Published online Dec 15, 2021. doi: 10.4239/wjd.v12.i12.2073
Peer-review started: April 21, 2021
First decision: July 15, 2021
Revised: August 3, 2021
Accepted: November 3, 2021
Article in press: October 31, 2021
Published online: December 15, 2021
Processing time: 238 Days and 14.1 Hours
Type 1 diabetes (T1D) is a severe and prevalent metabolic disease. Due to its high heredity, an increasing number of genome-wide association studies have been performed, most of which were from hospital-based case-control studies with a relatively small sample size. The association of single nucleotide polymorphisms (SNPs) and T1D has been less studied and is less understood in natural cohorts.
To investigate the significant variants of T1D, which could be potential biomarkers for T1D prediction or even therapy.
A genome-wide association study (GWAS) of adult T1D was performed in a nested case-control study (785 cases vs 804 controls) from a larger 5-year cohort study in Suzhou, China. Potential harmful or protective SNPs were evaluated for T1D. Subsequent expression and splicing quantitative trait loci (eQTL and sQTL) analyses were carried out to identify target genes modulated by these SNPs.
A harmful SNP for T1D, rs3117017 [odds ratio (OR) = 3.202, 95% confidence interval (CI): 2.296-4.466, P = 9.33 × 10-4] and three protective SNPs rs55846421 (0.113, 0.081-0.156, 1.76 × 10-9), rs75836320 (0.283, 0.205-0.392, 1.07 × 10-4), rs362071 (0.568, 0.495-0.651, 1.66 × 10-4) were identified. Twenty-two genes were further identified as potential candidates for T1D onset.
We identified a potential genetic basis of T1D, both protective and harmful, using a GWAS in a larger nested case-control study of a Chinese population.
Core Tip: Type 1 diabetes (T1D) is a severe and prevalent metabolic disease. Due to its high heredity, an increasing number of genome-wide association studies have been performed, most of which were from hospital-based case-control studies with a relatively small sample size. The aim of this study was to investigate the significant variants of T1D, which could be potential biomarkers for T1D prediction or even therapy. The effects of different polymorphisms in Chinese T1D patients were determined in a healthy population cohort study. The results showed 4 novel variants highly associated with the onset of T1D, namely rs3117017, rs55846421, rs75836320, and rs362071.
- Citation: Gao Y, Chen S, Gu WY, Fang C, Huang YT, Gao Y, Lu Y, Su J, Wu M, Zhang J, Xu M, Zhang ZL. Genome-wide association study reveals novel loci for adult type 1 diabetes in a 5-year nested case-control study. World J Diabetes 2021; 12(12): 2073-2086
- URL: https://www.wjgnet.com/1948-9358/full/v12/i12/2073.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i12.2073
Diabetes mellitus, a prevalent endocrine system disease worldwide, is characterized by high blood glucose level and can be life-threatening. Diabetes can be pathologically classified as type 1 diabetes (T1D), type 2 diabetes (T2D), gestational diabetes and other unclassified diabetes[1]. According to the International Diabetes Federation (IDF), T1D accounts for approximately 5%-10% of all diabetes[2]. Globally, the incidence of T1D is increasing, with an overall annual increase of 2%-5%[3,4]. There is also substantial regional disparity, ranging from 0.5% in the Caribbean to 7.0% in Africa[5]. In the IDF 9th edition, the assumption of T1D prevalence to incidence ratio was 6.3 for countries with no available age-specific incidence rates[2]. In developed countries, severe burden in patients and the entire healthcare system are expected as the incidence of T1D is estimated to double every 20 years[6,7]. According to current research, T1D is associated with increased mortality in the population: Its standardized mortality ratio ranges from 0 to 854 in different countries[8]. This implies that T1D patients have 3- to 4-times higher mortality than the general population[4,9].
With regard to the autoimmune feature of T1D, insulin-producing β cells in the pancreas are destroyed by the body due to insufficient islet secretion instead of insulin resistance in T2D[10]. T1D pathobiology is multifactorial, which includes genetic factors, environmental factors, and their potential interactions[11]. It has been demonstrated that first degree relatives of T1D patients had an approximately 6% risk of T1D, which is 15-times higher than the risk in the general population[12]. This finding suggests that genetic factors can be strong determinants of T1D. In China, existing observational studies indicated that approximately two-thirds of new T1D cases were reported in adults over 20 years old. T1D onset in adults is not rare in the Chinese population[13]. However, the exact mechanism of T1D onset in adults remains unknown[14].
Single nucleotide polymorphisms (SNPs) are potential genetic factors of T1D. SNPs have been investigated in population studies of both Caucasian and Chinese populations[15-18]. SNPs of the human leucocyte antigen (HLA) class II gene are identified as major risk factors for T1D[15]. However, these studies are mostly hospital-based case-control studies, which are inevitably prone to sampling bias[19,20]. The profile of T1D-related SNPs in a larger population is rarely addressed or investigated. Consequently, sampling bias in hospital-based studies can have a substantial impact on the subsequent genome-wide association study (GWAS), and may lead to unreliable results.
In order to overcome the challenge due to sampling bias, we performed a GWAS of adult T1D in a nested case-control study from a large 5-year cohort study in Suzhou, China. We aim to comprehensively investigate and quantify the association between T1D and SNPs in the general population. In addition, we will identify novel potential genes that are regulated by the identified SNPs, and evaluate their roles in the pathogenesis of adult T1D.
Ethical compliance: This study was reviewed and approved by the Research Ethics Committee of Jiangsu Provincial Center for Disease Control and Prevention (No. 2012025). The study was in full compliance with the Declaration of Helsinki.
Each participant, either in the case or control group, was informed of the nature of this study. Blood samples from the participants were collected after they signed the written informed consents.
Participants and background information on the study: A nested case-control study within a larger cohort study was performed. The original larger cohort included 3466780 residents in Suzhou, China. The complete demographic, clinical, and epidemiological data of the cohort were collected from the Suzhou medical and social insurance system between January 2012 and December 2018. To exclude latent autoimmune diabetes in adults, T2D and other types of diabetes, all corresponding medical records were cross validated by physicians at the First Affiliated Hospital of Soochow University and the Second Affiliated Hospital of Soochow University. Follow-up visits were performed at the Suzhou Center for Disease Prevention and Control, following the procedures recommended by Park et al[21].
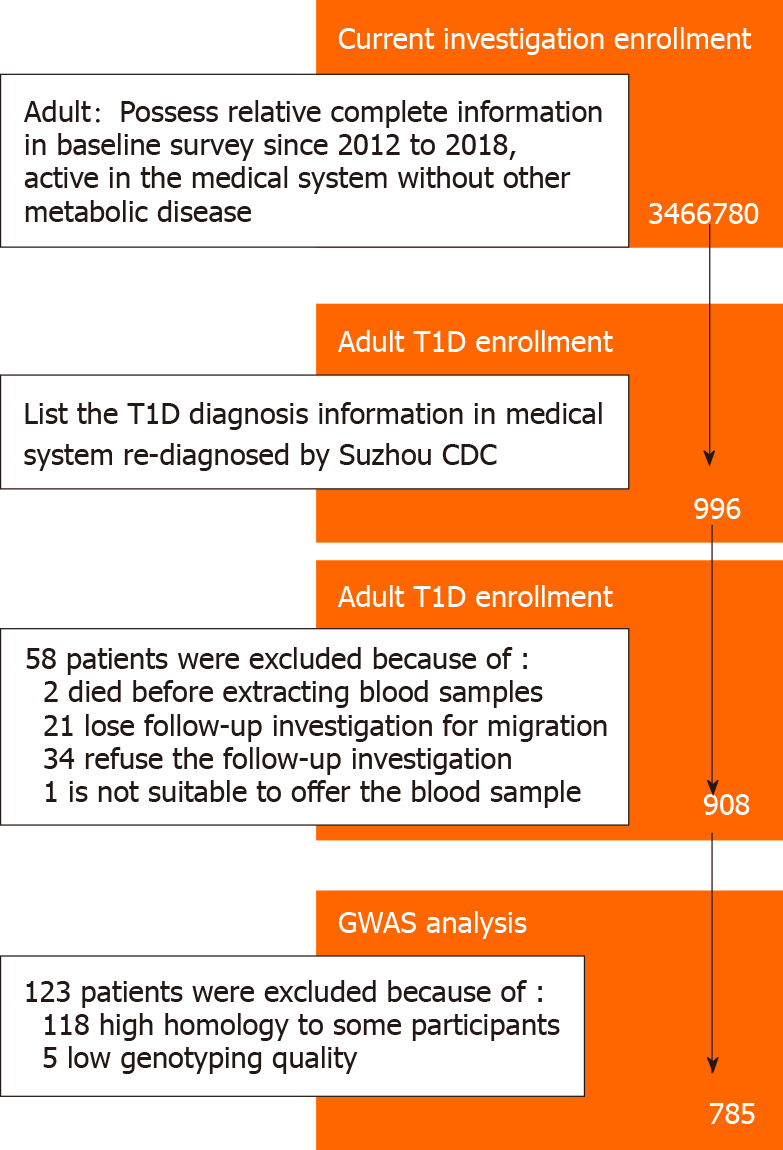
According to the medical records, a total of 1088 patients were diagnosed with T1D during this period, of whom 966 were adult patients more than 20 years old at the time of diagnosis. Fifty-eight patients were further excluded in the pre-collection stage for the following reasons: (1) 2 patients passed away before blood sample collection; (2) 21 patients moved and were unable to be contacted; (3) 34 patients declined to participate in the follow-up investigation; and (4) 1 patient was diagnosed with systemic lupus erythematosus with severe hematopoietic dysfunction, and was not eligible for blood sample collection. The T1D patients were divided into the testing and validation groups (414 and 494 patients, respectively), according to their residence.
For GWAS analysis, routine quality control steps included removing SNPs with imputation quality (INFO) scores < 0.4, with minor allele frequency (MAF) < 0.05, and without a valid Hardy-Weinberg test result (P < 10-6). SNPs on mitochondrial DNA and sex chromosomes were also excluded. Five patients were removed from the group. In addition, we further excluded 118 participants with close familial relationship after checking their genetic relationships. Finally, 785 adult T1D patients were included in the study and formed the nested case group. The complete screening process and quality control of T1D patients enrolled in the study are shown in Figure 1.
In the control group, a similar number of age-, gender- and residence-matched participants without metabolic system diseases were randomly selected. A total of 804 participants were further divided into the testing (377) and validation (427) groups.
Genotype imputation: Ungenotyped data were imputed using IMPUTE2 software (V2.3.2)[22]. 920636 SNPs were eventually filtered for the GWAS analysis, after excluding invalid imputed SNPs with INFO < 0.4. SHAPEIT V2 software was used to improve the imputation performance following the execution of IMPUTE2. The 1000 Genomes Project Phase III database (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html) was used as the reference dataset to determine population bias.
Gene regulatory network construction: Gene ontology functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of host genes of the polymorphisms were carried out with the R package clusterProfiler (R version 3.6.0). The Bayesian algorithm was chosen to optimize the regulatory network model and to identify the most probable gene regulation pathway in diabetes.
STRING software (version 11.0) was used to analyze functional interaction networks of potential downstream regulated genes of potential SNPs.
In Silico bioinformatics analysis: Expression quantitative trait loci (eQTLs) in GTEx (http://gtexportal.org/home/) was used to assess the impacts of SNPs on T1D. QTL overlapping was applied when the SNPs had a high linkage disequilibrium (LD) (r2> 0.8) with the top QTL genes for SNPs.
Statistical analyses: The χ2 test was performed to evaluate gender differences in the nested case-control study. Age and body mass index (BMI) were compared using the student t-test. After adjusting for age, gender, and BMI, the R package GWAS tools was for statistical analyses. Following existing GWAS publications, P < 5 × 10-8 was set as the detection limit to account for Bonferroni adjustment in this study[23].
Detailed demographic, clinical, and epidemiological characteristics of the 785 adult T1D cases and 804 control participants are provided in Table 1. There were no statistically significant differences in age, gender and BMI between the two groups in both the testing and validation stage of GWAS.
| Discovery stage | Validation stage | Combination | |||||||||
| Cases (n = 381) | Controls (n = 377) | P | Cases (n = 404) | Controls (n = 427) | P | Cases (n = 785) | Controls (n = 804) | P | |||
| Sex | |||||||||||
| Male | 197 | 200 | 0.711a | 209 | 223 | 0.887a | 406 | 423 | 0.722a | ||
| Female | 184 | 177 | 195 | 204 | 379 | 381 | |||||
| Age (yr) | |||||||||||
| mean ± SD | 28.45 ± 7.70 | 28.99 ± 8.01 | 0.338b | 28.53 ± 7.95 | 28.91 ± 7.67 | 0.496b | 28.49 ± 7.51 | 28.94 ± 7.98 | 0.248b | ||
| < 25 | 109 | 101 | 0.637a | 118 | 114 | 0.704a | 227 | 215 | 0.494a | ||
| [25, 35) | 180 | 174 | 188 | 203 | 368 | 377 | |||||
| ≥ 35 | 92 | 102 | 98 | 110 | 190 | 212 | |||||
| BMI (kg/m2) | |||||||||||
| mean ± SD | 21.77 ± 4.10 | 22.10 ± 4.00 | 0.266b | 21.93 ± 4.26 | 21.52 ± 3.70 | 0.146b | 21.85 ± 4.18 | 21.79 ± 3.85 | 0.773b | ||
| < 18.5 | 60 | 46 | 0.371a | 51 | 66 | 0.500a | 111 | 112 | 0.985a | ||
| [18.5, 25) | 270 | 279 | 303 | 311 | 573 | 590 | |||||
| ≥ 25 | 51 | 52 | 50 | 50 | 101 | 102 | |||||
| Glucose level (mmol/L) | |||||||||||
| mean ± SD | 12.83 ± 2.35 | 5.43 ± 1.33 | < 0.0001b | 13.42 ± 2.48 | 5.53 ± 1.30 | < 0.0001b | 13.13 ± 2.44 | 5.49 ± 81.32 | < 0.0001b | ||
Among all SNPs examined, four novel loci significantly associated with T1D were identified (Table 2): rs55846421 at 14q21 (PBonferroni-adjust = 4.28 × 10-11), rs3117017 at 6q13 (PBonferroni-adjust = 6.36 × 10-5), rs75836320 at 12q14 (PBonferroni-adjust = 0.011), and rs362071 at 22q11.2 (PBonferroni-adjust = 4.5 × 10-4). P-values of these four SNPs were significantly lower than the threshold of genome-wide significance (P-value after Bonferroni adjust should be less than 0.05, which usually means original P-value < 10-8). LD analysis further confirmed the independence of these SNPs. In the validation stage, the diabetic susceptibilities of these four loci between 404 adult T1D cases and 427 controls showed similar trends as in the testing stage. After combining participants in both stages, these four SNPs were still significantly associated with the onset of adult T1D: rs55846421 (PBonferroni-adjust = 1.76 × 10-9), rs3117017 (PBonferroni-adjust = 9.33 × 10-4), rs75836320 (PBonferroni-adjust = 1.07 × 10-4), and rs362071 (PBonferroni-adjust = 1.66 × 10-4).
| SNPs | Location | Stage | Genotype distribution | ORadd (95%CI) | Pa for Bonferroni | |
| Cases (n = 785) | Controls (n = 804) | |||||
| rs55846421 | Chr14. 50310401 | Discovery | 10/149/222 | 0/6/371 | 0.024 (0.010-0.056) | 4.28 × 10-11 |
| Validation | 8/124/272 | 2/37/385 | 0.232 (0.159-0.337) | 8.73 × 10-8 | ||
| Combination | 18/273/494 | 2/43/756 | 0.113 (0.081-0.156) | 1.76 × 10-9 | ||
| rs3117017 | Chr6. 33095275 | Discovery | 2/40/339 | 36/117/224 | 4.958 (3.107-7.912) | 6.36 × 10-5 |
| Validation | 6/32/365 | 32/98/297 | 3.202 (2.296-4.466) | 4.51 × 10-4 | ||
| Combination | 8/72/704 | 68/215/521 | 3.794 (2.984-4.823) | 9.33 × 10-4 | ||
| rs75836320 | Chr12. 65156128 | Discovery | 8/77/296 | 1/12/356 | 0.153 (0.082-0.285) | 0.011 |
| Validation | 8/63/333 | 4/27/396 | 0.429 (0.289-0.638) | 3.3 × 10-4 | ||
| Combination | 16/140/629 | 5/39/752 | 0.283 (0.205-0.392) | 1.07 × 10-4 | ||
| rs362071 | Chr22. 20811645 | Discovery | 82/193/104 | 46/101/230 | 0.445 (0.348-0.570) | 4.5 × 10-4 |
| Validation | 100/173/131 | 79/149/199 | 0.705 (0.588-0.846) | 0.0002 | ||
| Combination | 182/366/235 | 125/250/429 | 0.568 (0.495-0.651) | 1.66 × 10-4 | ||
As there were no specific data for islet tissue or diabetes in the GTEx database, we comprehensively analyzed these SNPs in all types of tissues (Table 3). Of the four novel SNPs, rs55846421 is located on the intron of NEMF gene. However, based on our eQTL results, rs55846421 showed a potential causal effect on LRR1, RP11-596C23.6, RHOQP1, KLHDC1 and VCPKMT genes, instead of NEMF.
| SNP name | Influence type | Gene symbol | Expression tissues | P value |
| rs55846421 | eQTLs | ARF6 | Cells-cultured fibroblasts | 0.00015 |
| KLHDC1 | Brain-cerebellum | 0.0000022 | ||
| Lung | 0.000034 | |||
| Thyroid | 0.00021 | |||
| LRR1 | Adipose-subcutaneous | 8.00 × 10-10 | ||
| Adipose-visceral | 5.20 × 10-7 | |||
| Breast-mammary tissue | 8.70 × 10-7 | |||
| Esophagus-muscularis | 0.000054 | |||
| Lung | 0.000077 | |||
| Nerve-tibial | 0.00013 | |||
| Thyroid | 0.00016 | |||
| RHOQP1 | Adipose-subcutaneous | 3.30 × 10-8 | ||
| RP11-596C23.6 | Artery-tibial | 1.1 × 10-9 | ||
| Esophagus-mucosa | 2.3 × 10-9 | |||
| Artery-aorta | 6.0 × 10-8 | |||
| Testis | 2.9 × 10-7 | |||
| Esophagus-muscularis | 0.0000019 | |||
| Esophagus-gastroesophageal junction | 0.0000025 | |||
| Muscle-skeletal | 0.0000027 | |||
| Small intestine-terminal ileum | 0.0000042 | |||
| Lung | 0.000019 | |||
| Colon-sigmoid | 0.00002 | |||
| Spleen | 0.000024 | |||
| Artery-coronary | 0.000031 | |||
| Thyroid | 0.000061 | |||
| Heart-atrial appendage | 0.000062 | |||
| sQTLs | RPL36AL | Liver | 0.0000063 | |
| Brain-nucleus accumbens | 0.0000071 | |||
| rs3117017 | eQTLs | COL11A2 | Thyroid | 2.0 × 10-8 |
| HCG24 | Brain-nucleus accumbens | 5.0 × 10-25 | ||
| Brain-putamen | 6.4 × 10-17 | |||
| Brain-cortex | 3.0 × 10-14 | |||
| Brain-caudate | 3.4 × 10-14 | |||
| Brain-frontal cortex | 4.3 × 10-11 | |||
| Brain-anterior cingulate cortex | 0.000022 | |||
| Brain-hippocampus | 0.000025 | |||
| HLA-DOA | Whole blood | 0.00005 | ||
| HLA-DPA1 | Cells-cultured fibroblasts | 0.000071 | ||
| HLA-DPB1 | Testis | 0.000028 | ||
| HLA-DPB2 | Brain-nucleus accumbens | 1.3 × 10-18 | ||
| Brain-caudate | 2.4 × 10-13 | |||
| Brain-putamen | 3.4 × 10-11 | |||
| Brain-cortex | 3.5 × 10-8 | |||
| Brain-hypothalamus | 3.6 × 10-7 | |||
| Brain-cerebellum | 0.000057 | |||
| HSD17B8 | Thyroid | 1.0 × 10-13 | ||
| Muscle-skeletal | 3.5 × 10-11 | |||
| Esophagus-muscularis | 1.3 × 10-8 | |||
| Artery-tibial | 5.6 × 10-7 | |||
| Esophagus-gastroesophageal junction | 0.0000011 | |||
| Colon-sigmoid | 0.0000018 | |||
| Heart-left ventricle | 0.0000028 | |||
| Testis | 0.0000032 | |||
| Adipose-visceral | 0.000016 | |||
| Artery-aorta | 0.000018 | |||
| Esophagus-mucosa | 0.000029 | |||
| Skin-not sun exposed | 0.00003 | |||
| Cells-cultured fibroblasts | 0.000065 | |||
| RING1 | Pancreas | 0.0000059 | ||
| RPS18 | Artery-tibial | 0.000032 | ||
| Whole blood | 0.000084 | |||
| WDR46 | Lung | 0.000018 | ||
| sQTLs | COL11A2 | Thyroid | 3.5 × 10-8 | |
| Brain-spinal cord | 9.0 × 10-7 | |||
| Pituitary | 0.0000084 | |||
| HLA-DPB1 | Skin-sun exposed (lower leg) | 6.6 × 10-7 | ||
| Muscle-skeletal | 8.5 × 10-7 | |||
| Skin-not sun exposed (suprapubic) | 8.6 × 10-7 | |||
| HLA-DPB2 | Brain-cerebellum | 1.9 × 10-12 | ||
| Skin-sun exposed (lower leg) | 6.6 × 10-7 | |||
| Muscle-skeletal | 8.5 × 10-7 | |||
| Skin-not sun exposed (suprapubic) | 8.6 × 10-7 | |||
| Brain-spinal cord | 0.000004 | |||
| Colon-transverse | 0.000014 | |||
| Artery-tibial | 0.000063 | |||
| Testis | 0.00012 | |||
| rs75836320 | - | - | - | - |
| rs362071 | eQTLs | MED15 | Artery-tibial | 3.1 × 10-13 |
| Nerve-tibial | 1.2 × 10-11 | |||
| Esophagus-muscularis | 5.4 × 10-11 | |||
| Lung | 1.4 × 10-9 | |||
| Adipose-subcutaneous | 6.5 × 10-9 | |||
| Thyroid | 8.2 × 10-9 | |||
| Spleen | 1.4 × 10-8 | |||
| Whole blood | 3.6 × 10-8 | |||
| Artery-aorta | 9.0 × 10-8 | |||
| Colon-transverse | 1.2 × 10-7 | |||
| Adrenal gland | 2.0 × 10-7 | |||
| Adipose-visceral | 5.0 × 10-7 | |||
| Prostate | 5.8 × 10-7 | |||
| Liver | 9.7 × 10-7 | |||
| Esophagus-gastroesophageal junction | 0.0000046 | |||
| Pancreas | 0.0000063 | |||
| Skin-sun exposed (lower leg) | 0.000015 | |||
| Cells-cultured fibroblasts | 0.000052 | |||
| sQTLs | KLHL22 | Skin-sun exposed (lower leg) | 0.000003 | |
| Skin-not sun exposed (suprapubic) | 0.0000049 | |||
| MED15 | Muscle-skeletal | 3.0 × 10-7 |
rs3117017 is located on a haplotype block, including part of the HLA-DPB2 gene. HLA-DPB2 is considered the key gene in both T1D and T2D metabolism[24]. In addition, expression of COL11A2, HSD17B8, HCG24, HLA-DOA, HLA-DPB2, HLA-DPA1, RPS18, and RING1 genes are also associated with rs3117017 in different tissues.
rs75836320 is located on the promoter region of TBC1D30 gene, a key gene for insulin processing and secretion. However, no GTEx data were available for this SNP due to its low frequency (MAF < 1%) in the samples.
rs362071 is located on the intron of KLHL22 gene, which has not been previously reported to be associated with diabetes. The expression of MED15 and KLHL22 genes may be highly correlated with this SNP, according to the outcome of eQTL prediction.
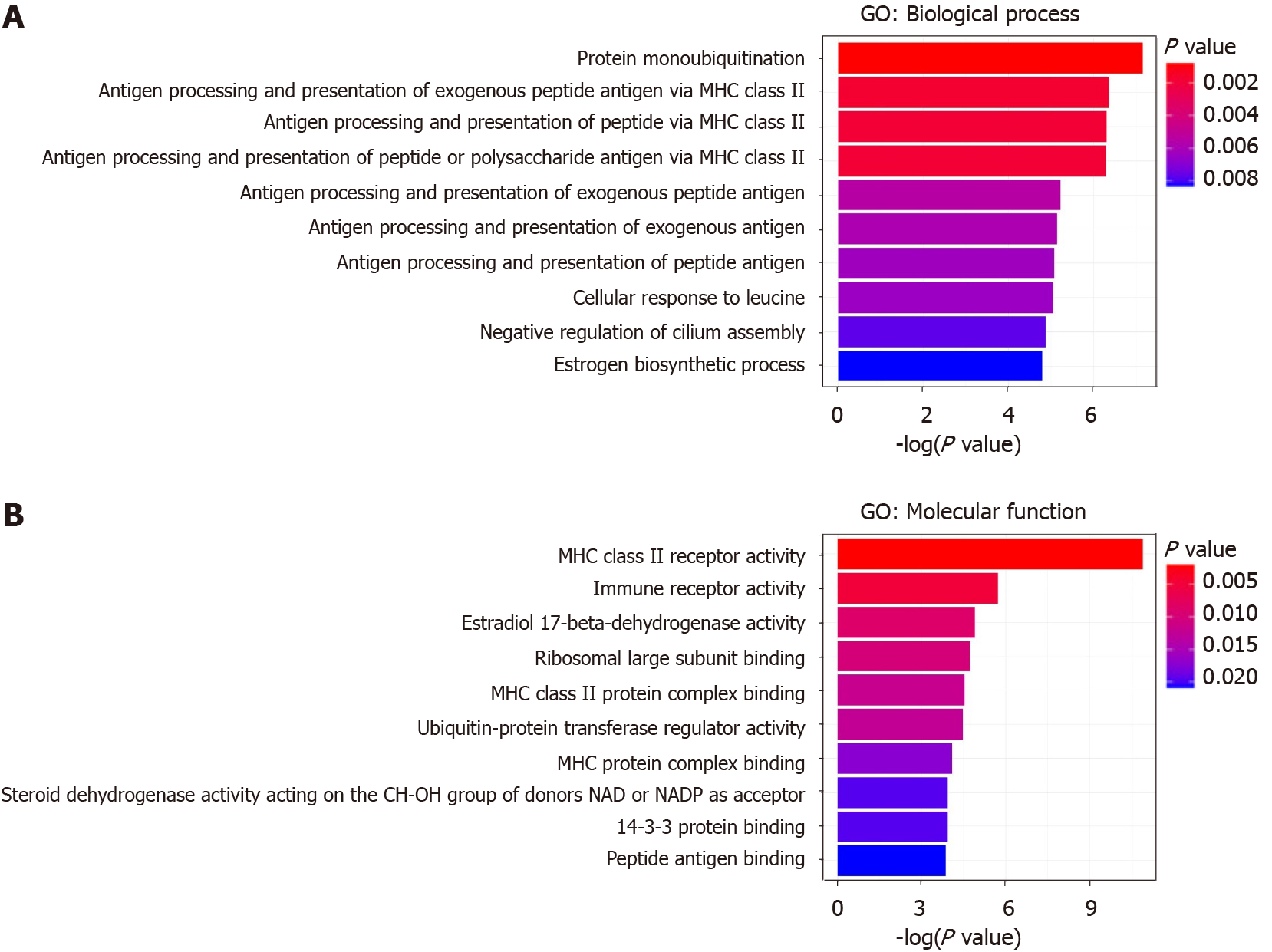
Gene ontology (GO) and KEGG pathway analyses were performed on the target genes listed in Table 3. As shown in Figure 2, these genes are mainly involved in enriched GO terms of “protein monoubiquitination”, “antigen processing and presentation of exogenous peptide antigen via major histocompatibility complex (MHC) class II”, “antigen processing and presentation of peptide antigen via MHC class II”, “antigen processing and presentation of peptide or polysaccharide antigen via MHC class II” in biological process (Figure 2A), and “MHC class II receptor activity”, “immune receptor activity”, “estradiol 17-beta-dehydrogenase activity”, and “Ribosomal large subunit binding” in molecular function (Figure 2B).
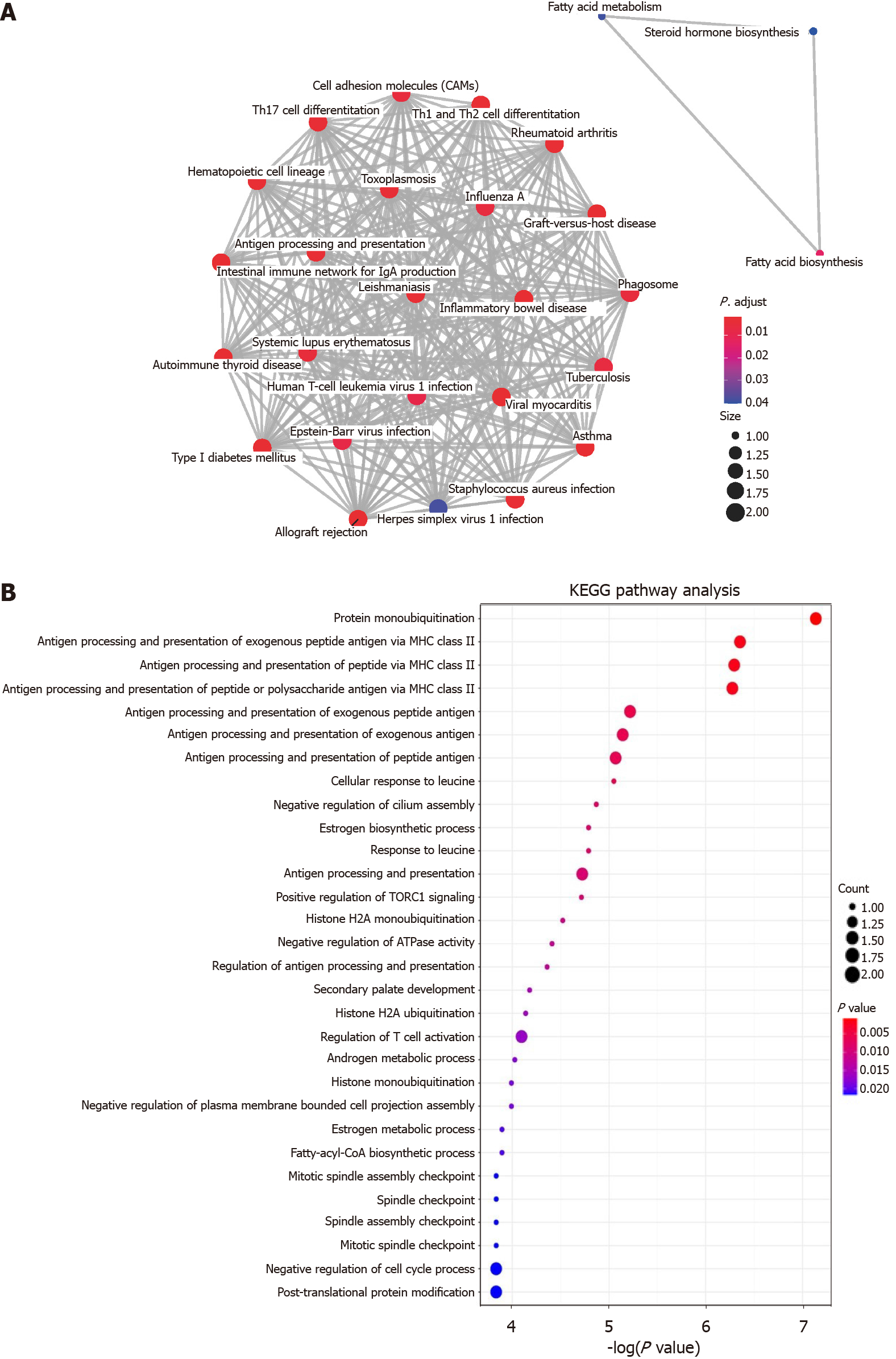
The results of KEGG analysis are shown in Figure 3. The top five pathways were “Asthma”, “Allograft rejection”, “Graft-versus-host diseases”, “Type 1 diabetes mellitus” and “intestinal immune network for IgA production” shown in Figure 3A. The enrichment map of KEGG in potential target genes is shown in Figure 3B.
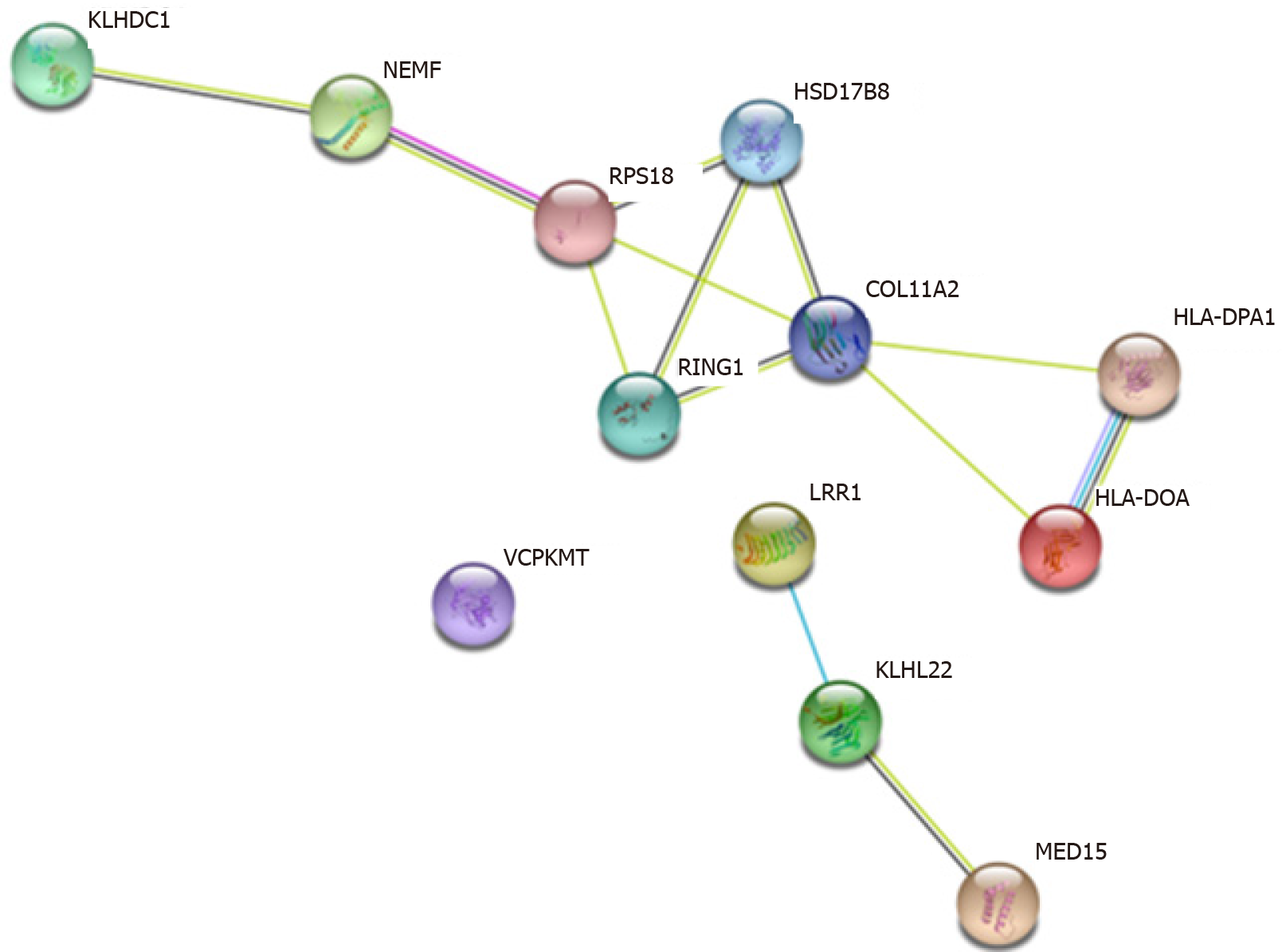
For the association among potential downstream regulated genes of these SNPs, the protein-protein interaction network is shown in Figure 4. KLHDC1, NEMF, RPS18, HSD17B8, RING1, COL11A2, HLA-DPA1 and HLA-DOA genes form a complicated regulation pathway. The corresponding SNPs on this pathway may have an interactive effect in T1D. In addition, LRR1, KLHL22 and MED15 genes also form a straightforward regulation chain, echoing the eQTL finding of rs362071 in both diabetes-related genes KLHL22 and MED15.
Through a comprehensive nested case-control study in a Chinese cohort, we identified 4 novel SNPs (one harmful and three protective) with a significant association with T1D via GWAS. This study has established the genetic basis of T1D in the Chinese Han population in Suzhou, one of the largest and fastest growing cities in southeast China.
According to the eQTL and sQTL results, rs55846421, the most significant SNP in this study, is highly associated with the expression of six genes (ARF6, KLHDC1, LRR1, RHOQP1, RP11-596C23.6 and VCPKMT) and splicing quantity of one gene (RPL36AL). However, only ARF6 is currently known to be associated with diabetes or diabetes-related diseases[25-27]. As a type of small G-protein, ARF6 is involved in transporting and docking insulin granules on plasma membrane for exocytotic insulin secretion[27]. rs3117017 is located on the HLA-DPB2 gene region and is highly associated with HLA-DPB2 gene expression, which belongs to the HLA genes. Several studies have demonstrated the importance of HLA class II genes, including HLA-DQA1, HLA-DR, and HLA-DQB1[28,29]. A recent hospital-based GWAS analysis of T1D identified a novel T1D-related SNP rs1770 on the 6p21.3 region of HLA-DQB1[15]. rs1770 is especially important for T1D in the Chinese population. Our findings on rs3117017 also suggest that another HLA class II gene - HLA-DPB2 - is associated with T1D susceptibility and pathogenesis in the Chinese population. As of now, there is no record of rs75836320 regulating downstream gene expression in either the GTEx database or GEO database. The last SNP identified in this study, rs362071, belongs to the KLHL22 gene as an intron variant, and we first report its association with T1D. KLHL22 is usually not associated with diseases, but the product of KLHL22 could be polymerized with CUL3. CUL3/KLHL2 is a notable E3 ubiquitin ligase gene[30-32] and its expression will trigger amino acid-dependent mTORC1 signaling, which may be involved in aging, cancer and diabetes[33,34].
In addition to genetic basis, we suggest that economic growth in the study region (Suzhou, China) may have influenced people’s diet preference, which increased the incidence of T1D. Studies have shown that T1D incidence in adolescents may be associated with gross domestic product (GDP) in Poland[17]. While GDP showed a steady increasing trend in Suzhou from 2012 to 2018, the incidence of adult T1D remained relatively stable during the same period (Supplementary Figure 1, provided by the Suzhou Statistical Yearbook since 2012 to 2018). Therefore, adult T1D is not significantly associated with economic growth in Suzhou, or may have a lag effect which warrants further investigation. In addition, diet may not play an important role in adult T1D in the Chinese population.
Furthermore, during the preprocessing stage of this study, we excluded 118 patients due to their close familial heredity to other T1D patients. Genetic correlation of T1D was more than 13% in our study sample. However, according to studies based on the Caucasian population[16,35], the degree of familial cluster was much lower than in the Chinese population. Another study confirmed a significant difference in GWAS results between Chinese and Caucasian populations[15]. Based on previous studies in both populations and our current study, we suggest that the genetic basis and pathogenesis of T1D may vary between Chinese and Caucasian populations. These distinctions can have profound implications on T1D risk estimation, diagnosis, and treatment across different ethnic groups.
There are some limitations in our study. First, unlike a hospital-based case-control study, the sample size of T1D patients (cases) in our nested case-control study was relatively small due to the low incidence of T1D in the natural cohort. Therefore, the results of this study need to be further validated in larger studies. Second, we only investigated the contribution of genetic factors (SNPs) in this study. Other factors, for example, dietary preferences and habits, could also be important in T1D pathogenesis. We will investigate the influence of these factors on T1D in future studies. Third, T1D is a disease with high heritability. In our study, we excluded individuals with high homology, following the protocol of GWAS. However, this exclusion may conceal the actual effects of some relevant SNPs in T1D.
To the best of our knowledge, this is the first study to conduct a GWAS for adult T1D in a nested case-control study. We identified 4 novel SNPs as genetic biomarkers for adult T1D onset. Subsequent basic pedigree can further complement and strengthen our research.
The genome-wide association study (GWAS) of type 1 diabetes (T1D) is valuable.
We hoped to add data from a natural cohort to fill gaps in the knowledge of T1D susceptibility.
We conducted a cohort study to evaluate the variants of genes in order to adjust the bias in hospital-based research.
The GWAS analysis used in our research reflected the associations between single nucleotide polymorphisms (SNPs) and T1D with high-throughput sequencing.
In T1D patients, rs3117017 displayed its damaging role in the onset of T1D, while rs55846421, rs75836320, and rs362071 displayed their protective roles in T1D.
A larger nested case-control study of a Chinese population will be helpful in validating our findings.
The GWAS analysis from a hospital-based population was a little different to that from normal population.
The specific function of these SNPs should be investigated in the future.
We thank the participants enrolled in our study, the Suzhou Bureau of Social Security, and the China National Center for Disease Control and Prevention. We would also like to thank Professor Yao-Rong Ge of the University of North Carolina, Charlotte and Mr. Chao Jiang at Nanjing University of Chinese Medicine for their help. We also appreciate the technical support on statistics from Target-Gene Biotechnology Co., Ltd., Nanjing, and the recommendation of eQTL from Biolight Biotechnology Co., Ltd, Nanjing.
Provenance and peer review: Unsolicited article; Externally peer reviewed
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ata F S-Editor: Wang JJ L-Editor: Webster JR P-Editor: Wang JJ
| 1. | Paneni F, Beckman JA, Creager MA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur Heart J. 2013;34:2436-2443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 592] [Cited by in RCA: 740] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 2. | International Diabetes Federation (IDF). IDF diabetes atlas, 9th ed, 2019. |
| 3. | DiMeglio LA, Evans-Molina C, Oram RA. Type 1 diabetes. Lancet. 2018;391:2449-2462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 964] [Article Influence: 137.7] [Reference Citation Analysis (0)] |
| 4. | Maffi P, Secchi A. The Burden of Diabetes: Emerging Data. Dev Ophthalmol. 2017;60:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 5. | Patterson CC, Karuranga S, Salpea P, Saeedi P, Dahlquist G, Soltesz G, Ogle GD. Worldwide estimates of incidence, prevalence and mortality of type 1 diabetes in children and adolescents: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 335] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 6. | Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb Perspect Med. 2012;2:a007799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1881] [Cited by in RCA: 1790] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 8. | Morgan E, Cardwell CR, Black CJ, McCance DR, Patterson CC. Excess mortality in Type 1 diabetes diagnosed in childhood and adolescence: a systematic review of population-based cohorts. Acta Diabetol. 2015;52:801-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 9. | Lind M, Svensson AM, Kosiborod M, Gudbjörnsdottir S, Pivodic A, Wedel H, Dahlqvist S, Clements M, Rosengren A. Glycemic control and excess mortality in type 1 diabetes. N Engl J Med. 2014;371:1972-1982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 647] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 10. | Laron Z. Interplay between heredity and environment in the recent explosion of type 1 childhood diabetes mellitus. Am J Med Genet. 2002;115:4-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Rodríguez-Rodríguez I, Chatzigiannakis I, Rodríguez JV, Maranghi M, Gentili M, Zamora-Izquierdo MÁ. Utility of Big Data in Predicting Short-Term Blood Glucose Levels in Type 1 Diabetes Mellitus Through Machine Learning Techniques. Sensors (Basel). 2019;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 12. | Dorman JS, Bunker CH. HLA-DQ locus of the human leukocyte antigen complex and type 1 diabetes mellitus: a HuGE review. Epidemiol Rev. 2000;22:218-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 13. | Weng J, Zhou Z, Guo L, Zhu D, Ji L, Luo X, Mu Y, Jia W; T1D China Study Group. Incidence of type 1 diabetes in China, 2010-13: population based study. BMJ. 2018;360:j5295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 212] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 14. | Chiang JL, Kirkman MS, Laffel LM, Peters AL; Type 1 Diabetes Sourcebook Authors. Type 1 diabetes through the life span: a position statement of the American Diabetes Association. Diabetes Care. 2014;37:2034-2054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 614] [Cited by in RCA: 613] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 15. | Zhu M, Xu K, Chen Y, Gu Y, Zhang M, Luo F, Liu Y, Gu W, Hu J, Xu H, Xie Z, Sun C, Li Y, Sun M, Xu X, Hsu HT, Chen H, Fu Q, Shi Y, Xu J, Ji L, Liu J, Bian L, Zhu J, Chen S, Xiao L, Li X, Jiang H, Shen M, Huang Q, Fang C, Huang G, Fan J, Jiang Z, Jiang Y, Dai J, Ma H, Zheng S, Cai Y, Dai H, Zheng X, Zhou H, Ni S, Jin G, She JX, Yu L, Polychronakos C, Hu Z, Zhou Z, Weng J, Shen H, Yang T. Identification of Novel T1D Risk Loci and Their Association With Age and Islet Function at Diagnosis in Autoantibody-Positive T1D Individuals: Based on a Two-Stage Genome-Wide Association Study. Diabetes Care. 2019;42:1414-1421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 16. | Garyu JW, Meffre E, Cotsapas C, Herold KC. Progress and challenges for treating Type 1 diabetes. J Autoimmun. 2016;71:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7741] [Cited by in RCA: 7155] [Article Influence: 397.5] [Reference Citation Analysis (0)] |
| 18. | Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, Farber E, Bonnie JK, Szpak M, Schofield E, Achuthan P, Guo H, Fortune MD, Stevens H, Walker NM, Ward LD, Kundaje A, Kellis M, Daly MJ, Barrett JC, Cooper JD, Deloukas P; Type 1 Diabetes Genetics Consortium, Todd JA, Wallace C, Concannon P, Rich SS. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 562] [Cited by in RCA: 520] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 19. | Rudant J, Clavel J, Infante-Rivard C. Selection bias in case-control studies on household exposure to pesticides and childhood acute leukemia. J Expo Sci Environ Epidemiol. 2010;20:299-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Nishimoto IN, Pintos J, Schlecht NF, Torloni H, Carvalho AL, Kowalski LP, Franco EL. Assessment of control selection bias in a hospital-based case-control study of upper aero-digestive tract cancers. J Cancer Epidemiol Prev. 2002;7:131-141. [PubMed] |
| 21. | Park Y, Wintergerst KA, Zhou Z. Clinical heterogeneity of type 1 diabetes (T1D) found in Asia. Diabetes Metab Res Rev. 2017;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2978] [Cited by in RCA: 3169] [Article Influence: 198.1] [Reference Citation Analysis (0)] |
| 23. | Meng W, Adams MJ, Palmer CNA; 23andMe Research Team, Shi J, Auton A, Ryan KA, Jordan JM, Mitchell BD, Jackson RD, Yau MS, McIntosh AM, Smith BH. Author Correction: Genome-wide association study of knee pain identifies associations with GDF5 and COL27A1 in UK Biobank. Commun Biol. 2020;3:149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Lie BA, Todd JA, Pociot F, Nerup J, Akselsen HE, Joner G, Dahl-Jørgensen K, Rønningen KS, Thorsby E, Undlien DE. The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am J Hum Genet. 1999;64:793-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 132] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Duan Y, Prasad R, Feng D, Beli E, Li Calzi S, Longhini ALF, Lamendella R, Floyd JL, Dupont M, Noothi SK, Sreejit G, Athmanathan B, Wright J, Jensen AR, Oudit GY, Markel TA, Nagareddy PR, Obukhov AG, Grant MB. Bone Marrow-Derived Cells Restore Functional Integrity of the Gut Epithelial and Vascular Barriers in a Model of Diabetes and ACE2 Deficiency. Circ Res. 2019;125:969-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 26. | Hu J, Yang Q, Chen Z, Liang W, Feng J, Ding G. Small GTPase Arf6 regulates diabetes-induced cholesterol accumulation in podocytes. J Cell Physiol. 2019;234:23559-23570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Kowluru A. Role of G-proteins in islet function in health and diabetes. Diabetes Obes Metab. 2017;19 Suppl 1:63-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Nerup J, Platz P, Andersen OO, Christy M, Lyngsoe J, Poulsen JE, Ryder LP, Nielsen LS, Thomsen M, Svejgaard A. HL-A antigens and diabetes mellitus. Lancet. 1974;2:864-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 508] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 29. | Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134-1148. [PubMed] |
| 30. | Chen J, Ou Y, Yang Y, Li W, Xu Y, Xie Y, Liu Y. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature. 2018;557:585-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 31. | Metzger T, Kleiss C, Sumara I. CUL3 and protein kinases: insights from PLK1/KLHL22 interaction. Cell Cycle. 2013;12:2291-2296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 32. | Beck J, Peter M. Regulating PLK1 dynamics by Cullin3/KLHL22-mediated ubiquitylation. Cell Cycle. 2013;12:2528-2529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5794] [Cited by in RCA: 6593] [Article Influence: 507.2] [Reference Citation Analysis (1)] |
| 34. | Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2855] [Cited by in RCA: 3174] [Article Influence: 211.6] [Reference Citation Analysis (0)] |
| 35. | Ikegami H, Ogihara T. Genetics of insulin-dependent diabetes mellitus. Endocr J. 1996;43:605-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |