Published online Jul 15, 2021. doi: 10.4239/wjd.v12.i7.1116
Peer-review started: March 2, 2021
First decision: April 6, 2021
Revised: April 11, 2021
Accepted: May 20, 2021
Article in press: May 20, 2021
Published online: July 15, 2021
Processing time: 132 Days and 1.6 Hours
As one of the major microvascular complications of diabetes, diabetic retinopathy (DR) is the leading cause of blindness in the working age population. Because the extremely complex pathogenesis of DR has not been fully clarified, the occurrence and development of DR is closely related to tissue ischemia and hypoxia and neovascularization The formation of retinal neovascularization (RNV) has great harm to the visual acuity of patients.
To investigate the expression of P-element-induced wimpy testis-interacting RNA (piRNA) in proliferative DR mice and select piRNA related to RNV.
One hundred healthy C57BL/6J mice were randomly divided into a normal group as control group (CG) and proliferative DR (PDR) group as experimental group (EG), with 50 mice in each group. Samples were collected from both groups at the same time, and the lesions of mice were evaluated by hematoxylin and eosin staining and retinal blood vessel staining. The retinal tissues were collected for second-generation high-throughput sequencing, and the differentially expressed piRNA between the CG and EG was detected, and polymerase chain reaction (PCR) was conducted for verification. The differentially obtained piRNA target genes and expression profiles were enrichment analysis based on gene annotation (Gene Ontology) and Kyoto Encyclopedia of Genes and Genomes.
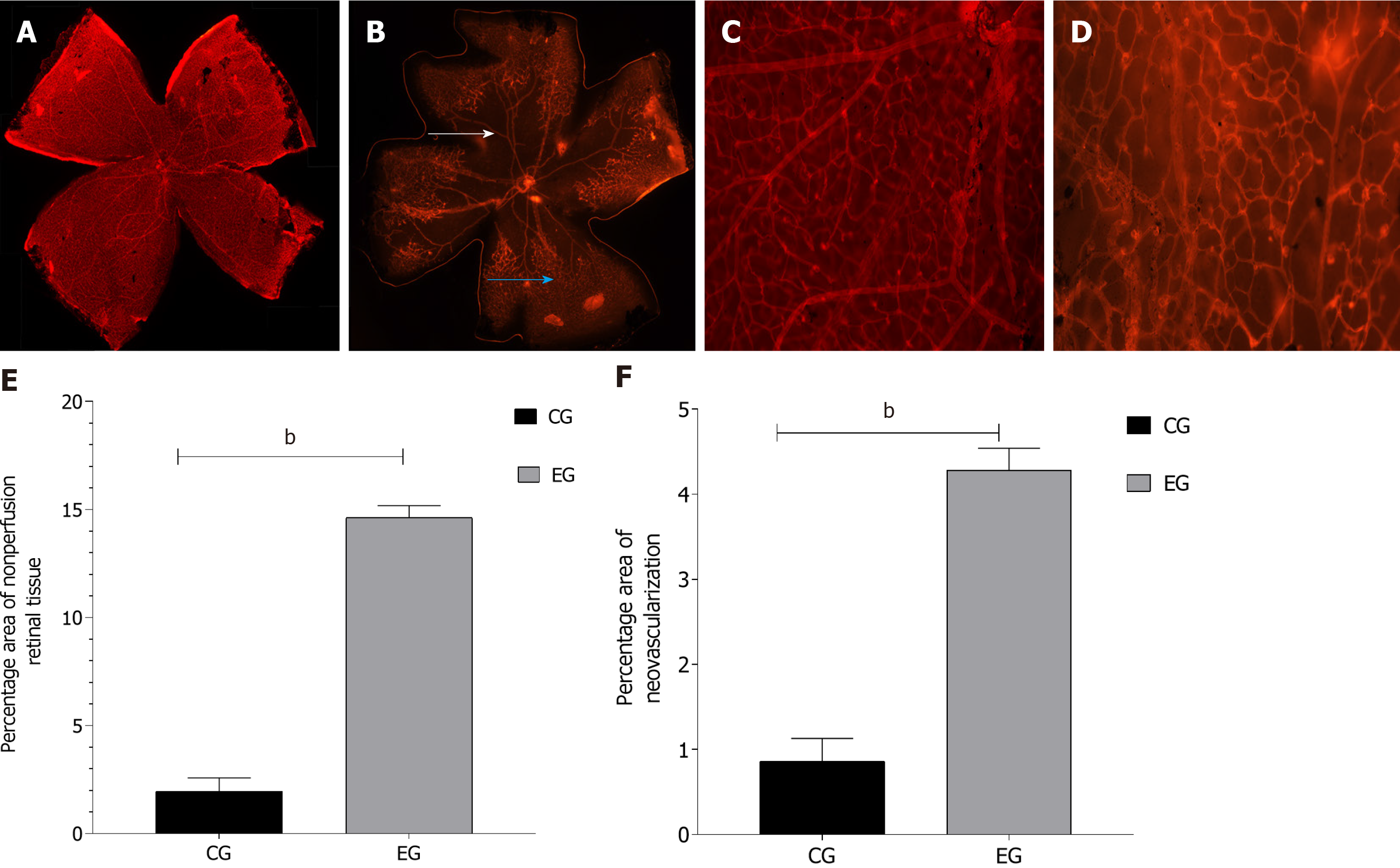
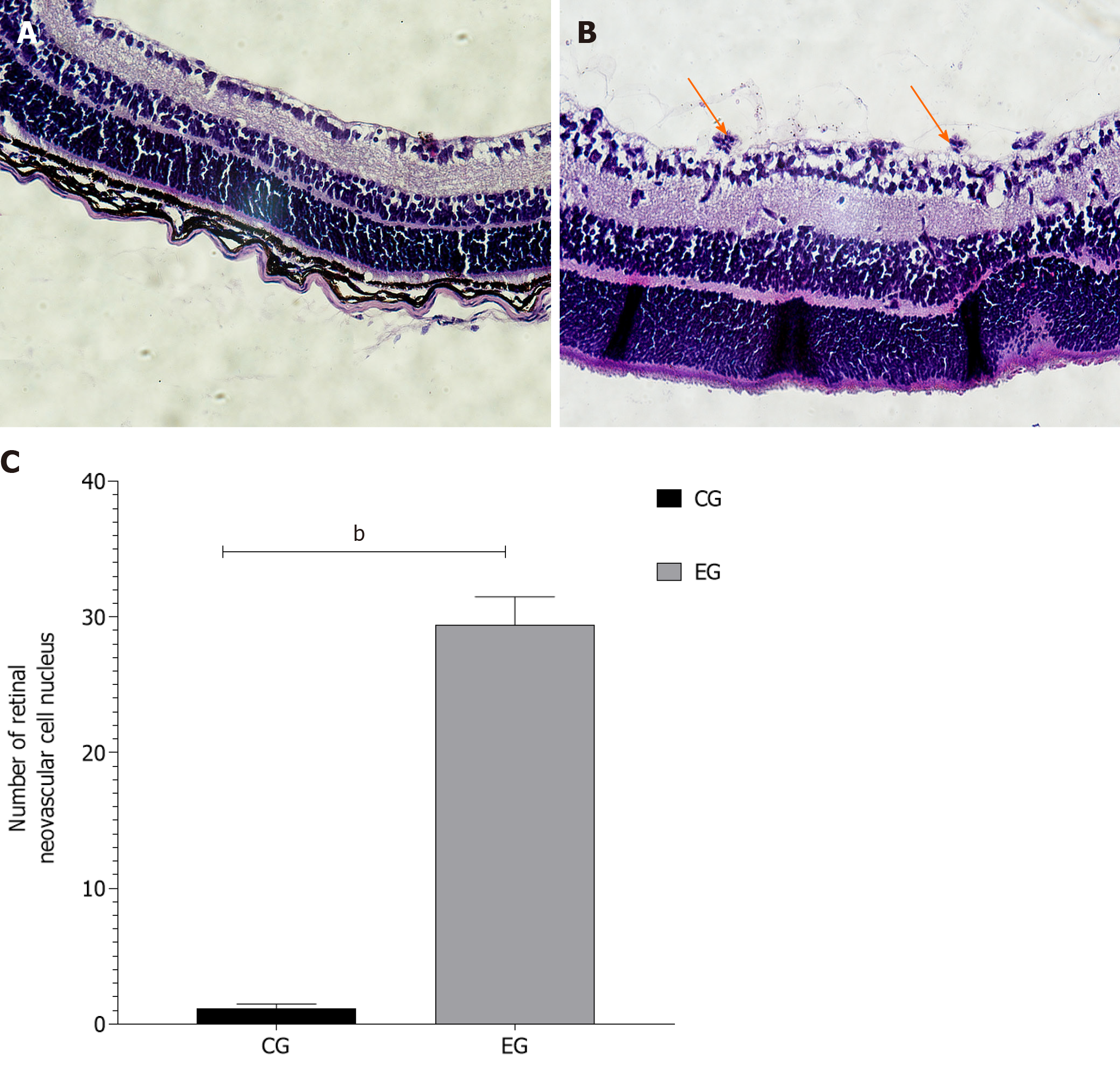
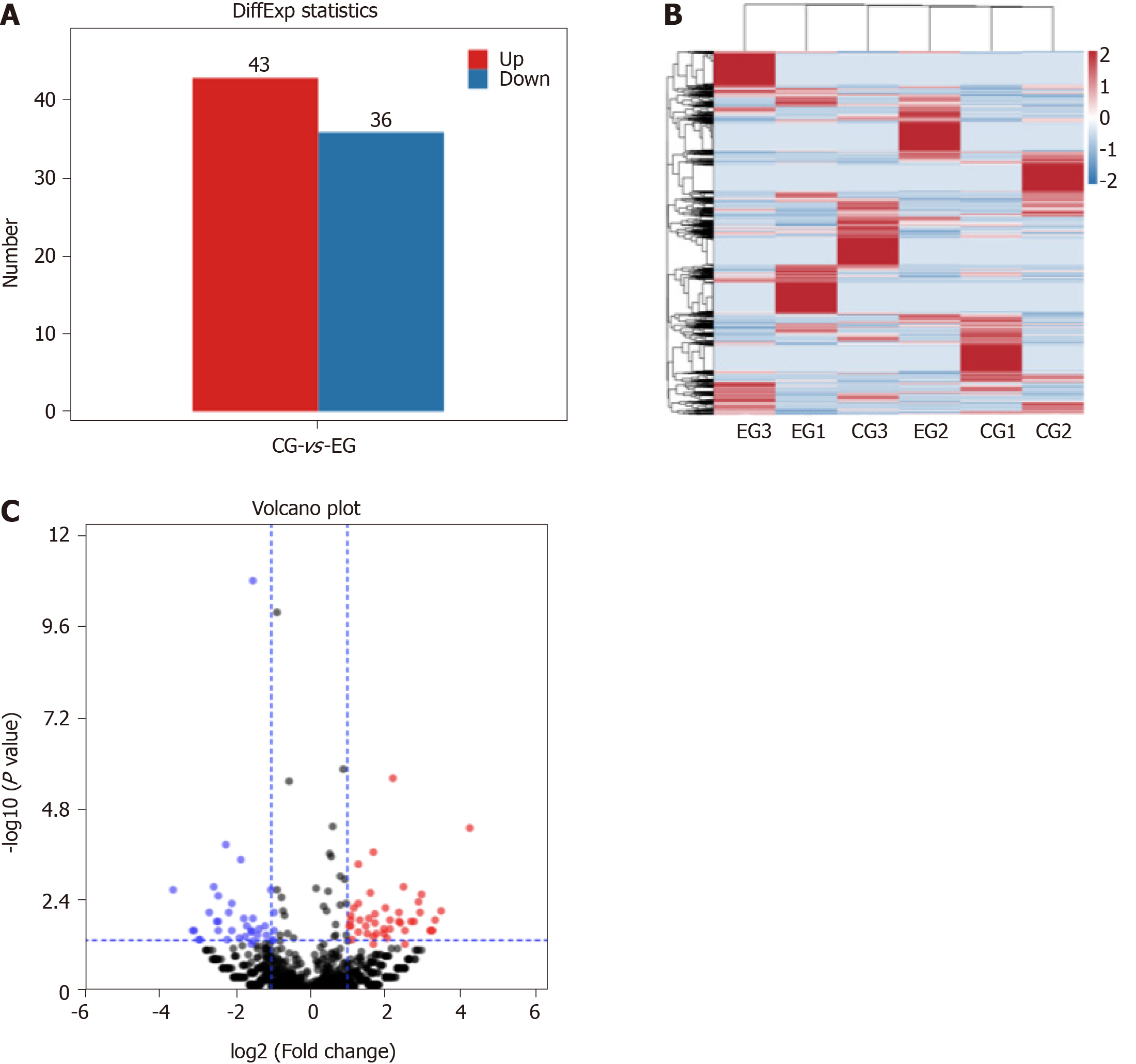
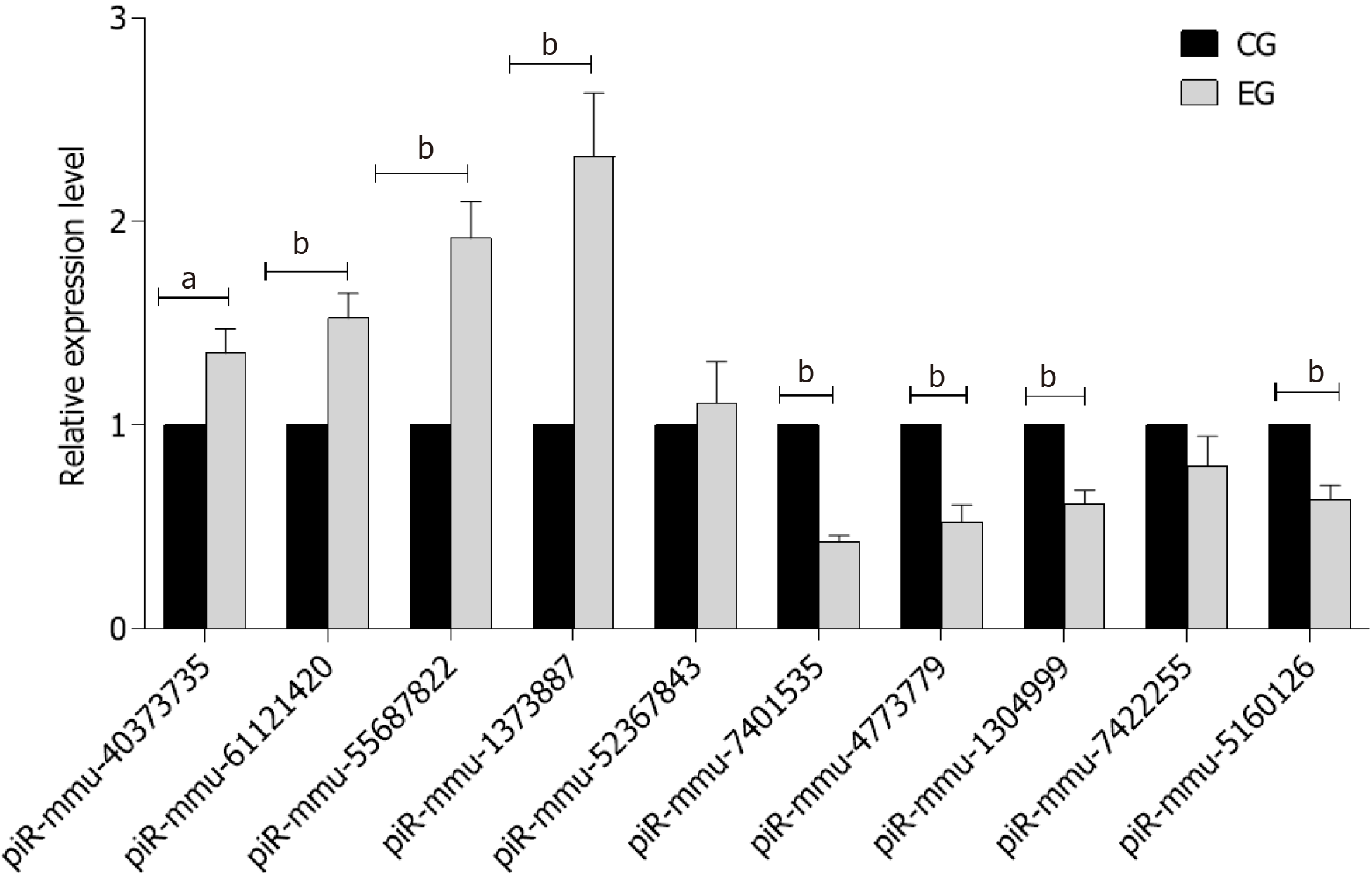
In the CG there was no perfusion area, neovascularization and endothelial nucleus broke through the inner boundary membrane of retinap. In the EG, there were a lot of nonperfused areas, new blood vessels and endothelial nuclei breaking through the inner boundary membrane of the retina. There was a statistically significant difference in the number of vascular endothelial nuclei breaking through the inner retinal membrane between the two groups. High-throughput sequencing analysis showed that compared with the CG, a total of 79 piRNAs were differentially expressed in EG, among which 43 piRNAs were up-regulated and 36 piRNAs were down-regulated. Bioinformatics analysis showed that the differentially expressed piRNAs were mainly concentrated in the signaling pathways of angiogenesis and cell proliferation. Ten piRNAs were selected for PCR, and the results showed that the expression of piR-MMU-40373735, piR-MMU-61121420, piR-MMU-55687822, piR-MMU-1373887 were high, and the expression of piR-MMU-7401535, piR-MMU-4773779, piR-MMU-1304999, and piR-MMU-5160126 were low, which were consistent with the sequencing results.
In the EG, the abnormal expression of piRNA is involved in the pathway of angiogenesis and cell proliferation, suggesting that piRNAs have some regulatory function in proliferative diabetic-retinopathy.
Core Tip: As one of the major microvascular complications of diabetes, diabetic retinopathy is the leading cause of blindness in the working age population. In the diabetic retinopathy model, the abnormal expression of P-element-induced wimpy testis-interacting RNAs (piRNAs) are involved in angiogenesis and cell proliferation pathways, suggesting that piRNAs have a certain regulatory function in diabetic retinopathy.
- Citation: Yu Y, Ren KM, Chen XL. Expression and role of P-element-induced wimpy testis-interacting RNA in diabetic-retinopathy in mice. World J Diabetes 2021; 12(7): 1116-1130
- URL: https://www.wjgnet.com/1948-9358/full/v12/i7/1116.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i7.1116
As one of the major microvascular complications of diabetes, diabetic retinopathy (DR) is the leading cause of blindness in working age population. Because the extremely complex pathogenesis of DR has not been fully clarified, the occurrence and development of DR is closely related to tissue ischemia and hypoxia and neovascularization[1,2]. The formation of retinal neovascularization (RNV) has great harm to the visual acuity of patients. Patients who are refractory to treatment may experience serious complications, such as neovinal glaucoma, vitreous hemorrhagia, and retinal detachment which can lead to permanent blindness. RNV is a key link in proliferative DR (PDR) and is a complex pathological process. Intravitreal injection of anti-vascular endothelial growth factor (VEGF) is an effective method for treating ocular neovascular diseases. However, the procedure can result in complications, side effects, and only short-term effects. Previous studies reported that anti-VEGF treatment cannot prevent the formation of RNV in some patients[3,4]. Therefore, it is particularly important to find other VEGF-independent pathways that promote angiogenesis to identify safe therapeutic targets against DR.
P-element-induced wimpy testis (PIWI)-interacting RNA (piRNA) was first found in the ovarian germ cells of Drosophila melanogaster in 2006. This novel short non-coding small RNA has an average length of 26-31 nucleotides[5] and functions by binding with PIWI protein to form a piRNA-induced silencing complex (PIRISC)[6-8]. piRNA plays an important role in transposon silencing, epigenetic regulation, protein regulation of genome rearrangement, spermatogenesis, and germ stem cell maintenance[9,10]. Additionally, piRNA participates in tumor formation, human aging, and neural axon regeneration[11-13]. Based on existing studies and literature reports, which highlight the role of piRNA in neovascularization-related diseases[14,15], piRNA may play an important role in the formation and development of DR. However, to the best of our knowledge, no previous study has explored this hypothesis. In the present study, the role of piRNA in RNV diseases was evaluated by sequencing of RNA obtained from the retinal tissues of mice with PDR mice and normal mice. The study aimed to provide theoretical support for the possible alternative clinical treatment of DR.
We used 100 7-d-old C57 mice of either sex in this study. All experimental animals were purchased from Changsheng Biology Co., Ltd. (Shenyang, China). The animals were fed and related operations were conducted in the animal laboratory of Shengjing Hospital in a stable, specific pathogen-free environment; the temperature was maintained at 23 ± 2 °C, with a 12 h light cycle. The experimental study was approved by the animal ethics committee of Shengjing Hospital of China Medical University (2020PS078K).
The animals were randomly divided into two groups, each consisting of 50 animals: normal group as control group (CG) and PDR group as experimental group (EG). Mice in CG were fed with mice and their mothers under normal conditions without any treatment. Briefly, 7-d-old EG mice and their mothers were housed and fed in a closed glass container with an oxygen concentration of 75% ± 2% for 5 d. Next, 12-d-old mice were fed under normal conditions, and the closed container was opened once per day to replace the bedding material, add water, and replace the mother mice[16,17]. All animals were euthanized at the age of 17 d for subsequent histopathological examination and total RNA extraction.
Mice aged 17 d from the two groups were anesthetized and sacrificed, after which their eyeballs were removed. The eyeballs were fixed in 4% paraformaldehyde for 12 h. The contents of the anterior segment and vitreous cavity were removed, and the retina was carefully separated. The retinas were incubated in isolectin B4-594 (Invitrogen, Carlsbad, CA, United States) in a shaker at 4 °C overnight. Next, the glass slide was covered with an anti-radiation agent, and this agent was also applied to the retina[18]. The retinas were observed under a microscope (Eclipse NI, Nikon, Tokyo, Japan) and images were collected.
After the eyeballs of mice were removed, they were fixed in 4% paraformaldehyde for 12 h. Next, they were embedded in paraffin and cut along the sagittal plane of the optic nerve to obtain serial sections with a thickness of 4 μm. Non-continuous sections were acquired from each eye for hematoxylin and eosin (HE) staining, and sections containing the optic nerve were excluded. The number of endothelial cells breaking through the inner limiting membrane in the vitreous cavity was counted, and the average of this number was calculated in each section[19]. Only the vascular nuclei located closely to the retina were counted; thus, vascular nuclei not close to the internal limiting membrane in the vitreous cavity were excluded[20,21].
The total RNA of each retinal tissue sample was extracted using Trizol reagent according to the manufacturer’s instructions (Takara, Japan). The quality of RNA was analyzed using NanoDrop ND-2000 (Thermo Fisher Scientific, United States). A small RNA library was constructed by real-time polymerase chain reaction (RT-PCR) using 5' and 3' linkers. Agilent 2100 and Applied Biosystems StepOnePlus Real-Time PCR systems were used to assess the quality and yield of the constructed library (Life Technologies). Finally, the RNA was sequenced by Illumina Hiseq 2000 (Illumina, San Diego, CA, United States).
After standardization and quality control of the sequencing data, 26-31 nt piRNAs were selected from small RNA reads, and the differential expression of piRNA was analyzed. Fold change in piRNA expression ≥ 1.5 (P < 0.05) was used as the threshold for determining gene upregulation or downregulation. Gene Ontology (GO) enrichment (http://www.geneontology.org/) was used to analyze the abnormal expression of genes, and KO enrichment (https://www.genome.jp/kegg/pathway) was utilized to determine the biological function of the differentially expressed piRNA and investigate its possible involvement in the disease mechanism[22].
RT-PCR was performed on total RNA extracted from retina samples using Trizol reagent (Takara, Shiga, Japan) according to the manufacturer’s instructions.
SPSS 22.0 software (SPSS, Inc., Chicago, IL, United States) was used for all statistical analyses. The mean ± SD of relative piRNA expression in PCR analyses was calculated. Student's t-test was used to analyze the difference between the two groups. P < 0.05 indicated a statistically significant difference.
The samples obtained from the normal control and EGs of mice were stained with isolectin B4-594 to observe the retinal vascular structure. In the normal CG, the retinal blood vessels were intact and clear, and large blood vessels were characterized by an even radial distribution around the optic disc reaching the periphery of the retina. In the DR group, there was no perfusion in the large vessel area and no decomposition of the perfusion area. The large vessels near the optic disc were tortuous and irregular, and they were mainly visible in the middle and periphery. A large number of disordered new vessels and new vascular buds were detected at the retina boundary along with vascular leakage (Figure 1).
HE staining images were used to quantitatively analyze the number of retinal vascular endothelial nuclei. In the CG, the structure of the retinal internal limiting membrane was intact and smooth with occasional vascular endothelial nuclei breaking through the internal limiting membrane on the vitreous side (average number 1.163 ± 0.31). In the EG, the morphology of the internal limiting membrane was irregular, and cells under the internal limiting membrane proliferated and were arranged in a disorderly manner. A large number of clusters of vascular endothelial cells broke through the inner limiting membrane and formed the neovascular lumen (average number 29.42 ± 1.07). There was a significant difference in the number of retinal vascular endothelial cells that broke through the inner limiting membrane between the two groups (P < 0.01) (Figure 2).
There were 79 piRNAs differentially expressed in EG compared to in CG mice, among which 43 were upregulated and 36 were downregulated (Figure 3).
We selected 10 differentially expressed genes according to their gene expression levels determined by high-throughput sequencing and verified the expression levels in two groups of retina samples by RT-PCR. The primer sequences used in this study are shown in Table 1. The results of quantitative verification by RT-PCR are shown in Figure 4.
| Gene ID | Fold-change | Up/down | Primer (5′-3′) |
| piR-mmu-40373735 | 17.4426 | Up | F: CCGGACCTCAAGCAGCC; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-61121420 | 17.5967 | Up | F: GGCCAGCCGGGGTACA; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-55687822 | 17.5982 | Up | F: CGCACTGCTTCACTTGACCAG; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-1373887 | 18.0930 | Up | F: AATGATGAACCTTTTGACGGG; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-52367843 | 18.4806 | Up | F: CGAGGACAGCCTGGTCTACACA; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-7401535 | -18.5696 | Down | F: CAGCCCTCGACACAAGGG; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-4773779 | -18.2665 | Down | F: GCACCATGATGACGGAAATT; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-1304999 | -17.8006 | Down | F: GCATGCAGGAGCATCAGTAGAC; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-7422255 | -17.5913 | Down | F: CCACATGATGATCCATAACGAGAT; R: AGTGCAGGGTCCGAGGTATT |
| piR-mmu-5160126 | -17.4335 | Down | F: GCTGAGACAGGAGGATCGCT; R: AGTGCAGGGTCCGAGGTATT |
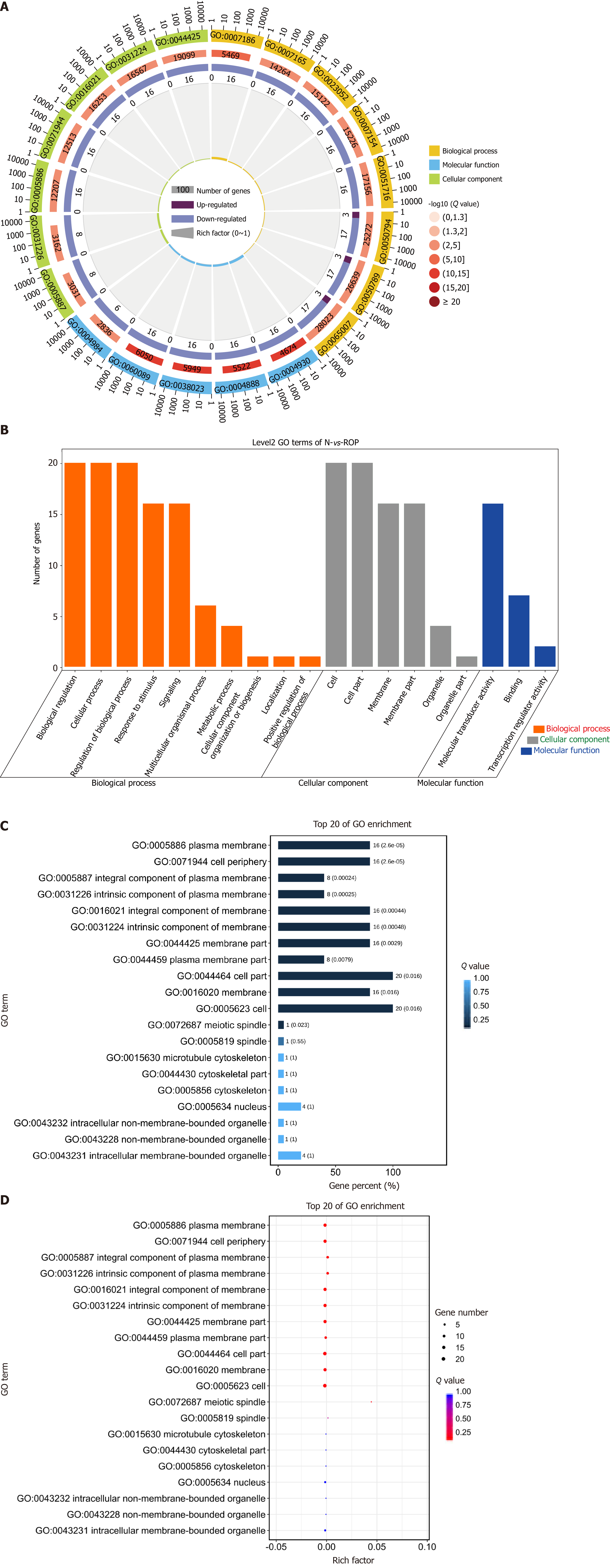
To further analyze the biological functions of differentially expressed genes in EG and CG mice, the GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses was performed. The results showed that the differentially expressed piRNAs were involved in many biological processes, such as sensory development, G-protein-coupled receptor signaling pathway, regulation of inflammatory factors, and visual development, which are potentially related to RNV (Figure 5).
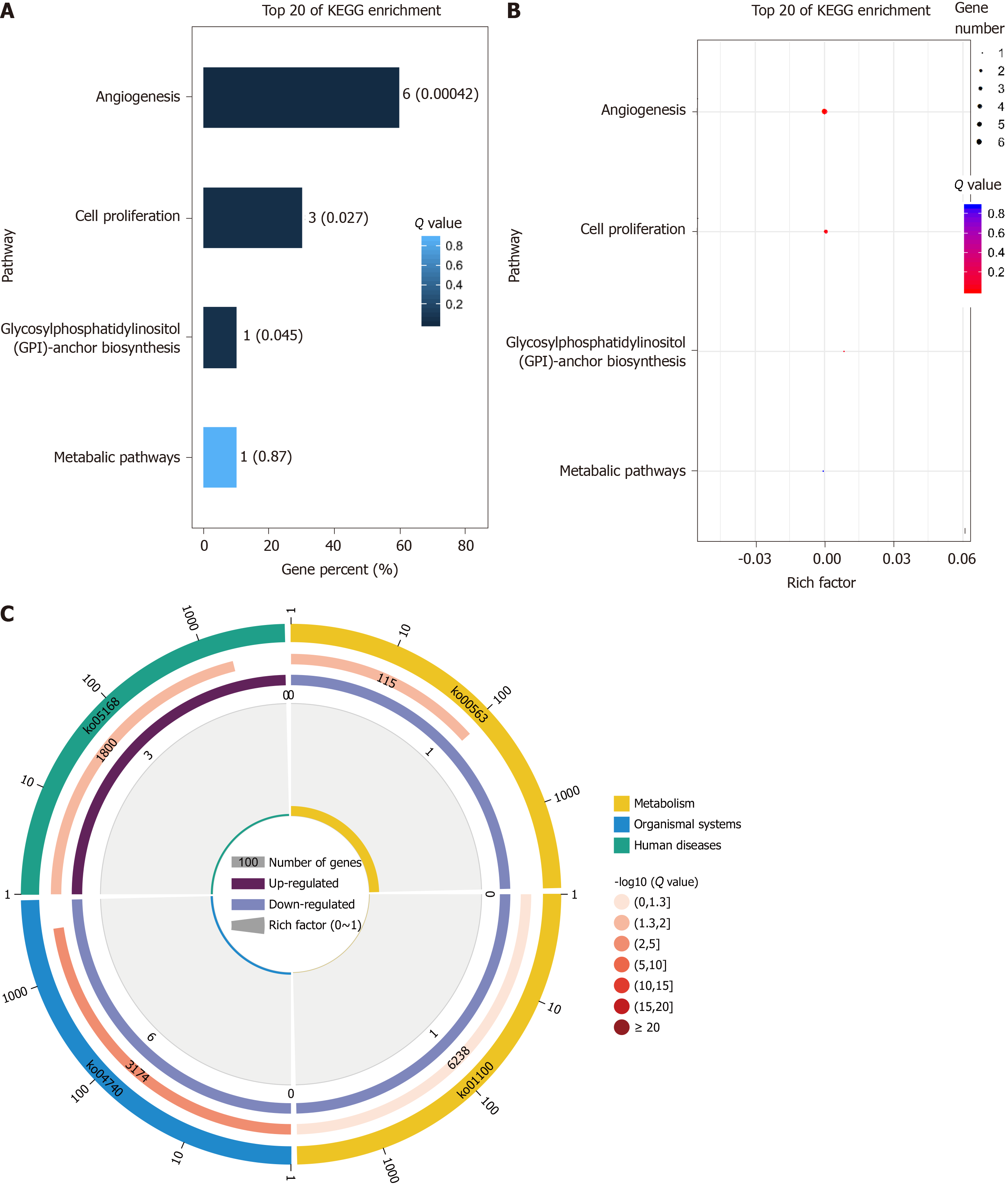
KEGG analysis of differentially expressed genes and proteins showed that the enriched pathways were mainly related to angiogenesis and cell proliferation and were strongly correlated with RNV (Figure 6).
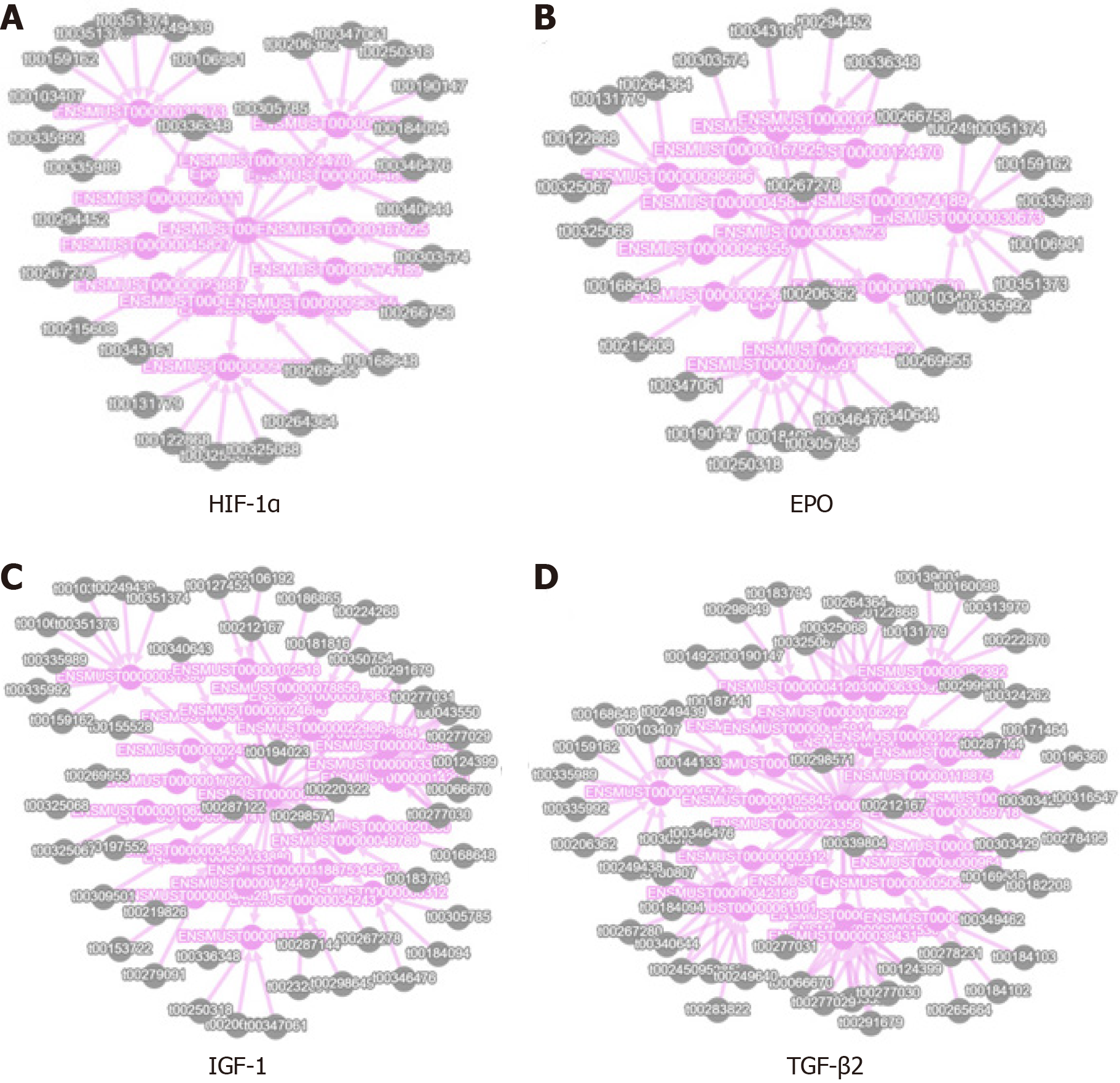
We obtained piRNA-target gene pairs correlated with expression level to construct the network analysis diagram. We selected the EPO, HIF-1α, IGF1, and TGF-β2 genes, which are potentially related to RNV, to construct the interaction analysis diagram (Figure 7).
Non-coding RNA is involved in the occurrence and development of several diseases. Numerous studies have shown that non-coding RNA is an important factor in the pathophysiological changes leading to malignant tumors and vascular diseases[23]. piRNA is a type of small non-coding RNA with an important role in animal development, reproduction, and gene regulation[10,24]. To date, 23439 piRNAs have been identified in the human genome, which is equivalent to the number of proteins encoded by mRNA (about 20000) and more than the number of miRNAs. These numbers indicate that piRNAs play an important role in regulating gene transcription[25,26].
After transcription, PIWI protein cleavage and amplification as well as piRNA clusters eventually form mature piRNAs having biological activity by forming PIRISC[27]. PIRISC, a silencing complex formed by piRNA and PIWI protein, can inhibit its target by transcriptional gene silencing and post-transcriptional gene silencing to maintain the genomic integrity of the germline[27,28]. PIRSIC regulates transposon expression at the transcriptional level by inducing epigenetic repression via histone H3K9me3 and DNA methylation[29]. Some studies report a larger number of allowed mismatches between target mRNA and piRNA as opposed to between target mRNA and miRNA[30]. siRNA and miRNA are easily and rapidly degraded by nucleases, whereas piRNA is relatively stable in the serum, and thus has the potential to serve as a marker for diagnosis and prediction of disease progression[31].
piRNAs are presently thought to act mainly in somatic cells and cancer tissues and are involved in cell proliferation, apoptosis, cell cycle arrest, angiogenesis, invasion, and metastasis[32]. Moreover, studies have shown that PIRISC may participate in tumorigenesis by inducing abnormal DNA methylation, which leads to genomic silencing[33]. Further, the piRNA-30473/WTAP/HK2 axis promotes the occurrence of breast cancer by regulating the methylation of m6A RNA in diffuse large B-cell lymphoma[34]. piRNA-19166 inhibits cell migration and metastasis through the cortactin/mitochondrial membrane potential pathway in prostate cancer[35]. piRNA-823 promotes the angiogenesis of endothelial cells by promoting the secretion of VEGF and interleukin -6. It also enhances the invasion of endothelial cells by inducing the expression of ICAM-1 and CXCR4[36]. Downregulation of piRNA-36712 expression is known to result in upregulation of SEPW1 expression, which in turn inhibits the expression of its downstream gene p53 and therefore it suppresses the formation of malignant tumors[37]. In addition, piRNA plays an important role in gastric cancer, liver cancer, glioma, and other diseases. Based on these correlations, piRNA is expected to serve as a new target for cancer treatment[6,10,38].
There are many models for the study of DR. At present, the model of RNV and vascular leakage without hyperglycemia has been applied. This model simulates PDR or the process to be developed from non-PDR (NPDR) to PDR. The retinopathy injury caused by hypoxia is due to the occurrence of the release of angiogenesis actors, presented with microhemangiomas, vascular leakage, venous occlusion, capillary in perfusion, neovascularization, and even vitreous hemorrhage, and retinal detachment[16]. There are notable nonperfusion areas in the center of the retina, the central great vessels are tortuous, and the number of vascular nuclei breaking through the inner limiting membrane is significantly increased, which are important factors indicating the success of the EG[39]. HE staining of retinal slices and paraffin sections of EG samples showed that there were large areas of nonperfusion, neovascularization, and vascular endothelial cells breaking through the inner limiting membrane in the retina.
DR is one of the most common microvascular diseases of diabetes and also the main cause of blindness in diabetic patients[40]. Studies have shown that DR occurs in both type 1 diabetes and type 2 diabetes[41]. With the increase in the number of diabetic patients, DR has become the main cause of visual impairment in diabetic patients[42]. The whole pathological process of DR includes important pathological changes such as loss of retinal capillary pericytes, thickening of basement membrane, loss of endothelial barrier function, destruction of blood-retinal barrier, and lead to retinal ischemia, which will increase the level of VEGF. Studies have shown that overexpression of VEGF is associated with RNV, which can cause retinal hemorrhage, macular edema, retinal detachment, and neovascularization glaucoma, etc., leading to severe visual impairment and eventually blindness.
The development of DR involves two stages: early NPDR and advanced PDR. The former is mainly characterized by increased retinal permeability and intraretinal hemorrhage, while the latter is mainly manifested by RNV. In NPDR, high glucose induced retinopathy mainly includes loss of capillary pericytes, thinning of the vascular layer and destruction of the blood-retinal barrier, which further leads to retinal ischemia and hypoxia. When the disease progresses to PDR, neovascularization occurs and eventually leads to severe visual impairment. RNV is a common pathological change in many retinopathies, including DR, retinopathy of prematurity, and age-related macular degeneration.
The common feature of clinical treatment of RNV as well as malignant tumors is treatment with targeted drugs, mainly anti-VEGF drugs. However, targeted drugs against RNV have short action time and require multiple intraocular injections, creating safety risks. Based on of the findings of previous studies on the role of piRNA in cancers and neovascularization-related diseases, we compared piRNA expression levels between the DR model and CGs. The results revealed 79 piRNAs with differential expression in EG and CG. Through GO and KEGG analysis, we established that the mRNA of the differentially expressed piRNAs was involved in processes such as angiogenesis, optic nerve development, inflammation, and proliferation of cells. Among them, EPO, HIF-1α, IGF1, TGF-β2, and other genes are closely related to angiogenesis. Interestingly, a change in EPO expression is considered as an important factor affecting the retinopathy of prematurity. EPO treatment can effectively protect the nervous system and optic nerve development of premature infants[42,43]. HIF-1α is stably expressed under hypoxia; it can regulate EPO and VEGF expression and promote RNV[44,45]. Inhibiting the VEGF/VEGFR2 and HIF-1α/VEGF signaling pathways can prevent angiogenesis[46]. TGF-β2 is a pro-inflammatory cytokine precursor related to the pathogenesis of DR. IGF1 has been identified as the direct target of miR-142-5p. It can reduce the level of miR-142-5p by activating the IGF1/IGF1R median signaling pathway (involving p-PI3K, p-ERK, p-Akt, and VEGF activation), eventually leading to cell proliferation and is involved in the pathological process of DR[47,48].
piRNA plays an important role in various diseases. By interacting with PIWI protein, piRNA can participate in cancer formation and neovascularization through DNA methylation. It can also affect the expression of target genes. We examined the differential expression of piRNA in an oxygen-induced retinopathy mouse model and the potential cellular pathways involved in this process. The study identified a set of target genes that can enhance the theoretical understanding of the role of piRNA in RNV. Because the specific mechanism of action has not been studied in detail, studies are needed to explore its mechanism in a larger sample size. Moreover, it can drive further research on new strategies for clinical treatment. We plan to predict the downstream targets of each differentially expressed piRNA and verify the predicted targets using molecular biology methods. These results provide a foundation for further exploration of the molecular mechanism underlying the development of PDR.
Abnormal expression of the piRNAs are involved in pathways of angiogenesis and cell proliferation, which suggests that piRNAs may regulate some functions in proliferative DR.
Retinal neovascularization is caused by the progression of ischemic retinal diseases, including diabetic retinopathy, retinopathy of prematurity, age-related macular degeneration, and retinal vein occlusion. Complications of retinal neovascularization can severely impair vision or lead to permanent blindness.
To explore the upstream molecules of vascular endothelial growth factor or rate-limiting steps of angiogenesis, and to reveal new approaches to the treatment of diabetic retinal.
The research on the role of piRNAs (p-Element-induced wimpy testis-interacting RNAs) in retinal neovascularization disease is expected to provide theoretical support for the clinical treatment of diabetic retina.
A diabetic retinopathy model was established. The differentially expressed piRNA was screened by high-throughput sequencing, and the differentially expressed piRNA was selected according to the sequencing results, and verified by polymerase chain reaction.
A total of 79 piRNAs were differentially expressed in experimental group, of which 43 were upregulated and 36 were down-regulated.
piRNAs were differentially expressed in DR model. Differentially expressed piRNAs is involved in the formation of retinal neovascularization. Differentially expressed piRNAs can regulate retinal development and retinal angiopathy through a variety of signaling pathways.
Drugs targeting piRNAs may be novel candidates for the treatment of diabetic retinopathy.
Manuscript source: Unsolicited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Armellini E, Fires D S-Editor: Gao CC L-Editor: Filipodia P-Editor: Wang LL
| 1. | Hou L, Shi Y, Wang S, Chen Q, Li Q, Zhao M, Zhou X. Associations of serum uric acid level with diabetic retinopathy and albuminuria in patients with type 2 diabetes mellitus. J Int Med Res. 2020;48:300060520963980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Park W, Kim J, Choi S, Kim T, Park M, Kim S, You JC, Kim JH, Ha KS, Lee JH, Kwon YG, Kim YM. Human plasminogen-derived N-acetyl-Arg-Leu-Tyr-Glu antagonizes VEGFR-2 to prevent blood-retinal barrier breakdown in diabetic mice. Biomed Pharmacother. 2021;134:111110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Delu S, Peiying H, Brian L V, Joshua L D, Juan E G, Ebenezer D, Maureen G M, Daniel F M, Gui-Shuang Y; and the CATT Research Group. Systemic Medication Use and the Incidence and Growth of Geographic Atrophy in the Comparison of Age-related Macular Degeneration Treatments Trials (CATT). Retina. 2021;41:1455-1462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Infeld DA, O'Shea JG. Diabetic retinopathy. Postgrad Med J. 1998;74:129-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1182] [Cited by in RCA: 1273] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 6. | Tamtaji OR, Behnam M, Pourattar MA, Hamblin MR, Mahjoubin-Tehran M, Mirzaei H, Asemi Z. PIWI-interacting RNAs and PIWI proteins in glioma: molecular pathogenesis and role as biomarkers. Cell Commun Signal. 2020;18:168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Vinasco-Sandoval T, Moreira FC, F Vidal A, Pinto P, Ribeiro-Dos-Santos AM, Cruz RLS, Fonseca Cabral G, Anaissi AKM, Lopes KP, Ribeiro-Dos-Santos A, Demachki S, de Assumpção PP, Ribeiro-Dos-Santos Â, Santos S. Global Analyses of Expressed Piwi-Interacting RNAs in Gastric Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Zou GL, Zhang XR, Ma YL, Lu Q, Zhao R, Zhu YZ, Wang YY. The role of Nrf2/PIWIL2/purine metabolism axis in controlling radiation-induced lung fibrosis. Am J Cancer Res. 2020;10:2752-2767. [PubMed] |
| 9. | Rayford KJ, Cooley A, Arun A, Rachakonda G, Kleschenko Y, Villalta F, Pratap S, Lima MF, Nde PN. Trypanosoma cruzi Modulates PIWI-Interacting RNA Expression in Primary Human Cardiac Myocytes during the Early Phase of Infection. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Beltran T, Pahita E, Ghosh S, Lenhard B, Sarkies P. Integrator is recruited to promoter-proximally paused RNA Pol II to generate Caenorhabditis elegans piRNA precursors. EMBO J. 2021;40:e105564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 195] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 12. | Onishi R, Sato K, Murano K, Negishi L, Siomi H, Siomi MC. Piwi suppresses transcription of Brahma-dependent transposons via Maelstrom in ovarian somatic cells. Sci Adv. 2020;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Lee YJ, Moon SU, Park MG, Jung WY, Park YK, Song SK, Ryu JG, Lee YS, Heo HJ, Gu HN, Cho SJ, Ali BA, Al-Khedhairy AA, Lee I, Kim S. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials. 2016;101:143-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 14. | Supe S, Upadhya A, Singh K. Role of small interfering RNA (siRNA) in targeting ocular neovascularization: A review. Exp Eye Res. 2021;202:108329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Masjedi A, Ahmadi A, Atyabi F, Farhadi S, Irandoust M, Khazaei-Poul Y, Ghasemi Chaleshtari M, Edalati Fathabad M, Baghaei M, Haghnavaz N, Baradaran B, Hojjat-Farsangi M, Ghalamfarsa G, Sabz G, Hasanzadeh S, Jadidi-Niaragh F. Silencing of IL-6 and STAT3 by siRNA loaded hyaluronate-N,N,N-trimethyl chitosan nanoparticles potently reduces cancer cell progression. Int J Biol Macromol. 2020;149:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 16. | Olivares AM, Althoff K, Chen GF, Wu S, Morrisson MA, DeAngelis MM, Haider N. Animal Models of Diabetic Retinopathy. Curr Diab Rep. 2017;17:93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 17. | Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376:124-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1781] [Cited by in RCA: 2292] [Article Influence: 152.8] [Reference Citation Analysis (0)] |
| 18. | Geng W, Qin F, Ren J, Xiao S, Wang A. Mini-peptide RPL41 attenuated retinal neovascularization by inducing degradation of ATF4 in oxygen-induced retinopathy mice. Exp Cell Res. 2018;369:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Wang X, Ma Y, Wang YX, Di Y. Expression profiles of long noncoding RNAs in retinopathy of prematurity. Neural Regen Res. 2020;15:1962-1968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Pelikánová T. [Diabetic retinopathy: pathogenesis and therapeutic implications]. Vnitr Lek. 62:620-628. [PubMed] |
| 21. | Scott A, Powner MB, Fruttiger M. Quantification of vascular tortuosity as an early outcome measure in oxygen induced retinopathy (OIR). Exp Eye Res. 2014;120:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Li Y, Min Cheng, Zhao Y, Wang J. Effects of fluoride on PIWI-interacting RNA expression profiling in testis of mice. Chemosphere. 2021;269:128727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Kumar S, Gonzalez EA, Rameshwar P, Etchegaray JP. Non-Coding RNAs as Mediators of Epigenetic Changes in Malignancies. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 24. | Xin J, Du M, Jiang X, Wu Y, Ben S, Zheng R, Chu H, Li S, Zhang Z, Wang M. Systematic evaluation of the effects of genetic variants on PIWI-interacting RNA expression across 33 cancer types. Nucleic Acids Res. 2021;49:90-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 25. | Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KS, Pikor LA, Becker-Santos DD, Brown CJ, Lam S, Lam WL. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5:10423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 26. | Moyano M, Stefani G. piRNA involvement in genome stability and human cancer. J Hematol Oncol. 2015;8:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 105] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 27. | Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41:324-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 339] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 28. | Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem. 2015;84:405-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 525] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 29. | Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 30. | Watanabe T, Lin H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol Cell. 2014;56:18-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 31. | Balaratnam S, West N, Basu S. A piRNA utilizes HILI and HIWI2 mediated pathway to down-regulate ferritin heavy chain 1 mRNA in human somatic cells. Nucleic Acids Res. 2018;46:10635-10648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Lin Y, Zheng J, Lin D. PIWI-interacting RNAs in human cancer. Semin Cancer Biol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 33. | Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113:373-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 34. | Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W, Su Q, Xue X, Zhuang W, Li B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2021;137:1603-1614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 122] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 35. | Qi T, Cao H, Sun H, Feng H, Li N, Wang C, Wang L. piR-19166 inhibits migration and metastasis through CTTN/MMPs pathway in prostate carcinoma. Aging (Albany NY). 2020;12:18209-18220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 36. | Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, Wu XF, Xu J, Hu Y. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29:196-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 37. | Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, Ye Y, Li M, Pan L, Su J, Zheng Y, Liu Z, Zuo Z, Zhao Q, Li X, Huang X, Yang J, Tan W, Zheng J, Lin D. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 38. | Zhou J, Zhou W, Zhang R. The potential mechanisms of piRNA to induce hepatocellular carcinoma in human. Med Hypotheses. 2021;146:110400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Solomon SD, Chew E, Duh EJ, Sobrin L, Sun JK, VanderBeek BL, Wykoff CC, Gardner TW. Diabetic Retinopathy: A Position Statement by the American Diabetes Association. Diabetes Care. 2017;40:412-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 596] [Article Influence: 74.5] [Reference Citation Analysis (0)] |
| 40. | Recchia FM, Xu L, Penn JS, Boone B, Dexheimer PJ. Identification of genes and pathways involved in retinal neovascularization by microarray analysis of two animal models of retinal angiogenesis. Invest Ophthalmol Vis Sci. 2010;51:1098-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 41. | Yoshida S, Nakama T, Ishikawa K, Arima M, Tachibana T, Nakao S, Sassa Y, Yasuda M, Enaida H, Oshima Y, Kono T, Ishibashi T. Antiangiogenic shift in vitreous after vitrectomy in patients with proliferative diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53:6997-7003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 42. | Natalucci G, Latal B, Koller B, Rüegger C, Sick B, Held L, Fauchère JC; Swiss EPO Neuroprotection Trial Group. Neurodevelopmental Outcomes at Age 5 Years After Prophylactic Early High-Dose Recombinant Human Erythropoietin for Neuroprotection in Very Preterm Infants. JAMA. 2020;324:2324-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 43. | Arellano-Buendía AS, Castañeda-Lara LG, Loredo-Mendoza ML, García-Arroyo FE, Rojas-Morales P, Argüello-García R, Juárez-Rojas JG, Tapia E, Pedraza-Chaverri J, Sánchez-Lozada LG, Osorio-Alonso H. Effects of Allicin on Pathophysiological Mechanisms during the Progression of Nephropathy Associated to Diabetes. Antioxidants (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 44. | Wang H, Xu X, Yin Y, Yu S, Ren H, Xue Q. Catalpol protects vascular structure and promotes angiogenesis in cerebral ischemic rats by targeting HIF-1α/VEGF. Phytomedicine. 2020;78:153300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 45. | Lazzara F, Trotta MC, Platania CBM, D'Amico M, Petrillo F, Galdiero M, Gesualdo C, Rossi S, Drago F, Bucolo C. Stabilization of HIF-1α in Human Retinal Endothelial Cells Modulates Expression of miRNAs and Proangiogenic Growth Factors. Front Pharmacol. 2020;11:1063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 46. | Han JM, Choi YS, Dhakal D, Sohng JK, Jung HJ. Novel Nargenicin A1 Analog Inhibits Angiogenesis by Downregulating the Endothelial VEGF/VEGFR2 Signaling and Tumoral HIF-1α/VEGF Pathway. Biomedicines. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 47. | Bonfiglio V, Platania CBM, Lazzara F, Conti F, Pizzo C, Reibaldi M, Russo A, Fallico M, Ortisi E, Pignatelli F, Longo A, Avitabile T, Drago F, Bucolo C. TGF-β Serum Levels in Diabetic Retinopathy Patients and the Role of Anti-VEGF Therapy. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 48. | Liu X, Li J, Li X. miR-142-5p regulates the progression of diabetic retinopathy by targeting IGF1. Int J Immunopathol Pharmacol. 2020;34:2058738420909041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |