Published online Aug 15, 2021. doi: 10.4239/wjd.v12.i8.1164
Peer-review started: February 8, 2021
First decision: April 20, 2021
Revised: May 20, 2021
Accepted: July 7, 2021
Article in press: July 7, 2021
Published online: August 15, 2021
Processing time: 182 Days and 1.3 Hours
Gestational diabetes mellitus (GDM) is one of the most common metabolic disorders of pregnancy and can cause short- and long-term adverse effects in both pregnant women and their offspring. However, the etiology and pathogenesis of GDM are still unclear. As a metabolic disease, GDM is well suited to meta
Core Tip: Gestational diabetes mellitus (GDM) is one of the most common metabolic disorders of pregnancy. As a metabolic disease, GDM is well suited to metabolomics study, which can monitor the changes in small molecular metabolites induced by maternal stimuli or perturbation in real time. This review provides comprehensive documentation of metabolomics research methods and techniques as well as the current progress in GDM research. We anticipate that the review will contribute to identifying gaps in the current knowledge or metabolomics technology, provide evidence-based information, and inform future research directions in GDM.
- Citation: Wang QY, You LH, Xiang LL, Zhu YT, Zeng Y. Current progress in metabolomics of gestational diabetes mellitus. World J Diabetes 2021; 12(8): 1164-1186
- URL: https://www.wjgnet.com/1948-9358/full/v12/i8/1164.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i8.1164
Gestational diabetes mellitus (GDM) is one of the most common metabolic disorders of pregnancy and is defined as any degree of carbohydrate intolerance that occurs or is first recognized during pregnancy[1]. The prevalence of GDM has been rising worldwide, including in China, in the past few decades[2] and its prevalence ranges from 9.3% to 25.5% worldwide[3]. GDM can cause short-term and long-term adverse effects in pregnant women, including an increased risk of macrosomia, preterm delivery, preeclampsia, cesarean section, neonatal hypoglycemia, hyperbilirubinemia, respiratory distress syndrome, type 2 diabetes (T2D), hypertension, metabolic di
Systems biology inaugurated a new era of biological and biomedical research, which is different from the traditional one-gene-at-a-time approach, which discovers detailed molecular functions of individual genes or proteins[13]. Systems biology includes genomics, transcriptomics, proteomics, and metabolomics to comprehensively research the changes in biological systems under physiological and pathological conditions[14]. Among them, the concept of metabolomics was first put forward by Nicholson et al[15] in 1999. Metabolomics is the qualitative or quantitative analysis of a large number of small molecular metabolites (MW < 1000 Da) that are intermediate or final products of all the metabolic pathways in a living organism[16]. Therefore, the biggest difference between metabolomics and other ‘omics’ disciplines is that it directly reflects the activities that are occurring or have already taken place in the body, while genomics, transcriptomics, and proteomics explain the possible activities and processes in the body.
Metabolomics has the following characteristics: (1) Small changes in gene and protein expression will be magnified in metabolites, which makes the detection much easier; (2) Nonfunctional changes in genes and proteins will not be reflected in metabolites, so they play a “noise filtering” role in the process of transmitting information from upstream to downstream; (3) The number of metabolite varieties is far less than that of genes and proteins, and the molecular structure is much simpler; and (4) The structure of metabolites is similar in various organisms, so the study technology used in metabolomics is in common[17]. In the past decade, metabolomics research has resulted in rapid progress in various fields of life science, such as detecting biomarkers, diagnosing and treating diseases, interpreting life phenomena, exploring disease mechanisms, developing drugs, and promoting human cognition of life phenomena[18].
In previous studies, many risk factors for GDM have been identified, including advanced maternal age, family history of diabetes, prepregnancy overweight and obesity, and genetic components[14]. However, the etiology and pathogenesis of GDM are still unclear. As a metabolic disease, GDM is very suitable to be studied by metabolomics, which can monitor the changes in small molecular metabolites induced by maternal stimuli or perturbation. The application of metabolomics in GDM can discover diagnostic biomarkers, evaluate the prognosis of the disease, guide the application of diet or drugs, evaluate the curative effect, and explore the mechanism. This article reviews the metabolomics research methods and techniques as well as the current progress in metabolomics in GDM research. We anticipate that the review will contribute to identifying gaps in current knowledge or in metabolomics technology, provide evidence-based information, and inform future research directions in GDM.
Metabolomics detects a complete set of small molecular metabolites in cells, body fluids, or tissues based on modern analytic techniques with high throughput, high sensitivity, and high accuracy and analyzes the association of metabolites with pathophysiological changes under the influence of internal or external factors, which reflect the “end result” of environmental exposures, disease invasion, drug treatment, and genetic variation at a certain time in the organism[19,20].
At present, the two most commonly used strategies for metabolomics analysis are untargeted metabolomics and targeted metabolomics[21]. Untargeted metabolomics, also known as global metabolomics profiling, means that all or most possible metabolites are unbiasedly qualitatively or quantitatively analyzed in given biological samples[22,23]. This approach will be applied when it is not known which metabolites are of importance in the research question or when the aim is to discover unknown physiological patterns since it can produce a large number of complex molecules, provide directions for identifying biological markers and metabolic pathways, and offer a better understanding of internal metabolic physiology by high-throughput methods[24,25]. In contrast, targeted metabolomics refers to dozens to hundreds of metabolites, usually a specific class of metabolites with similar physicochemical properties (such as carbohydrates, lipids, and amino acids) or a class of metabolites involved in the same biochemical pathways (such as gluconeogenesis, beta-oxidation, or the citric acid cycle), being analyzed[26,27]. It is usually used to identify new biomarkers or deeply investigate metabolite functions and pathways as well as the relationship between metabolites and diseases[28]. However, though widely applied, this approach is often biased, artificially amplifying the effects of selected metabolites on the performance of the biological system and neglecting the metabolites not in the selection[24,28,29].
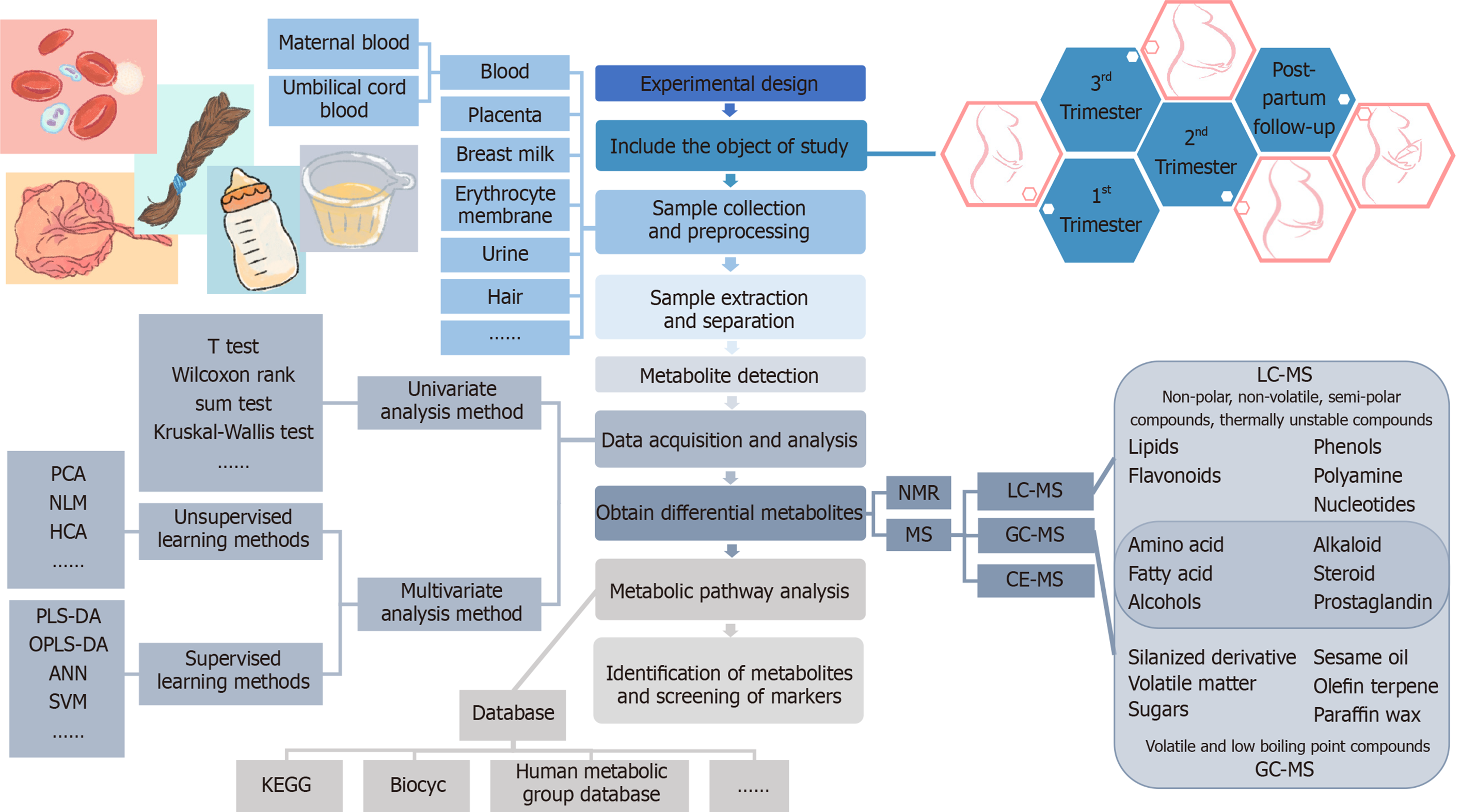
Due to the comprehensive nature of metabolomics, careful consideration of the following aspects is required: Selection and storage of the sample specimens, technology for the extraction of the metabolites, quality control of the analyses, and statistical methods to determine the metabolites undergoing significant change[30]. The basic procedures of metabolomics research consist of the experimental design, sample collection and pretreatment, metabolite extraction and detection, original data acquisition and processing, bioinformatics and statistical analysis, and annotation or identification of metabolites[31]. The main analysis workflow of the metabolomics analysis is shown in Figure 1.
Human metabolites include thousands of known and unknown small molecular metabolites whose properties of polarity, size, and concentration vary enormously, ranging from low-molecular-weight, hydrophilic, polar metabolites (e.g. amino acids) to higher-molecular-weight, hydrophobic, nonpolar metabolites (e.g. lipids)[32]. This diversity brings great technical challenges to the detection, identification, and quantification of metabolites[33]. At present, there is no analytical method that can simultaneously detect or quantify all metabolites in human biological samples. Scientists try to use a variety of analytical techniques to analyze various complicated metabolites, among which nuclear magnetic resonance (NMR) spectroscopy and mass spectro
NMR technology is a spectral technique that converts resonant frequencies into molecular chemistry and structure information by making use of the different resonance frequencies produced by different nuclear absorption radiation[35]. It does not need chromatographic processing, and the sample preparation is simple. It can detect tissue samples in situ, providing accurate structural information on metabolites[36], so it is suitable for the qualitative and quantitative study of metabolites[24,30]. However, its deficiency lies in its low sensitivity and incomplete metabolite coverage in biomedical research. However, in recent years, with the progress of high magnetic field magnets, pulse sequences, and freezing probe technology[37], the sensitivity and resolution of NMR approaches have been significantly improved.
MS analysis is a method to separate and detect moving ions according to the ratio of mass to charge (m/z) by electric and magnetic fields[29]. Metabolomics based on MS analysis usually involves separation by chromatography combined with chromatography to reduce the matrix effect and ion inhibition effect. At present, MS hyphenated chromatography can be divided into three categories: Liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), and capillary electrophoresis-mass spectrometry (CE-MS). The choice of different chromatographic separation methods depends on the properties of the analyte (such as molecular weight and hydrophilicity/hydrophobicity): LC-MS is mainly used to identify nonpolar, nonvolatile, semipolar, and thermally unstable compounds[38]. LC-MS technology separates compounds in LC through the interaction between the mobile phase (solvent) and stationary phase and identifies the separated compounds in the mass spectrometer. LC-MS can cover a wider range of metabolites and is the mainstream method for metabolomic analysis at present. GC-MS is mainly used for volatile and low boiling point compounds. The derivation method can increase the volatility and thermal stability of nonvolatile compounds for GC-MS analysis. The separated analyte must be ionized before entering the MS instrument, and the ionization method is related to the type of MS detector[29,39].
Compared with NMR technology, the high sensitivity and high resolution of the MS method make it possible to detect thousands of metabolites in a wide dynamic range[40], which makes MS the first choice for researchers who need low abundance metabolite data or to separate large amounts of metabolites at the same time[41,42]. Both NMR and MS have obvious advantages and disadvantages (shown in Table 1), and they are highly complementary. The combination of these two technologies can improve the overall quality of the research. In recent years, researchers have begun to use NMR and MS to analyze the metabolic spectrum of samples, drug efficacy and toxicology, disease pathophysiology, and other aspects[30,43], and to explore the compatibility of NMR and MS analysis platforms[30].
| Comparison item | NMR | MS |
| Sensitivity | Low, less than 100 metabolites at a time | High, more than 1000 metabolites at a time |
| Selectivity | Poor | Good |
| Targeting analysis ability | Usually used for nontargeted analysis | Targeted and non-targeted analysis |
| Sample preparation | Simple | Complex |
| Sample measurement | Completed in a single measurement | Require different chromatographic techniques |
| Sample recovery | Nondestructive, recyclable, and long-term preservation | Unrecyclable, less sample consuming |
| Repeatability | High | Low |
| Tissue sample | Directly without pretreatment. | Need pretreatment such as tissue extraction |
In recent years, metabolomics studies in GDM have shown the characteristics of universality and diversity. For example, there are a variety of biological samples, including blood plasma or serum from the maternal or fetal umbilical cord, erythrocyte membrane, urine, amniotic fluid, breast milk, placenta, and hair, but most samples used are plasma or serum. The time periods of study also have different variations from the first trimester, second trimester, third trimester, at delivery, to postpartum periods following delivery. At present, GDM metabolomics research is mainly focused on carbohydrates, amino acids, lipid metabolites [including fatty acids (FAs), phospholipids, sphingomyelin, etc.], purines, uric acid, bile acid, and other small molecular metabolites, as well as related metabolic pathways. The results of the papers published in recent years are shown in Table 2[44-58].
| Study design | Study methods | GDM diagnostic guidelines | Sample collection time | Biological samples | Analytical platform (s) | Changes in direction (vs control) | Main differential metabolites | Ref. |
| Biological samples collected before GDM diagnosis | ||||||||
| Prospective study (overweight and obese), n = 82 GDM, n = 275 controls | Targeted NMR-based metabolomics approach | IADPSG | 12.8–15.6 wk | Maternal serum (fasting) | NMR | Up | All-sized VLDL particles; medium-sized HDL particles; small-sized HDL particles; glucose; lactate; pyruvate; isoleucine; leucine; alanine; phenylalanine; GlycA | Mokkala et al[44], Finland, 2020 |
| Down | Very large HDL particles | |||||||
| Nested case-control study, n = 243 GDM, n = 243 controls | Targeted Lysophosphatidylcholines and bile acids | IADPSG | 9-11 wk | Maternal plasma (fasting) | LC-MS/MS | Up | LPC egg; LPC 15:0; LPC 17:0; LPC 18:0; LPC 18:1 | Liu et al[45], China, 2020 |
| Down | Deoxycholic acid; glycoursodeoxycholic acid | |||||||
| Case-control study, n = 65 GDM, n = 366 controls | Targeted amino acids | IADPSG | 12-16 wk | Maternal serum | UHPLC-MS/MS | Up | Alanine; glutamate; isoleucine; phenylalanine; tyrosine | Jiang et al[46], China, 2020 |
| Down | None | |||||||
| Case-control study, n = 121 p-GDM, n = 121 control | Targeted metabolomics | multi-step screening | 16-19 wk | Maternal serum(randomly) | HILIC-MS/MS | Up | None | Sakurai et al[47], Japan, 2019 |
| Down | Glutamine; pyrophosphate; octulose-1,8-bisphosphate | |||||||
| Case-controls study, n = 36 p-GDM, n = 31 controls | Maternal urine (randomly) | HILIC-MS/MS | Up | Shikimate-3-phosphate; 1,3-diphosphoglycerate; N-acetyl-L-alanine | ||||
| Down | Ethanolamine; methionine | |||||||
| Nested case-control study, n = 131 GDM, n = 138 controls | Nontargeted metabolomics | IADPSG | 12 wk | Maternal serum (fasting) | UPLC-QTOFMS, UPLC-TQMS, GC-TOFMS | Up | C14:1(trans-9); C16:1 (cis-7); C17:1 (cis-10); C18:1 (cis-9); C19:1 (cis-10); C20:1 (cis-11); C12:0; C16:0; C17:0; C20:0; C16:2 (cis-9_12); C18:2 (9_11&10_12-cis&trans-conjugated-99%); C18:3 (cis-9_12_15); C22:3 (cis-13_16_19); C18:3 (cis-6_9_12); C18:2 (cis-9_12); C20:4 (cis-5_8_11_14); C20:3 (cis-8_11_14); C20:2 (cis-11_14); C22:5 (cis-7_10_13_16_19); THDCA; LCA; HDCA; isoDCA; 6_7_diketoLCA; leucine; isoleucine; valine; acetylaspartic acid; alanine; glutamic acid; 2-aminobutanoic acid; 2-oxo-4-methylvaleric acid; gamma-aminobutanoic acid; Aminomalonic acid; pyruvic acid; 1-monooleoylglycerol; 2-ethylhexanoic acid; mannose; threitol | Hou et al[48], China, 2018 |
| Down | CA; dehydro_LCA; cysteine; 2,3,4-trihydroxybutyric acid; maltose; threonic acid | |||||||
| Biological samples collected after or simultaneously with GDM diagnosis | ||||||||
| Nested case-control study, n = 23 GDM, n = 77 controls | Targeted polar metabolites and lipids | IADPSG | 24-28 wk | Maternal serum (2 h OGTT) | LC-MS | Up | Adenosine; taurolithocholic acid; glycoli thocholic acid; glycochenodeoxycholic acid | Geiaye et al[49], Peru, 2019 |
| Down | Methionine sulfoxide; C58:10 triacylglycerol; C58:9 triacylglycerol; C8 carnitine; C14:2 carnitine; C14:1 carnitine; C12 carnitine; C4-OH carnitine | |||||||
| Case-control study (overweight), n = 100 GDM, n = 252 controls | Targeted NMR-based metabolomics approach | IADPSG | 34.6-35.9 wk | Maternal serum (fasting) | NMR | Up | VLDL particles; glucose; citrate; isoleucine; leucine; phenylalanine; acetoacetate; GlycA | Mokkala et al[50], Finland, 2020 |
| Down | Small HDL particles | |||||||
| Case-control study (overweight), n = 19 GDM with medication, n = 76 GDM with diet | Up | VLDL particles; lactate; isoleucine; leucine; GlycA | ||||||
| Down | Small HDL particles | |||||||
| Case-control study, n = 45 GDM, n = 98 controls | Targeted lipids metabolomics approach | IADPSG | At admission, after admission, and up to 24 h after delivery | Maternal plasma (fasting) | LC-IMS-MS | Up | PE(P-20:0_18:1); PE(P-18:0_22:6); PE(P-20:0_22:6); PE(P-18:1_22:6); PC(P-18:0_22:6); PC(17:0_22:6) | Odenkerk et al[51], United States, 2020 |
| Down | PEBP1; TG(12:0_16:0_18:1); TG(14:0_16:0_18:1); TG(14:0_16:0_18:2); PDIA6; PDIA5;LYAG; EXT1; B4GA1 | |||||||
| Case-control study, n = 11 GDM only, n = 11 hyperlipidemia only, n = 12 GDM plus hyperlipidemia, n = 11 controls | Targeted lipids metabolomics approach | IADPSG | 27–33 wk | Maternal serum (fasting) | GC-MS/MS | Up | 3-Dehydrocarnitine; 6-hexanoyl-D-erythro-sphingosine; arachidoyl ethanolamide; CER; Che; DG; dMePE; eicosanoicacid; FA; hexadecanamide; LdMePE; LPA; LPC; LPEt; LPG; LPI; MG; N-hexadecyl-ethanolamine; OAHFA; octadecanoicacid; PAF; PC; PE; Pet; PG; phSM; PI; PIP,SM; So; TG | Liu et al[52], China, 2019 |
| Down | None | |||||||
| Case-control study, n = 30 GDM, n = 30 controls | Nontargeted metabolomics | IADPSG | 24-28 wk | Maternal serum | LC-MS | Up | TXB2; traumatic acid; PGC2; PGJ2; PGB2; PGA2; pravastatin; PGD2-d4; PGE2-d4; crotonoyl-CoA; methacrylyl-CoA; 2S-hydroxybutanoic acid; D(-)-beta-hydroxy butyric acid; 4-hydroxy-butyric acid; DPA; oleic acid; rumenic acid; linoleic acid; urocortisone; corticosterone; 11-deoxycortisol; tetrahydrocortisol; 2-hydroxyestrone; dehydroepiandrosterone sulfate; tetrahydrocorticosterone; LPA (0:0/16:0); LysoPC (20:4); psychosine | Li et al[53], China, 2019 |
| Down | PGG2; 6-keto PGE1; 11-dehydro-TXB2; cholesterol; lathosterol; coenzyme Q10; lutein; zeaxanthin | |||||||
| Case-control study, n = 32 GDM, n = 11 controls | Targeted lipidomic profile | WHO—2013 criteria | 24-28 wk | Maternal erythrocyte membrane (fasting) | GC-MS | Up | Oleic acid; vaccenic acid; sapienic acid | Bukowiecka-Matusiak et al[54], Poland, 2018 |
| Down | Myristic, palmitic, and stearic acids | |||||||
| Biological samples collected in longitudinal cohort study | ||||||||
| Nested case–control studyBaseline (6–9 wk postpartum), n = 173 future T2D, n = 485 non-T2D | Targeted metabolomics | Carpenter and Coustan criteria | 6–9 wk postpartum (baseline); | Maternal plasma (fasting) | FIA-MS/MSLC-MS/MS | Up (baseline) | Hexose; histidine; isoleucine; serine; tyrosine; leucine; methionine; glutamate; lysine; tryptophan; threonine; proline; acylcarnitines AC3; acylcarnitines AC10; acylcarnitines AC16; spermidine; diacyl-glycerophospholipids | Lai et al[55], United States, 2020 |
| Down (baseline) | Glutamine; kynurenine; sphingomyelins; lysophosphatidylcholines; acyl-alkyl-glycerophospholipids | |||||||
| Follow-up (2~8 yr postbaseline), n = 98 future T2D, n = 239 non-T2D | Up to 2 yr postbaseline (follow-up). | Up (follow-up) | Hexose; glutamate; isoleucine; tyrosine; leucine; valine; alanine; PC; aa C32:1; acylcarnitines AC5 | |||||
| Down (follow-up) | Glycine; PC aa C38:1; PC aa C38:6; PC ae C36:2; PC ae C40:6; PC ae C34:3; PC ae C34:2; PC ae C36:3; PC ae C38:4; lysoPC a C17:0; lysoPC a C18:1; lysoPC a C20:4; lysoPC a C18:2; lysoPC a C16:0; lysoPC a C18:0; SM(OH)C22:2; SM(OH)C14:1; creatinine | |||||||
| Case-control study, n = 90 GDM, n = 94 controls | Nontargeted metabolomics | IADPSG | 1–3 d postpartum | Colostrum | GC-MS | Up | None | Wen et al[3], China, 2019 |
| Down | Heneicosane; G\glycine, N-(methoxyoxoacetyl)-methyl ester; 3-aminoisobutyric acid; glutamine; oxaloacetic acid; 4-aminobutyric acid | |||||||
| 7-10 d postpartum | Transition milk | GC-MS | Up | Asparagine; malic acid | ||||
| Down | D-Proline; glycine, N-(methoxyoxoacetyl)-, methyl ester; hydroxybenzoic acid; malonic acid; 9-heptadecenoic acid | |||||||
| 4 wk postpartum | Mature milk | GC-MS | Up | Heneicosane; cysteine; lignoceric; malic acid | ||||
| Down | Nonacosane; glycine; N-ethyl-N-(2-ethoxyethoxycarbonyl)-; 2-methoxyethyl ester; pyroglutamic acid; beta-alanine; 2-oxoadipic acid; 3-methyl- 2-oxovaleric acid; 4-aminobutyric acid; glutamine; oxalic acid; oxaloacetic acid; pimelic acid; 9-heptadecenoic acid; 10-pentadece- noic acid; 2-hydroxyglutaramic acid; nervonic acid | |||||||
| Case-control study, n = 50 Healthy, overweight/obese, n = 45 GDM, n = 67 Healthy, normal weight | Targeted metabolomics | National Diabetes Data Group criteria | At delivery | Maternal plasma | LC–MS/MS | Up | Hexoses; Asn/Asp | Shokry et al[56], Spain, 2019 |
| Down | Phospholipids (PL); LPC16:0; PCaa38:3; PCaa 38:5; SM32.2 | |||||||
| Case-control study, n = 40 Healthy, overweight/obese, n = 27 GDM, n = 49 Healthy, normal weight | Cord plasma of offspring | Up | Hexoses; Asn/Asp; 3-methyl-2-oxobutanoic acid | |||||
| Down | PCae38:0; Carn; short-chain AC; acetyl carnitine; NEFA26:1, malic; succinic acids | |||||||
| Case-control study, n = 107 GDM, n = 107 controls | Nontargeted metabolomics | Criteria of the American Diabetes Association | First trimester (12.8 wk median) | Maternal serum | UHPLC-MS | Up | Proline; ornithine; glycerophosphocholine; glutamic acid; taurine; acetylcarnitine; uracil; hypoxanthine; aspartyl-isoleucine; pantothenic acid; lysoPC (14:0); LysoPC [16:1(9Z)]; linoleoyl carnitine; palmitoylcarnitine; LysoPC (16:0); histidine; succinic acid semialdehyde; malic acid; xanthosine | Zhao et al[57], China, 2019 |
| Down | Threoninyl-phenylalanine; asparaginyl-tryptophan; phenylalanyl-gamma-glut amate; valyl-isoleucine; aspartyl –phenylalanine; phenylalanyl-valine; DL-2-aminooctanoic acid; phenylalanyl-isoleucine; acetylglycine | |||||||
| Second trimester (26.1 wk median) | Up | Guanidoacetic acid; acetylcarnitine; propionylcarnitine; 2-octenoylcarnitine; LysoPC (14:0); LysoPC [16:1(9Z)]; palmitoylcarnitine; vaccenyl carnitine; LysoPC (16:0); LysoPE [0:0/18:1(11Z)]; LysoPC (17:0) | ||||||
| Down | Threoninyl-phenylalanine; phenylalanyl-gamma-glut-amate; phenylalanyl-isoleucine | |||||||
| Case-control study, n = 15 GDM, n = 50 controls | Nontargeted metabolomics | IADPSG | 6-8 wk | Maternal urine | UPLC-MS/MS | Up | Levoglucosan; polyethylene glycol; 6-hydroxy-5-methoxyindole glucuronide | Liu et al[58], China, 2019 |
| Down | 1'-acetoxyeugenol acetate; 3,4-dimethyl-5-pentyl-2-furanundecanoic acid; 2-hydroxylauroylcarnitine; L-phenylalanyl-L-proline | |||||||
It is well known that the characteristic clinical biochemical index of GDM is an increase in blood glucose content, which continues to be high with the development of pregnancy. A recent study tracked the metabolic changes in individuals from baseline (6-9 wk postpartum) to a 2-year follow-up (2 years postpartum) in 1035 women diagnosed with GDM and studied the trajectory of T2D progression with a longitudinal analysis[55]. The researchers found that hexose, which represents the sum of all 6-carbon monosaccharides including glucose and fructose, was the only significantly increased metabolite in all three analyses (at baseline, at follow-up, and longitudinal analysis)[55]. Shokry’s study has also found that the sum of hexose (approximately 90%-95% glucose, 5% other hexoses) was significantly increased in GDM in both maternal plasma and, especially, umbilical cord plasma[56]. The association between hexose and future T2D risk was highly significant at baseline[55,59] and within 2 years of follow-up[60,61]. However, the increase in hexose (likely glucose) is not surprising, since diabetes and its severity are defined by the circulating blood glucose level. Although there is a continuous interaction between various nutrient metabolism pathways, in most cases, carbohydrate metabolism takes precedence over protein and fat in energy production. Hexose metabolism is the most regulated, which is also most closely related to the occurrence of T2D. Disorders in circulating hexoses may indicate problems with carbohydrate metabolism but may also be potential problems with amino acid or lipid metabolism.
In addition to perturbations in hexose metabolism, some disorders of glucose metabolism pathway molecules have also been detected in women with GDM. Mokkala et al[50] found that the level of citrate, which is an intermediate in the tricarboxylic acid cycle synthesized from acetyl-CoA and oxaloacetate, increased in women with GDM. Citric acid inhibits glycolysis and conversely stimulates gluconeogenesis and lipid synthesis, i.e., energy storage. Thus citric acid is an important regulator of energy metabolism[62]. Therefore, in addition to its relationship with GDM, citric acid may be a determinant of adiposity in overweight and obese women, as observed in a study of nonpregnant participants with obesity[63]. Early studies, designed as case-control studies, showed that the circulating concentration of β-hydroxybutyric acid in GDM patients was higher than that in controls[64,65]. β-hydroxybutyric acid is a classic ketone body found alongside acetone and acetoacetate. Mokkala et al[50] also found elevated levels of the ketone body acetoacetate in patients with gestational diabetes. Previous studies have also detected increased concentrations of ketone bodies in pregnant women with high fasting blood glucose levels[66] and found that they are associated with GDM[67-69].
Similar to T2D, a failure of glucose utilization will lead to the formation of these ketone bodies. It is well known that in the latter half of normal pregnancy, maternal metabolism is transformed into catabolism, during which increased lipolysis provides FAs that can be utilized in ketogenesis[70]. The produced ketone bodies are used as maternal energy to replace glucose, which is mainly utilized by the fetus[71]. Elevated pyruvate and glucose anaerobic decomposition products (3.27 times higher than that in the normal group) were observed in the urine of GDM patients at 17 wk of pregnancy, in the research from Michelle A. Willims’ group[72]. Such results on pyruvate are consistent with other reports[73,74]. These results indicate that abnormalities in glycolysis and tricarboxylic acid cycle metabolic pathways result in disturbed energy metabolism and dysregulation of glucose metabolism associated with GDM.
The association of increased serum amino acid levels with obesity and IR was observed more than 40 years ago[75]. Advances in metabolomic analysis have renewed interest in this field. By using metabolomic techniques, a large number of amino acid metabolites related to GDM have been detected, including amino acids (alanine, glutamine, and glycine), branched-chain amino acids (isoleucine, leucine, and valine), and aromatic amino acids (phenylalanine, tyrosine, and histidine). Among all these amino acids, the relationship between branched-chain amino acids (BCAAs) and IR has been deeply studied.
Abnormal amino acid metabolism can occur in the early stage of pregnancy. Mokkala et al[44] analyzed fasting serum samples via a targeted NMR approach in early pregnancy (median: 14.3 wk of gestation), and found that the concentrations of two branched-chain amino acids, namely, isoleucine and leucine, as well as phenylalanine and alanine, were already increased in women who developed GDM. Similar findings have been reported previously in which elevated BCAA levels including valine and phenylalanine, but not alanine, were detected in obese women with GDM[67]. In contrast, surveys of women with heterogeneous body mass indexes (BMIs) have reported that increased concentrations of valine[76] and alanine[76,77] in early pregnancy or no changes in BCAAs[77,78] were associated with the onset of GDM. Other studies have reported that glutamine[47], glutamic acid, and serine[79] in serum significantly differed between the prior to diagnosis of GDM (p-GDM) and control groups in the first trimester. Glutamine in serum showed an area under the receiver operating characteristic curve of 0.81 and may be a predictive metabolite for GDM[47]. Combined metabolomic analysis of plasma and urine revealed that serotonin, 5-HIAA, L-tryptophan, melatonin, and 6-hydroxymelatonin may be effective predictors of GDM[80]. However, the results of studies on the relationship between the concentrations of amino acids in early pregnancy (prior to diagnosis of GDM) and the risk of GDM are not consistent[67,77,81]. Based on findings from previous studies and considering that amino acids can induce IR[82], it can be proposed that amino acids may become effective indexes for the prediction and early diagnosis of GDM[76-78]. There is no consensus on amino acids for early screening of GDM[83]; however, many scholars still suggest that the measurement of amino acid concentrations in early pregnancy should be taken as part of routine testing in the future, so that clinicians can identify high-risk pregnant women with GDM as early as possible, to take effective intervention measures to prevent GDM[46,60,84,85].
Perturbed amino acid metabolism can not only predict the risk of GDM, but it also can predict the transition from GDM to T2D[86]. Allalou et al[59] enrolled 1010 GDM women without T2D at 6-9 wk postpartum (baseline) who were screened for T2D annually for 2 years. They found that 113 women progressed to T2D within 2 years and another 17 women developed T2D between 2 and 4 years. By a metabolomics study with baseline fasting plasma, it was found that 21 metabolites such as free FAs and amino acids (including isoleucine, leucine, tryptophan, tyrosine, alanine, and amino phenylalanine) were identified and could effectively predict the transition from GDM to T2D 2-4 years after delivery. Clinical trials have also demonstrated that the concentrations of BCAAs, such as leucine, isoleucine, and valine, are increased up to 7 years before the onset of T2D[84]. The latest study found that higher levels of BCAAs and 3-hydroxyisobutyric acid in plasma were associated with IR in the transition from GDM to T2D[86].
Disturbance of BCAAs and their metabolites is closely related to IR and decreased islet β-cell function in women with GDM[60,73,84,87-93]. It is the most common and most important abnormality of amino acid metabolism in GDM. The levels of leucine and isoleucine were associated with IR and decreased insulin sensitivity[82], while the tyrosine concentration was positively correlated with IR and insulin secretion[94]. The elevated levels of BCAAs in plasma increased the transport of FAs through endothelial cells to skeletal muscle, increased intracellular lipid accumulation, and weakened the insulin signaling pathway mediated by 3-hydroxyisobutyric acid[95]. BCAAs are involved in IR through several pathways, including fatty acid oxidation and the mTOR, JNK, and IRS1 pathways[96,97]. In addition, the increased BCAA concentrations may imply an increase in the absorption or production of intestinal microflora or a decrease in utilization/decomposition/degradation. In Mokkala et al’s study, higher concentrations of BCAAs (isoleucine and leucine) and phenylalanine were detected in women with GDM even after adjusting for pregestational BMI[44]. Other studies have reported an increase in leucine[98], isoleucine[46], and other amino acids such as alanine[99] and a decrease in L-valine[100] in patients with GDM compared with control patients. However, several studies have failed to detect differences in BCAA concentrations between patients with GDM and non-GDM in the first, second, or third trimester of pregnancy[77,101]. In addition, animal and human studies supplementing or restricting BCAA levels have reported conflicting effects[90,102-107]. Therefore, we still need to further explore and study amino acids, especially BCAAs, to track the individual changes in metabolism during disease progression, and explore the underlying molecular mechanisms.
Lipids are involved in regulating a variety of life processes, including energy conversion, material transport, information recognition and transmission, cell development and differentiation, and apoptosis. However, abnormal lipid metabolism is closely related to some diseases, such as diabetes, arteriosclerosis, obesity, Alzheimer’s disease, and tumorigenesis. In women with GDM, the physiological changes in lipids are magnified, which may indicate potential metabolic disorders during pregnancy[108]. At present, the understanding of the structures and functions of lipids still lags far behind that of genes and proteins, mainly due to the diversity and complexity of lipid molecular structures. In addition, the lag of analytical technology has hindered the systematic research on whole lipids and their complex metabolic network and functional regulation. The common lipids are fat, phospholipid, and sterol. The lipid differences closely related to GDM are mainly found in FAs, phospholipids, glycerolipids, glycerophospholipids, and sphingolipids.
FAs are the simplest lipid molecules and the main components of many complex lipids such as phospholipids, triacylglycerols, and sphingolipids. According to the saturation degree of the hydrocarbon chain, FAs can be divided into three categories: Saturated FAs (SFAs), monounsaturated FAs (MUFAs), and polyunsaturated FAs (PUFAs).
The SFA and MUFA families are the most abundant fatty acid species involved in metabolic disorders and are related to IR and related diseases[109,110] as well as diabetes-related pathways[111]. They are molecules of important biological and pathophysiological significance that are responsible for cell membrane fluidity, cell proliferation, lipid-mediated cytotoxicity, programmed cell death, unfolded protein response, the pathogenesis of obesity and cancer, and especially for metabolic diseases[112-114]. In the early studies of women with overweight and GDM, the free MUFAs in blood were greatly influenced by diet[115] and obesity[116]. Chen et al[117] studied circulating (serum) FAs and detected impaired fatty acid composition, not only in women with GDM but also in women with impaired glucose tolerance. There is a strong correlation between the severity of maternal hyperglycemia and the concentration of serum FAs in the third trimester; however, plasma lipids are more dependent on dietary intake than tissues or cell membranes[117]. Bukowiecka-Matusiak et al[54] investigated the erythrocyte membrane fatty acid profiles of 32 pregnant women with GDM and 11 pregnant women with normal glucose tolerance by GC-MS and found that of the 14 measured FAs representing the characteristics of membrane lipids, three kinds of SFAs (myristic acid, palmitic acid, and stearic acid) of erythrocyte membranes in patients with GDM showed a downward trend. The relative content of MUFAs in the erythrocyte membrane of the GDM group was higher than that of the NGT group, especially the oleic acid and vaccenic acid contents, which were significantly increased[54]. There was no significant change in PUFAs in erythrocytes between the GDM and NGT groups. Based on the differences between the GDM and NGT lipidomic profiles, the authors postulated that stearic acid and cis-acetic acid can be regarded as dual biomarkers of specific SFA-MUFA transformation pathways. Stearic acid and cis-acetic acid are involved in erythrocyte membrane remodeling in women with GDM by the coupling of δ-9 desaturase and elongase enzymes[54]. In addition, the higher concentration of vaccenic acids in red blood cells was related to lower fasting glucose, better insulin sensitivity, and a reduced risk of T2D[118]. Because erythrocyte membrane composition reflects the dietary intake for the past 2-3 mo, the erythrocyte membrane fatty acid state can be more reflective. The results from a study on breast milk found increased levels of FAs in mature milk compared to those in colostrum and transition milk, while there was a considerable amount of decreased FAs (9-heptadecenoic acid, pimelic acid, 10-pentadecenoic acid, 2-hydroxyglutaramic acid, and nervonic acid) in breast milk from GDM women. This may be due to the fact that free FAs are the cornerstone of adipogenesis and neurons. Differences in free FA profiles may play a more profound role in breastfeeding-related developmental disorders or future health risks in offspring. Therefore, functional lipidomics based on membrane FAs and other tissues has become a convenient and relevant molecular tool to examine the nutritional and metabolic status of patients.
FAs can be categorized according to the length of the carbon chain into short-chain FAs (less than 6 carbon atoms), medium-chain FAs (6-12 carbon atoms), and long-chain FAs (more than 12 carbon atoms). It has been reported that long-chain acylcarnitines[119,120] and their precursors long-chain acyl-CoAs are related to IR. In the Zhao et al’s [57] study, long-chain acylcarnitines (such as palmitoyl carnitine and vaccenyl carnitine) increased in women with GDM both in the first and second trimesters. Another study observed 201 metabolites in maternal plasma at delivery and cord plasma obtained from 325 participants by LC-MS/MS[56]. The long-chain NEFAs and Krebs metabolites in cord blood from GDM offspring showed an overall decrease, and the most significant metabolites were NEFA26:1, malic acid, succinic acid, and 3-methyl-2-oxybutyric acid. In an analysis of fatty acyl groups in GDM[51], lipids containing 12:0, 14:0, 15:0, 18:3, 22:4, or 24:1 fatty acyls were downregulated, while 20:0 and 22:6 fatty acyls were upregulated, such as 22:6 for PE P- and PC and PE O- species. Furthermore, myristic acid or 14:0, which has previously been thought to be negatively associated with 22:6[121], was also downregulated in GDM but not in the same lipid species[51]. As previously reported, a higher level of palmitoleic acid is beneficial to insulin sensitivity in the case of metabolic diseases, and its administration in animal models can effectively reduce IR and hepatic lipid accumulation[122]. Therefore, different FAs may play different roles in maternal and infant health, and even the same FAs may have different effects due to different doses and exposure times.
The complex relationships among phospholipids and sphingomyelin metabolites and the risk of diabetes have not been widely studied and some studies have conflicting results. A longitudinal analysis revealed that six diacyl-glycerolphospholipids (the PC aa C group) were positively correlated with T2D at baseline (6–9 wk postpartum) and follow-up (from baseline to 2 years), while 11 acyl-alkyl-glycerolphospholipids (the PC ae C group) were negatively correlated with T2D risk[55]. In a case-cohort study, some diacyl-phosphatidylcholines were associated with a higher risk of T2D, while sphingomyelin and 1-acyl-alkyl-phosphatidylcholine (C18:2) were associated with a lower risk[89]. Moreover, in a subset of the hyperglycemia and adverse pregnancy outcome study, disturbed lipid metabolism of certain classes of phospholipids and lysophospholipids was detected in the diabetes risk group prior to hyperglycemia at 2 years of postpartum[89]. Wheeler’s group found that several sphingomyelins decreased at baseline (6–9 wk postpartum) and follow-up (from baseline to 2 years)[55]. Interestingly, in contrast to the finding of a negative relationship between sphingomyelin metabolism and T2D risk by Wheeler’s group[55,123], another study[89] showed that sphingomyelins were increased in T2D patients and positively associated with future T2D[124,125]. Thus, further research is needed to investigate the role of phospholipids and sphingomyelins in T2D risk among different populations.
In short, the chain length and the degree of desaturation of FA moieties in lipid molecules increase the complexity of assigning biological roles to various lipid classes. Moreover, lipids that are synthesized endogenously or obtained through the diet influence their accumulation and/or metabolism and subsequent biological roles. Therefore, it is not surprising that opposing views exist on the pathogenicity and mechanisms of specific lipids during the development of IR. However, new high-resolution metabolomic technology provides bright prospects to identify lipid subclasses and novel families of lipids.
Purine is a key component of the cellular energy system. Hypoxanthine is a degradation product of adenosine triphosphate, which is a major bioenergy source, and is further converted to xanthine by xanthine oxidase (XO) and then converted to uric acid, the final oxide of purine metabolism[126]. The study found that hypoxanthine and xanthine in the GDM group were significantly higher than those in the control group during early pregnancy[57], which may be due to higher energy intake in women with GDM in early pregnancy. However, in the second trimester, the levels of hypoxanthine and xanthine in women with GDM were significantly lower than those in the first trimester, while the concentration of uric acid was significantly increased. This is because the activity of XO in the peripheral blood of pregnant women with GDM is higher than that of healthy women, resulting in excessive uric acid accumulation[127,128]. A high uric acid level is associated with IR[129] and is considered to be a risk factor for GDM[130] and T2D[131]. In addition, the levels of inflammatory markers such as glycoprotein acetylation (GlycA, a marker of low-grade inflammation)[44,67,68,132-134] and some sterol hormones such as 11-deoxycortisol, 17α-hydroxyprogesterone, progesterone[76], cortisol, androstenedione, dehydroepiandrosterone sulfate, and 11 deoxy-cortisol steroids[53]in pregnant women with GDM are different from those in normal pregnant women[135]. Disorders of these processes lead to metabolic and immune dysfunction and the development of pregnancy complications.
Li et al[136] previously reported that decreased concentrations of two secondary bile acids, deoxycholic acid (DCA) and glycoursodeoxycholic acid (GUDCA), in early pregnancy were associated with a significantly elevated risk of GDM. Recently, they investigated the associations between trimethylamine (TMA), trimethylamine nitrogen oxide (TMAO), and related metabolites (choline, betaine, L-carnitine) and the risk of GDM during early pregnancy[137]. The levels of TMAO and related metabolites decreased significantly in women with GDM, while the concentration of TMA was increased. The change in TMAO had a clear threshold effect, independent of DCA and GUDCA, to a great extent. It is commonly known that the conversion from betaine, choline, and L-carnitine to TMA depends on gut microflora, and the conversion of TMA to TMAO depends on the activity of flavin monooxygenase 3 (FMO3), especially FMO3 in the liver[138,139]. According to Rothman’s multicausality theory[140], the significant additive interaction between abnormal gut microflora and decreased FMO activity in the liver may be one of the causes of GDM. In addition, Liu et al[52] also conducted microbiomics coupled with lipidomics analyses to characterize gut microbiota and lipometabolism in pregnant women with GDM, hyperlipidemia, or GDM plus hyperlipidemia. This study and other studies[48,141,142] further indicate that changes in the fecal microbiota and plasma lipidome can predict and characterize the development of GDM with lipid metabolic abnormalities.
Huhtala et al[143] enrolled 217 GDM pregnant women treated randomly with metformin or insulin, and 126 GDM pregnant women who achieved sufficient glycemic control by diet and lifestyle modifications alone as a control. The levels of serum amino acids and lactic acid at diagnosis and 36 wk of gestation were measured by 1H NMR. It was found that the majority of the amino acid concentrations increased with gestational age. Compared with the insulin-treated group, the metformin-treated group had a greater increase in alanine (16% vs 8%), isoleucine (11% vs 5%), and lactic acid (29% vs 14%). Another study[144] explored 71 pregnant women with GDM (including 28 patients who received dietary treatment only, 20 patients who received metformin, and 23 patients who received insulin) at three different gestational time points: 15-18+6 and 27-28+6 wk (before treatment) and 34-36+0 wk (after treatment). The results showed that prior to the initiation of treatment, metabolic differences were the most obvious between the diet and insulin-treated groups, especially very-large-density lipoprotein (VLDL) and high-density lipoprotein (HDL) subclasses and components. Even after drug treatment, there were still differences in lipid composition and particle size among the three groups at 34-36 wk of gestation. In recent years, Mokkala et al[50] found no differences in lipids in VLDL, and very large and large HDL particles were detected in overweight or obese women who had been treated for GDM for approximately 10 wk. Variations among these results may be due to the inconsistency in population selection, sampling times, and the improvement in metabolic variables to a certain extent by treatment[67,145]. These results indicate that diet, metformin, or insulin therapy in pregnant women with GDM can affect amino acid or lipid metabolism-related IR. Thus, the pathophysiological process of blood glucose control can be revealed by metabolic profiles.
Metabolomics research in GDM focuses on the abnormalities of small molecule metabolites such as carbohydrate, amino acids, lipid, sterol hormones, as well as bile acids and disordered metabolic pathways. In addition, signaling pathways and metabolic pathways analysis on results obtained from detected substances is an important part of metabolomics research, which helps to further explore the related metabolites, enzymes, and genes, so as to deeply understand their underlying biological phenomena.
The clinical utilities of metabolomics in GDM depend on the different time periods of the subjects. The main clinical utilities of research focused on the first trimester period are to identify early diagnostic or predictive molecules related to GDM disease[44-48]. The main clinical utilities of the studies concentrated on the second and third trimesters of pregnancy are to accurately evaluate the condition of GDM, identify differential metabolic markers and metabolic pathways, predict pregnancy outcomes and maternal and fetal prognoses, or evaluate the efficacy of drug/diet therapy[49-54,144,145]. A postpartum follow-up study mainly predicts the long-term occurrences of T2D or other metabolic diseases in GDM pregnant women and their offspring as well as the metabolic impact of GDM on their offspring[3,55,146].
Although metabolomics is developing rapidly as a new discipline, it still has some limitations and faces some technical challenges.
First, as a new research method, metabolomics needs to be further improved with respect to analytical technology, data acquisition, and analysis. GC-MS and LC-MS are affected by the matrix effect and ion suppression/enhancement[147], which are potential threats to the accuracy of metabolomics experiments. In addition, the process of screening specific biomarkers by metabolomics technology is tedious and complicated, especially for metabolic pathway analysis. Thus, the coverage of metabolomics is still insufficient for global metabolomics experiments despite the expansion of the metabolomics database[148].
Second, in recent years, although large amounts of data have been obtained from metabolomics in GDM research, many data are contradictory. Dissimilarities among these findings may be due to differences in the GDM diagnostic criteria, ethnic origin, size of the study populations, specimens prepared for testing, and metabolite profiling platforms. It is important to highlight that future research needs to follow a strict experimental design to improve the repeatability and reliability of research results.
Finally, metabolomics research has strict requirements for sample quantity, quality, and sample processing technology. Metabolites are more affected by the environment and individual differences and are also very sensitive to the storage mode and time of biological sample collections[149,150], which greatly increases the difficulty in obtaining reliable samples. Thus, future studies, especially large-scale, multicenter, dynamic monitoring, and prospective cohort studies are required before metabolomics can be routinely applied in clinical practice.
Although the research task of metabolomics is arduous, the popularization of mass spectrometry has laid the foundation for clinical application. Future metabolomics research still has broad prospects, which are embodied in the following:
First, considering the complexity and individual differences in biological samples, as well as other relevant lifestyle factors, standardized sample collection and operational procedures should be developed to ensure the accuracy and repeatability of the experiment.
Second, cross-sectional designs limit the analysis of the relationship between metabolite disturbance and the risk of GDM, so it is necessary to increase the sample size, conduct longitudinal analysis, and establish reliable clinical cohort samples. Tracking the metabolic changes in the individual and describing the disease progression of each case can also provide a theoretical basis for early stratification and appropriate treatment.
Third, many studies have identified specific metabolites and their relative changes in the relative concentration without providing the absolute concentration of metabolites, as a result, metabolomics data from different laboratories could not be compared with each other. The absolute quantification of metabolites and the establishment of a general standard database closely related to GDM are important research directions in the future.
Finally, in the postgenomic era of modern high-throughput computational technologies, it is essential to integrate the increasing amount of data generated from metabolomics and its upstream genomics, including genomics, transcriptomics, and proteomics, by sophisticated computational biology tools. Thus, this valuable comprehensive analysis can provide further insight into the etiology and pathophysiology of GDM, which might help to clarify the mechanisms of the occurrence and development of GDM and provide precise medical treatments for GDM.
In brief, metabolomics related to GDM research is in a period of vigorous development. Given the rapid improvement in omics technologies such as metabolomics, together with updating metabolic databases, bioinformatics, and artificial intelligence, our understanding of metabolic regulation in GDM will rapidly advance and thus significantly benefit the prevention and treatment of GDM.
Manuscript source: Invited manuscript
Specialty type: Obstetrics and gynecology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sridharan G S-Editor: Zhang H L-Editor: Wang TQ P-Editor: Wang LYT
| 1. | Zhou M, Liu XH, Liu QQ, Chen M, Bai H, Guan LB, Fan P. Lactonase Activity, Status, and Genetic Variations of Paraoxonase 1 in Women with Gestational Diabetes Mellitus. J Diabetes Res. 2020;2020:3483427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 2. | Li J, Wang P, Zhang C, Leng J, Li N, Wang L, Li W, Liu H, Yu Z, Hu G, Chan JCN, Yang X. Short Body Height and Pre-pregnancy Overweight for Increased Risk of Gestational Diabetes Mellitus: A Population-Based Cohort Study. Front Endocrinol (Lausanne). 2018;9:349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Wen L, Wu Y, Yang Y, Han TL, Wang W, Fu H, Zheng Y, Shan T, Chen J, Xu P, Jin H, Lin L, Liu X, Qi H, Tong C, Baker P. Gestational Diabetes Mellitus Changes the Metabolomes of Human Colostrum, Transition Milk and Mature Milk. Med Sci Monit. 2019;25:6128-6152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 4. | Kramer CK, Campbell S, Retnakaran R. Gestational diabetes and the risk of cardiovascular disease in women: a systematic review and meta-analysis. Diabetologia. 2019;62:905-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 635] [Cited by in RCA: 581] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 5. | HAPO Study Cooperative Research Group; Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, Coustan DR, Hadden DR, McCance DR, Hod M, McIntyre HD, Oats JJ, Persson B, Rogers MS, Sacks DA. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2008;358:1991-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3783] [Cited by in RCA: 3691] [Article Influence: 217.1] [Reference Citation Analysis (0)] |
| 6. | Voormolen DN, de Wit L, van Rijn BB, DeVries JH, Heringa MP, Franx A, Groenendaal F, Lamain-de Ruiter M. Neonatal Hypoglycemia Following Diet-Controlled and Insulin-Treated Gestational Diabetes Mellitus. Diabetes Care. 2018;41:1385-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 7. | Bellamy L, Casas JP, Hingorani AD, Williams D. Type 2 diabetes mellitus after gestational diabetes: a systematic review and meta-analysis. Lancet. 2009;373:1773-1779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2141] [Cited by in RCA: 2332] [Article Influence: 145.8] [Reference Citation Analysis (1)] |
| 8. | Tobias DK, Hu FB, Forman JP, Chavarro J, Zhang C. Increased risk of hypertension after gestational diabetes mellitus: findings from a large prospective cohort study. Diabetes Care. 2011;34:1582-1584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Hillier TA, Pedula KL, Schmidt MM, Mullen JA, Charles MA, Pettitt DJ. Childhood obesity and metabolic imprinting: the ongoing effects of maternal hyperglycemia. Diabetes Care. 2007;30:2287-2292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 463] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 10. | Wright CS, Rifas-Shiman SL, Rich-Edwards JW, Taveras EM, Gillman MW, Oken E. Intrauterine exposure to gestational diabetes, child adiposity, and blood pressure. Am J Hypertens. 2009;22:215-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 11. | Tam WH, Ma RC, Yang X, Ko GT, Tong PC, Cockram CS, Sahota DS, Rogers MS, Chan JC. Glucose intolerance and cardiometabolic risk in children exposed to maternal gestational diabetes mellitus in utero. Pediatrics. 2008;122:1229-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 12. | Vounzoulaki E, Khunti K, Abner SC, Tan BK, Davies MJ, Gillies CL. Progression to type 2 diabetes in women with a known history of gestational diabetes: systematic review and meta-analysis. BMJ. 2020;369:m1361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 616] [Article Influence: 123.2] [Reference Citation Analysis (0)] |
| 13. | Chen C, McGarvey PB, Huang H, Wu CH. Protein Bioinformatics Infrastructure for the Integration and Analysis of Multiple High-Throughput "omics" Data. Adv Bioinformatics. 2010;423589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Zhao C, Ge J, Li X, Jiao R, Li Y, Quan H, Li J, Guo Q, Wang W. Integrated metabolome analysis reveals novel connections between maternal fecal metabolome and the neonatal blood metabolome in women with gestational diabetes mellitus. Sci Rep. 2020;10:3660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2918] [Cited by in RCA: 2721] [Article Influence: 104.7] [Reference Citation Analysis (0)] |
| 16. | Zhang AH, Ma ZM, Sun H, Zhang Y, Liu JH, Wu FF, Wang XJ. High-Throughput Metabolomics Evaluate the Efficacy of Total Lignans From Acanthophanax Senticosus Stem Against Ovariectomized Osteoporosis Rat. Front Pharmacol. 2019;10:553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Shi Q, Zhao H, Chen J, Li Y, Li Z, Wang J, Wang W. Study on qi deficiency syndrome identification modes of coronary heart disease based on metabolomic biomarkers. Evid Based Complement Alternat Med. 2014;2014:281829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Gao RR, Shi S, Zhu Y, Huang HL, Yao TM. A RET-supported logic gate combinatorial library to enable modeling and implementation of intelligent logic functions. Chem Sci. 2016;7:1853-1861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 19. | Trainor PJ, Yampolskiy RV, DeFilippis AP. Wisdom of artificial crowds feature selection in untargeted metabolomics: An application to the development of a blood-based diagnostic test for thrombotic myocardial infarction. J Biomed Inform. 2018;81:53-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1560] [Cited by in RCA: 1690] [Article Influence: 130.0] [Reference Citation Analysis (0)] |
| 21. | Anh NH, Long NP, Kim SJ, Min JE, Yoon SJ, Kim HM, Yang E, Hwang ES, Park JH, Hong SS, Kwon SW. Steroidomics for the Prevention, Assessment, and Management of Cancers: A Systematic Review and Functional Analysis. Metabolites. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Cajka T, Fiehn O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal Chem. 2016;88:524-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 584] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 23. | Priante E, Verlato G, Giordano G, Stocchero M, Visentin S, Mardegan V, Baraldi E. Intrauterine Growth Restriction: New Insight from the Metabolomic Approach. Metabolites. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 24. | Au A, Cheng KK, Wei LK. Metabolomics, Lipidomics and Pharmacometabolomics of Human Hypertension. Adv Exp Med Biol. 2017;956:599-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 25. | Shi X, Xi B, Jasbi P, Turner C, Jin Y, Gu H. Comprehensive Isotopic Targeted Mass Spectrometry: Reliable Metabolic Flux Analysis with Broad Coverage. Anal Chem. 2020;92:11728-11738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Struct Biol. 2010;80:45-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 27. | Jang WJ, Choi JY, Park B, Seo JH, Seo YH, Lee S, Jeong CH. Hair Metabolomics in Animal Studies and Clinical Settings. Molecules. 2019;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 28. | Klepacki J, Klawitter J, Karimpour-Fard A, Thurman J, Ingle G, Patel D, Christians U. Amino acids in a targeted vs a non-targeted metabolomics LC-MS/MS assay. Are the results consistent? Clin Biochem. 2016;49:955-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Tsiropoulou S, McBride M, Padmanabhan S. Urine Metabolomics in Hypertension Research. Methods Mol Biol. 2017;1527:61-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Marshall DD, Powers R. Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics. Prog Nucl Magn Reson Spectrosc. 2017;100:1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 31. | Petrova I, Xu S, Joesten WC, Ni S, Kennedy MA. Influence of Drying Method on NMR-Based Metabolic Profiling of Human Cell Lines. Metabolites. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | O'Brien KA, Griffin JL, Murray AJ, Edwards LM. Mitochondrial responses to extreme environments: insights from metabolomics. Extrem Physiol Med. 2015;4:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608-D617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2659] [Cited by in RCA: 2542] [Article Influence: 363.1] [Reference Citation Analysis (0)] |
| 34. | Heather LC, Wang X, West JA, Griffin JL. A practical guide to metabolomic profiling as a discovery tool for human heart disease. J Mol Cell Cardiol. 2013;55:2-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 35. | Bothwell JH, Griffin JL. An introduction to biological nuclear magnetic resonance spectroscopy. Biol Rev Camb Philos Soc. 2011;86:493-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 36. | Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ, Westerhoff HV, van Dam K, Oliver SG. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 644] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 37. | Emwas AH. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol Biol. 2015;1277:161-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 343] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 38. | Mikami T, Aoki M, Kimura T. The application of mass spectrometry to proteomics and metabolomics in biomarker discovery and drug development. Curr Mol Pharmacol. 2012;5:301-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 39. | Theodoridis GA, Gika HG, Want EJ, Wilson ID. Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Anal Chim Acta. 2012;711:7-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 338] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 40. | Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 295] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 41. | Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 706] [Cited by in RCA: 596] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 42. | Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr Opin Biotechnol. 2015;31:73-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 43. | Gil AM, Duarte D. Biofluid Metabolomics in Preterm Birth Research. Reprod Sci. 2018;25:967-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Mokkala K, Vahlberg T, Pellonperä O, Houttu N, Koivuniemi E, Laitinen K. Distinct Metabolic Profile in Early Pregnancy of Overweight and Obese Women Developing Gestational Diabetes. J Nutr. 2020;150:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 45. | Liu J, Li J, Li S, Leng J, Li W, Yang W, Huo X, Chen L, Ma RCW, Hu G, Fang Z, Yang X. Circulating Lysophosphatidylcholines in Early Pregnancy and Risk of Gestational Diabetes in Chinese Women. J Clin Endocrinol Metab. 2020;105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 46. | Jiang R, Wu S, Fang C, Wang C, Yang Y, Liu C, Hu J, Huang Y. Amino acids levels in early pregnancy predict subsequent gestational diabetes. J Diabetes. 2020;12:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 47. | Sakurai K, Eguchi A, Watanabe M, Yamamoto M, Ishikawa K, Mori C. Exploration of predictive metabolic factors for gestational diabetes mellitus in Japanese women using metabolomic analysis. J Diabetes Investig. 2019;10:513-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (1)] |
| 48. | Hou W, Meng X, Zhao A, Zhao W, Pan J, Tang J, Huang Y, Li H, Jia W, Liu F. Development of Multimarker Diagnostic Models from Metabolomics Analysis for Gestational Diabetes Mellitus (GDM). Mol Cell Proteomics. 2018;17:431-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Gelaye B, Clish CB, Denis M, Larrabure G, Tadesse MG, Deik A, Pierce K, Bullock K, Dennis C, Enquobahrie DA, Williams MA. Metabolomics signatures associated with an oral glucose challenge in pregnant women. Diabetes Metab. 2019;45:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 50. | Mokkala K, Vahlberg T, Houttu N, Koivuniemi E, Laitinen K. Distinct Metabolomic Profile Because of Gestational Diabetes and its Treatment Mode in Women with Overweight and Obesity. Obesity (Silver Spring). 2020;28:1637-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Odenkirk MT, Stratton KG, Gritsenko MA, Bramer LM, Webb-Robertson BM, Bloodsworth KJ, Weitz KK, Lipton AK, Monroe ME, Ash JR, Fourches D, Taylor BD, Burnum-Johnson KE, Baker ES. Unveiling molecular signatures of preeclampsia and gestational diabetes mellitus with multi-omics and innovative cheminformatics visualization tools. Mol Omics. 2020;16:521-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 52. | Liu H, Pan LL, Lv S, Yang Q, Zhang H, Chen W, Lv Z, Sun J. Alterations of Gut Microbiota and Blood Lipidome in Gestational Diabetes Mellitus With Hyperlipidemia. Front Physiol. 2019;10:1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 53. | Li G, Gao W, Xu Y, Xie M, Tang S, Yin P, Guo S, Chu S, Sultana S, Cui S. Serum metabonomics study of pregnant women with gestational diabetes mellitus based on LC-MS. Saudi J Biol Sci. 2019;26:2057-2063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 54. | Bukowiecka-Matusiak M, Burzynska-Pedziwiatr I, Sansone A, Malachowska B, Zurawska-Klis M, Ferreri C, Chatgilialoglu C, Ochedalski T, Cypryk K, Wozniak LA. Lipid profile changes in erythrocyte membranes of women with diagnosed GDM. PLoS One. 2018;13:e0203799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 55. | Lai M, Liu Y, Ronnett GV, Wu A, Cox BJ, Dai FF, Röst HL, Gunderson EP, Wheeler MB. Amino acid and lipid metabolism in post-gestational diabetes and progression to type 2 diabetes: A metabolic profiling study. PLoS Med. 2020;17:e1003112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 56. | Shokry E, Marchioro L, Uhl O, Bermúdez MG, García-Santos JA, Segura MT, Campoy C, Koletzko B. Impact of maternal BMI and gestational diabetes mellitus on maternal and cord blood metabolome: results from the PREOBE cohort study. Acta Diabetol. 2019;56:421-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 57. | Zhao H, Li H, Chung ACK, Xiang L, Li X, Zheng Y, Luan H, Zhu L, Liu W, Peng Y, Zhao Y, Xu S, Li Y, Cai Z. Large-Scale Longitudinal Metabolomics Study Reveals Different Trimester-Specific Alterations of Metabolites in Relation to Gestational Diabetes Mellitus. J Proteome Res. 2019;18:292-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Liu X, Wang X, Sun H, Guo Z, Liu X, Yuan T, Fu Y, Tang X, Li J, Sun W, Zhao W. Urinary metabolic variation analysis during pregnancy and application in Gestational Diabetes Mellitus and spontaneous abortion biomarker discovery. Sci Rep. 2019;9:2605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 59. | Allalou A, Nalla A, Prentice KJ, Liu Y, Zhang M, Dai FF, Ning X, Osborne LR, Cox BJ, Gunderson EP, Wheeler MB. A Predictive Metabolic Signature for the Transition From Gestational Diabetes Mellitus to Type 2 Diabetes. Diabetes. 2016;65:2529-2539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 60. | Menni C, Fauman E, Erte I, Perry JR, Kastenmüller G, Shin SY, Petersen AK, Hyde C, Psatha M, Ward KJ, Yuan W, Milburn M, Palmer CN, Frayling TM, Trimmer J, Bell JT, Gieger C, Mohney RP, Brosnan MJ, Suhre K, Soranzo N, Spector TD. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270-4276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 336] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 61. | Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes HW, Hrabé de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 477] [Cited by in RCA: 442] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 62. | Iacobazzi V, Infantino V. Citrate--new functions for an old metabolite. Biol Chem. 2014;395:387-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 211] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 63. | Gralka E, Luchinat C, Tenori L, Ernst B, Thurnheer M, Schultes B. Metabolomic fingerprint of severe obesity is dynamically affected by bariatric surgery in a procedure-dependent manner. Am J Clin Nutr. 2015;102:1313-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 64. | Maresh M, Gillmer MD, Beard RW, Alderson CS, Bloxham BA, Elkeles RS. The effect of diet and insulin on metabolic profiles of women with gestational diabetes mellitus. Diabetes. 1985;34 Suppl 2:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 65. | Pappa KI, Anagnou NP, Salamalekis E, Bikouvarakis S, Maropoulos G, Anogianaki N, Evangeliou A, Koumantakis E. Gestational diabetes exhibits lack of carnitine deficiency despite relatively low carnitine levels and alterations in ketogenesis. J Matern Fetal Neonatal Med. 2005;17:63-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 66. | Scholtens DM, Muehlbauer MJ, Daya NR, Stevens RD, Dyer AR, Lowe LP, Metzger BE, Newgard CB, Bain JR, Lowe WL Jr; HAPO Study Cooperative Research Group. Metabolomics reveals broad-scale metabolic perturbations in hyperglycemic mothers during pregnancy. Diabetes Care. 2014;37:158-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 67. | White SL, Pasupathy D, Sattar N, Nelson SM, Lawlor DA, Briley AL, Seed PT, Welsh P, Poston L; UPBEAT Consortium. Metabolic profiling of gestational diabetes in obese women during pregnancy. Diabetologia. 2017;60:1903-1912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 68. | Taylor K, Ferreira DLS, West J, Yang T, Caputo M, Lawlor DA. Differences in Pregnancy Metabolic Profiles and Their Determinants between White European and South Asian Women: Findings from the Born in Bradford Cohort. Metabolites. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 69. | Dudzik D, Zorawski M, Skotnicki M, Zarzycki W, Kozlowska G, Bibik-Malinowska K, Vallejo M, García A, Barbas C, Ramos MP. Metabolic fingerprint of Gestational Diabetes Mellitus. J Proteomics. 2014;103:57-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 70. | Zeng Z, Liu F, Li S. Metabolic Adaptations in Pregnancy: A Review. Ann Nutr Metab. 2017;70:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 151] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 71. | Bronisz A, Ozorowski M, Hagner-Derengowska M. Pregnancy Ketonemia and Development of the Fetal Central Nervous System. Int J Endocrinol. 2018;2018:1242901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 72. | Qiu C, Enquobahrie DA, Frederick IO, Sorensen TK, Fernandez MA, David RM, Bralley JA, Williams MA. Early pregnancy urinary biomarkers of fatty acid and carbohydrate metabolism in pregnancies complicated by gestational diabetes. Diabetes Res Clin Pract. 2014;104:393-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 73. | Tai ES, Tan ML, Stevens RD, Low YL, Muehlbauer MJ, Goh DL, Ilkayeva OR, Wenner BR, Bain JR, Lee JJ, Lim SC, Khoo CM, Shah SH, Newgard CB. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia. 2010;53:757-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 74. | Würtz P, Tiainen M, Mäkinen VP, Kangas AJ, Soininen P, Saltevo J, Keinänen-Kiukaanniemi S, Mäntyselkä P, Lehtimäki T, Laakso M, Jula A, Kähönen M, Vanhala M, Ala-Korpela M. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35:1749-1756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 171] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 75. | Felig P, Marliss E, Cahill GF Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 632] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 76. | Enquobahrie DA, Denis M, Tadesse MG, Gelaye B, Ressom HW, Williams MA. Maternal Early Pregnancy Serum Metabolites and Risk of Gestational Diabetes Mellitus. J Clin Endocrinol Metab. 2015;100:4348-4356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 77. | Bentley-Lewis R, Huynh J, Xiong G, Lee H, Wenger J, Clish C, Nathan D, Thadhani R, Gerszten R. Metabolomic profiling in the prediction of gestational diabetes mellitus. Diabetologia. 2015;58:1329-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 78. | Nevalainen J, Sairanen M, Appelblom H, Gissler M, Timonen S, Ryynänen M. First-Trimester Maternal Serum Amino Acids and Acylcarnitines Are Significant Predictors of Gestational Diabetes. Rev Diabet Stud. 2016;13:236-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Chen L, Cheng CY, Choi H, Ikram MK, Sabanayagam C, Tan GS, Tian D, Zhang L, Venkatesan G, Tai ES, Wang JJ, Mitchell P, Cheung CM, Beuerman RW, Zhou L, Chan EC, Wong TY. Plasma Metabonomic Profiling of Diabetic Retinopathy. Diabetes. 2016;65:1099-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 80. | Leitner M, Fragner L, Danner S, Holeschofsky N, Leitner K, Tischler S, Doerfler H, Bachmann G, Sun X, Jaeger W, Kautzky-Willer A, Weckwerth W. Combined Metabolomic Analysis of Plasma and Urine Reveals AHBA, Tryptophan and Serotonin Metabolism as Potential Risk Factors in Gestational Diabetes Mellitus (GDM). Front Mol Biosci. 2017;4:84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 81. | Roy C, Tremblay PY, Anassour-Laouan-Sidi E, Lucas M, Forest JC, Giguère Y, Ayotte P. Risk of gestational diabetes mellitus in relation to plasma concentrations of amino acids and acylcarnitines: A nested case-control study. Diabetes Res Clin Pract. 2018;140:183-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 82. | Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol. 2014;2:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 209] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 83. | Petrović O, Belci D. A critical appraisal and potentially new conceptual approach to screening and diagnosis of gestational diabetes. J Obstet Gynaecol. 2017;37:691-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 84. | Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O'Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448-453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2618] [Cited by in RCA: 2436] [Article Influence: 174.0] [Reference Citation Analysis (0)] |
| 85. | Chen T, Ni Y, Ma X, Bao Y, Liu J, Huang F, Hu C, Xie G, Zhao A, Jia W. Branched-chain and aromatic amino acid profiles and diabetes risk in Chinese populations. Sci Rep. 2016;6:20594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 136] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 86. | Andersson-Hall U, Gustavsson C, Pedersen A, Malmodin D, Joelsson L, Holmäng A. Higher Concentrations of BCAAs and 3-HIB Are Associated with Insulin Resistance in the Transition from Gestational Diabetes to Type 2 Diabetes. J Diabetes Res. 2018;2018:4207067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 87. | Sandler V, Reisetter AC, Bain JR, Muehlbauer MJ, Nodzenski M, Stevens RD, Ilkayeva O, Lowe LP, Metzger BE, Newgard CB, Scholtens DM, Lowe WL Jr; HAPO Study Cooperative Research Group. Associations of maternal BMI and insulin resistance with the maternal metabolome and newborn outcomes. Diabetologia. 2017;60:518-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 88. | Yoon MS. The Emerging Role of Branched-Chain Amino Acids in Insulin Resistance and Metabolism. Nutrients. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 300] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 89. | Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost HG, Fritsche A, Häring HU, Hrabě de Angelis M, Peters A, Roden M, Prehn C, Wang-Sattler R, Illig T, Schulze MB, Adamski J, Boeing H, Pischon T. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 707] [Cited by in RCA: 780] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 90. | Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311-326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2638] [Cited by in RCA: 2439] [Article Influence: 152.4] [Reference Citation Analysis (0)] |
| 91. | Würtz P, Soininen P, Kangas AJ, Rönnemaa T, Lehtimäki T, Kähönen M, Viikari JS, Raitakari OT, Ala-Korpela M. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 382] [Cited by in RCA: 425] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 92. | Shah SH, Crosslin DR, Haynes CS, Nelson S, Turer CB, Stevens RD, Muehlbauer MJ, Wenner BR, Bain JR, Laferrère B, Gorroochurn P, Teixeira J, Brantley PJ, Stevens VJ, Hollis JF, Appel LJ, Lien LF, Batch B, Newgard CB, Svetkey LP. Branched-chain amino acid levels are associated with improvement in insulin resistance with weight loss. Diabetologia. 2012;55:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 277] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 93. | Palmer ND, Stevens RD, Antinozzi PA, Anderson A, Bergman RN, Wagenknecht LE, Newgard CB, Bowden DW. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab. 2015;100:E463-E468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 191] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 94. | Park S, Park JY, Lee JH, Kim SH. Plasma levels of lysine, tyrosine, and valine during pregnancy are independent risk factors of insulin resistance and gestational diabetes. Metab Syndr Relat Disord. 2015;13:64-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 95. | Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, Baca LG, Kim E, Ghosh CC, Parikh SM, Jiang A, Chu Q, Forman DE, Lecker SH, Krishnaiah S, Rabinowitz JD, Weljie AM, Baur JA, Kasper DL, Arany Z. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med. 2016;22:421-426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 437] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 96. | Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55:2565-2582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 648] [Cited by in RCA: 738] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 97. | Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 736] [Cited by in RCA: 835] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 98. | Safai N, Suvitaival T, Ali A, Spégel P, Al-Majdoub M, Carstensen B, Vestergaard H, Ridderstråle M; CIMT Trial Group. Effect of metformin on plasma metabolite profile in the Copenhagen Insulin and Metformin Therapy (CIMT) trial. Diabet Med. 2018;35:944-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 99. | Lehmann R, Friedrich T, Krebiehl G, Sonntag D, Häring HU, Fritsche A, Hennige AM. Metabolic profiles during an oral glucose tolerance test in pregnant women with and without gestational diabetes. Exp Clin Endocrinol Diabetes. 2015;123:483-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 100. | Hajduk J, Klupczynska A, Dereziński P, Matysiak J, Kokot P, Nowak DM, Gajęcka M, Nowak-Markwitz E, Kokot ZJ. A Combined Metabolomic and Proteomic Analysis of Gestational Diabetes Mellitus. Int J Mol Sci. 2015;16:30034-30045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 101. | Cetin I, de Santis MS, Taricco E, Radaelli T, Teng C, Ronzoni S, Spada E, Milani S, Pardi G. Maternal and fetal amino acid concentrations in normal pregnancies and in pregnancies with gestational diabetes mellitus. Am J Obstet Gynecol. 2005;192:610-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 102. | White PJ, Lapworth AL, An J, Wang L, McGarrah RW, Stevens RD, Ilkayeva O, George T, Muehlbauer MJ, Bain JR, Trimmer JK, Brosnan MJ, Rolph TP, Newgard CB. Branched-chain amino acid restriction in Zucker-fatty rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Mol Metab. 2016;5:538-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 209] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 103. | Smith GI, Yoshino J, Stromsdorfer KL, Klein SJ, Magkos F, Reeds DN, Klein S, Mittendorfer B. Protein Ingestion Induces Muscle Insulin Resistance Independent of Leucine-Mediated mTOR Activation. Diabetes. 2015;64:1555-1563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 104. | Macotela Y, Emanuelli B, Bång AM, Espinoza DO, Boucher J, Beebe K, Gall W, Kahn CR. Dietary leucine--an environmental modifier of insulin resistance acting on multiple levels of metabolism. PLoS One. 2011;6:e21187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 105. | Zeanandin G, Balage M, Schneider SM, Dupont J, Hébuterne X, Mothe-Satney I, Dardevet D. Differential effect of long-term leucine supplementation on skeletal muscle and adipose tissue in old rats: an insulin signaling pathway approach. Age (Dordr). 2012;34:371-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 106. | Xiao F, Yu J, Guo Y, Deng J, Li K, Du Y, Chen S, Zhu J, Sheng H, Guo F. Effects of individual branched-chain amino acids deprivation on insulin sensitivity and glucose metabolism in mice. Metabolism. 2014;63:841-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 107. | Fu L, Bruckbauer A, Li F, Cao Q, Cui X, Wu R, Shi H, Zemel MB, Xue B. Leucine amplifies the effects of metformin on insulin sensitivity and glycemic control in diet-induced obese mice. Metabolism. 2015;64:845-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 108. | Carpenter MW. Gestational diabetes, pregnancy hypertension, and late vascular disease. Diabetes Care. 2007;30 Suppl 2:S246-S250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 187] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 109. | Ebbesson SO, Tejero ME, López-Alvarenga JC, Harris WS, Ebbesson LO, Devereux RB, MacCluer JW, Wenger C, Laston S, Fabsitz RR, Howard BV, Comuzzie AG. Individual saturated fatty acids are associated with different components of insulin resistance and glucose metabolism: the GOCADAN study. Int J Circumpolar Health. 2010;69:344-351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 110. | Boden G, Shulman GI. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur J Clin Invest. 2002;32 Suppl 3:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 890] [Cited by in RCA: 901] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 111. | Mozaffarian D, Cao H, King IB, Lemaitre RN, Song X, Siscovick DS, Hotamisligil GS. Circulating palmitoleic acid and risk of metabolic abnormalities and new-onset diabetes. Am J Clin Nutr. 2010;92:1350-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 112. | Ariyama H, Kono N, Matsuda S, Inoue T, Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J Biol Chem. 2010;285:22027-22035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 113. | Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31:1509-1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 114. | Sansone A, Tolika E, Louka M, Sunda V, Deplano S, Melchiorre M, Anagnostopoulos D, Chatgilialoglu C, Formisano C, Di Micco R, Faraone Mennella MR, Ferreri C. Hexadecenoic Fatty Acid Isomers in Human Blood Lipids and Their Relevance for the Interpretation of Lipidomic Profiles. PLoS One. 2016;11:e0152378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 115. | Ferreri C, Masi A, Sansone A, Giacometti G, Larocca AV, Menounou G, Scanferlato R, Tortorella S, Rota D, Conti M, Deplano S, Louka M, Maranini AR, Salati A, Sunda V, Chatgilialoglu C. Fatty Acids in Membranes as Homeostatic, Metabolic and Nutritional Biomarkers: Recent Advancements in Analytics and Diagnostics. Diagnostics (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 116. | Min Y, Ghebremeskel K, Lowy C, Thomas B, Crawford MA. Adverse effect of obesity on red cell membrane arachidonic and docosahexaenoic acids in gestational diabetes. Diabetologia. 2004;47:75-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 117. | Chen X, Scholl TO, Leskiw M, Savaille J, Stein TP. Differences in maternal circulating fatty acid composition and dietary fat intake in women with gestational diabetes mellitus or mild gestational hyperglycemia. Diabetes Care. 2010;33:2049-2054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 118. | Ma W, Wu JH, Wang Q, Lemaitre RN, Mukamal KJ, Djoussé L, King IB, Song X, Biggs ML, Delaney JA, Kizer JR, Siscovick DS, Mozaffarian D. Prospective association of fatty acids in the de novo lipogenesis pathway with risk of type 2 diabetes: the Cardiovascular Health Study. Am J Clin Nutr. 2015;101:153-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 119. | Schooneman MG, Vaz FM, Houten SM, Soeters MR. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 437] [Cited by in RCA: 487] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 120. | Virmani A, Pinto L, Bauermann O, Zerelli S, Diedenhofen A, Binienda ZK, Ali SF, van der Leij FR. The Carnitine Palmitoyl Transferase (CPT) System and Possible Relevance for Neuropsychiatric and Neurological Conditions. Mol Neurobiol. 2015;52:826-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 121. | Pschera H, Larsson B, Kjaeldgaard A. Changes in fatty acid composition of cervical mucus lecithin during pregnancy. Gynecol Obstet Invest. 1989;28:118-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 122. | Yang ZH, Miyahara H, Hatanaka A. Chronic administration of palmitoleic acid reduces insulin resistance and hepatic lipid accumulation in KK-Ay Mice with genetic type 2 diabetes. Lipids Health Dis. 2011;10:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 123. | Khan SR, Mohan H, Liu Y, Batchuluun B, Gohil H, Al Rijjal D, Manialawy Y, Cox BJ, Gunderson EP, Wheeler MB. The discovery of novel predictive biomarkers and early-stage pathophysiology for the transition from gestational diabetes to type 2 diabetes. Diabetologia. 2019;62:687-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 124. | Lemaitre RN, Yu C, Hoofnagle A, Hari N, Jensen PN, Fretts AM, Umans JG, Howard BV, Sitlani CM, Siscovick DS, King IB, Sotoodehnia N, McKnight B. Circulating Sphingolipids, Insulin, HOMA-IR, and HOMA-B: The Strong Heart Family Study. Diabetes. 2018;67:1663-1672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 125. | Alshehry ZH, Mundra PA, Barlow CK, Mellett NA, Wong G, McConville MJ, Simes J, Tonkin AM, Sullivan DR, Barnes EH, Nestel PJ, Kingwell BA, Marre M, Neal B, Poulter NR, Rodgers A, Williams B, Zoungas S, Hillis GS, Chalmers J, Woodward M, Meikle PJ. Plasma Lipidomic Profiles Improve on Traditional Risk Factors for the Prediction of Cardiovascular Events in Type 2 Diabetes Mellitus. Circulation. 2016;134:1637-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 200] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 126. | Law KP, Han TL, Mao X, Zhang H. Tryptophan and purine metabolites are consistently upregulated in the urinary metabolome of patients diagnosed with gestational diabetes mellitus throughout pregnancy: A longitudinal metabolomics study of Chinese pregnant women part 2. Clin Chim Acta. 2017;468:126-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 127. | Shang M, Dong X, Hou L. Correlation of adipokines and markers of oxidative stress in women with gestational diabetes mellitus and their newborns. J Obstet Gynaecol Res. 2018;44:637-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 128. | Biri A, Onan A, Devrim E, Babacan F, Kavutcu M, Durak I. Oxidant status in maternal and cord plasma and placental tissue in gestational diabetes. Placenta. 2006;27:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 129. | Zhu Y, Hu Y, Huang T, Zhang Y, Li Z, Luo C, Luo Y, Yuan H, Hisatome I, Yamamoto T, Cheng J. High uric acid directly inhibits insulin signalling and induces insulin resistance. Biochem Biophys Res Commun. 2014;447:707-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 213] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 130. | Wolak T, Sergienko R, Wiznitzer A, Paran E, Sheiner E. High uric acid level during the first 20 wk of pregnancy is associated with higher risk for gestational diabetes mellitus and mild preeclampsia. Hypertens Pregnancy. 2012;31:307-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 131. | van der Schaft N, Brahimaj A, Wen KX, Franco OH, Dehghan A. The association between serum uric acid and the incidence of prediabetes and type 2 diabetes mellitus: The Rotterdam Study. PLoS One. 2017;12:e0179482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 132. | Connelly MA, Otvos JD, Shalaurova I, Playford MP, Mehta NN. GlycA, a novel biomarker of systemic inflammation and cardiovascular disease risk. J Transl Med. 2017;15:219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 133. | Lorenzo C, Festa A, Hanley AJ, Rewers MJ, Escalante A, Haffner SM. Novel Protein Glycan-Derived Markers of Systemic Inflammation and C-Reactive Protein in Relation to Glycemia, Insulin Resistance, and Insulin Secretion. Diabetes Care. 2017;40:375-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 134. | Akinkuolie AO, Buring JE, Ridker PM, Mora S. A novel protein glycan biomarker and future cardiovascular disease events. J Am Heart Assoc. 2014;3:e001221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 182] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 135. | Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1580] [Article Influence: 83.2] [Reference Citation Analysis (0)] |
| 136. | Li J, Huo X, Cao YF, Li SN, Du Z, Shao P, Leng J, Zhang C, Sun XY, Ma RCW, Fang ZZ, Yang X. Bile acid metabolites in early pregnancy and risk of gestational diabetes in Chinese women: A nested case-control study. EBioMedicine. 2018;35:317-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 137. | Huo X, Li J, Cao YF, Li SN, Shao P, Leng J, Li W, Liu J, Yang K, Ma RCW, Hu G, Fang ZZ, Yang X. Trimethylamine N-Oxide Metabolites in Early Pregnancy and Risk of Gestational Diabetes: A Nested Case-Control Study. J Clin Endocrinol Metab. 2019;104:5529-5539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 138. | Velasquez MT, Ramezani A, Manal A, Raj DS. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins (Basel). 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 360] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 139. | Canyelles M, Tondo M, Cedó L, Farràs M, Escolà-Gil JC, Blanco-Vaca F. Trimethylamine N-Oxide: A Link among Diet, Gut Microbiota, Gene Regulation of Liver and Intestine Cholesterol Homeostasis and HDL Function. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 140. | Rothman KJ, Greenland S. Causation and causal inference in epidemiology. Am J Public Health. 2005;95 Suppl 1:S144-S150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 695] [Cited by in RCA: 688] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 141. | Cătoi AF, Pârvu A, Mureşan A, Busetto L. Metabolic Mechanisms in Obesity and Type 2 Diabetes: Insights from Bariatric/Metabolic Surgery. Obes Facts. 2015;8:350-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 142. | Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32 Suppl 2:S237-S245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 291] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 143. | Huhtala MS, Tertti K, Pellonperä O, Rönnemaa T. Amino acid profile in women with gestational diabetes mellitus treated with metformin or insulin. Diabetes Res Clin Pract. 2018;146:8-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 144. | White SL, Begum S, Vieira MC, Seed P, Lawlor DL, Sattar N, Nelson SM, Welsh P, Pasupathy D, Poston L; UPBEAT Consortium. Metabolic phenotyping by treatment modality in obese women with gestational diabetes suggests diverse pathophysiology: An exploratory study. PLoS One. 2020;15:e0230658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 145. | Wang Q, Würtz P, Auro K, Mäkinen VP, Kangas AJ, Soininen P, Tiainen M, Tynkkynen T, Jokelainen J, Santalahti K, Salmi M, Blankenberg S, Zeller T, Viikari J, Kähönen M, Lehtimäki T, Salomaa V, Perola M, Jalkanen S, Järvelin MR, Raitakari OT, Kettunen J, Lawlor DA, Ala-Korpela M. Metabolic profiling of pregnancy: cross-sectional and longitudinal evidence. BMC Med. 2016;14:205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 134] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 146. | Cosson E, Bihan H, Reach G, Vittaz L, Carbillon L, Valensi P. Psychosocial deprivation in women with gestational diabetes mellitus is associated with poor fetomaternal prognoses: an observational study. BMJ Open. 2015;5:e007120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 147. | Habran A, Commisso M, Helwi P, Hilbert G, Negri S, Ollat N, Gomès E, van Leeuwen C, Guzzo F, Delrot S. Roostocks/Scion/Nitrogen Interactions Affect Secondary Metabolism in the Grape Berry. Front Plant Sci. 2016;7:1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 148. | Crowther LM, Poms M, Plecko B. Multiomics tools for the diagnosis and treatment of rare neurological disease. J Inherit Metab Dis. 2018;41:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 149. | Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 655] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 150. | Shrestha A, Müllner E, Poutanen K, Mykkänen H, Moazzami AA. Metabolic changes in serum metabolome in response to a meal. Eur J Nutr. 2017;56:671-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |