549 cells. Onco Targets Ther. 2019;12:1401-1412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Ma F, Wu J, Jiang Z, Huang W, Jia Y, Sun W, Wu H. P53/NRF2 mediates SIRT1's protective effect on diabetic nephropathy. Biochim Biophys Acta Mol Cell Res. 2019;1866:1272-1281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 46. | Choi T, Lee JW, Kim SK, Yoo KH. Diabetes Mellitus Promotes Smooth Muscle Cell Proliferation in Mouse Ureteral Tissue through the P-ERK/P-JNK/VEGF/PKC Signaling Pathway. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 47. | Zhang J, Liu Y, Zheng Y, Luo Y, Du Y, Zhao Y, Guan J, Zhang X, Fu J. TREM-2-p38 MAPK signaling regulates neuroinflammation during chronic cerebral hypoperfusion combined with diabetes mellitus. J Neuroinflammation. 2020;17:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 48. | Ehlers MR, Rigby MR. Targeting memory T cells in type 1 diabetes. Curr Diab Rep. 2015;15:84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 49. | Soon MS, Engel JA, Lee HJ, Haque A. Development of circulating CD4(+) T-cell memory. Immunol Cell Biol. 2019;97:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 50. | Schreiner D, King CG. CD4+ Memory T Cells at Home in the Tissue: Mechanisms for Health and Disease. Front Immunol. 2018;9:2394. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 51. | Eichmann M, Baptista R, Ellis RJ, Heck S, Peakman M, Beam CA. Costimulation Blockade Disrupts CD4(+) T Cell Memory Pathways and Uncouples Their Link to Decline in β-Cell Function in Type 1 Diabetes. J Immunol. 2020;204:3129-3138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. The Human Transcription Factors. Cell. 2018;172:650-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1134] [Cited by in RCA: 1944] [Article Influence: 324.0] [Reference Citation Analysis (0)] |
| 53. | Liang B, He X, Zhao YX, Zhang XX, Gu N. Advances in Exosomes Derived from Different Cell Sources and Cardiovascular Diseases. Biomed Res Int. 2020;2020:7298687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 54. | Mead B, Tomarev S. The role of miRNA in retinal ganglion cell health and disease. Neural Regen Res. 2022;17:516-522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 55. | Pearson JA, Wong FS, Wen L. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J Autoimmun. 2016;66:76-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 203] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 56. | Guzik TJ, Cosentino F. Epigenetics and Immunometabolism in Diabetes and Aging. Antioxid Redox Signal. 2018;29:257-274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 57. | Raphael I, Joern RR, Forsthuber TG. Memory CD4(+) T Cells in Immunity and Autoimmune Diseases. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 58. | Oling V, Reijonen H, Simell O, Knip M, Ilonen J. Autoantigen-specific memory CD4+ T cells are prevalent early in progression to Type 1 diabetes. Cell Immunol. 2012;273:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 59. | Spanier JA, Sahli NL, Wilson JC, Martinov T, Dileepan T, Burrack AL, Finger EB, Blazar BR, Michels AW, Moran A, Jenkins MK, Fife BT. Increased Effector Memory Insulin-Specific CD4(+) T Cells Correlate With Insulin Autoantibodies in Patients With Recent-Onset Type 1 Diabetes. Diabetes. 2017;66:3051-3060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |