Published online Dec 15, 2020. doi: 10.4239/wjd.v11.i12.567
Peer-review started: June 26, 2020
First decision: September 24, 2020
Revised: October 3, 2020
Accepted: October 29, 2020
Article in press: October 29, 2020
Published online: December 15, 2020
Processing time: 169 Days and 20.4 Hours
MicroRNAs (miRNA) are recently discovered endogenous, small noncoding RNAs (of 22 nucleotides) that play pivotal roles in gene regulation. They are involved in post-transcriptional control of gene expression. miRNAs are emerging as important regulators of cell proliferation, development, cancer formation, stress responses, cell death and physiological conditions. Increasing evidence has demonstrated the human miRNAs bind to their target mRNA sequences with perfect or near-perfect sequence complementarily. This provides a powerful strategy for discovering potential type 2 diabetes mellitus (T2DM) targets and gives the probability to exploit them for diagnostic and therapeutic causes. About 6% of the world population is affected by T2DM, and it is recognized as a global epidemic by the World Health Organization. At present there is no valid biomarker to control or manage T2DM. Therefore, the present study applied a mature sequence of miRNAs from publicly accessible databases to identify the miRNA from T2DM expressed sequence tags, and the results are detailed and discussed below.
Core Tip: MicroRNAs (miRNA) are endogenous, small noncoding RNAs that play pivotal roles in gene regulation. They are involved in post-transcriptional control of gene expression and are important regulators of cell proliferation, development, cancer formation, stress responses, cell death and physiological conditions. About 6% of the world population is affected by type 2 diabetes mellitus. It is recognized as a global epidemic. At present there is no valid biomarker to control or manage type 2 diabetes mellitus. The present study applied a mature sequence of miRNAs from publicly accessible databases to identify miRNAs from type 2 diabetes mellitus expressed sequence tags.
- Citation: Rajkumar KV, Lakshmanan G, Sekar D. Identification of miR-802-5p and its involvement in type 2 diabetes mellitus. World J Diabetes 2020; 11(12): 567-571
- URL: https://www.wjgnet.com/1948-9358/full/v11/i12/567.htm
- DOI: https://dx.doi.org/10.4239/wjd.v11.i12.567
MicroRNAs (miRNA) are recently discovered endogenous, small noncoding RNAs (of 22 nucleotides) that play pivotal roles in gene regulations[1]. They are involved in post-transcriptional control of gene expression. miRNAs are emerging as important regulators of cell proliferation, development, cancer formation, stress responses, cell death and physiological conditions[2]. These circulating miRNAs are detected in body fluids including saliva, urine and blood. miRNAs regulate gene control and a variety of biological and metabolic processes[3]. Gaining insight into the miRNA targets will help us to understand the spectrum of miRNA regulation and elucidate the functional importance of miRNAs[4]. Increasing evidence has demonstrated that human miRNAs bind to their target mRNA sequences with perfect or near-perfect sequence complementarily. This provides a powerful strategy for discovering potential type 2 diabetes mellitus (T2DM) targets and gives the probability to exploit them for diagnostic and therapeutic causes[5].
T2DM is known as adult-onset diabetes, a systemic chronic disease of heterogeneous origin[6]. About 6% of the world population is affected by T2DM, and it is recognized as a global epidemic by the World Health Organization[7]. It is characterized by insulin resistance and delayed insulin secretion[8,9]. At present, the management and treatment strategies for T2DM are elusive, and the exact molecular mechanism is not yet completely discovered. Many reports suggest that miRNAs are a promising tool for the management and treatment of various diseases.
On the other side, expressed sequence tags (ESTs) are a simple segment of a sequence from a cDNA clone that correspond to an mRNA. ESTs longer than 150bp were found to be the most useful for similarity searches and mapping[10]. At present there is no valid biomarker to control or manage T2DM. The present study applied a mature sequence of microRNAs from publicly accessible databases to identify the microRNA from T2DM ESTs, and the results are detailed and discussed below.
EST sequence data was obtained through the National Center for Biotechnology Information web portal for International Nucleotide Sequence Database Consortium. The search term keyword “type-2 diabetes mellitus in Homo sapiens” (18271 ESTs as of April 2020) were extracted using this free search engine. Human mature miRNAs were selected out of 38589 entries from miRbase (http://www.mirbase.org/). After removing the low-quality sequences, local nucleotide database was formed for T2DM specific EST sequences[11]. The above-mentioned nucleotide database was searched for the homolog among the miRNAs dataset. The mature miRNAs were used as a source to search for similar T2DM ESTs.
Reference miRNA sequences were used as a query for homology search against the specific T2DM nucleotide sequence database at the e-value threshold < 0.01 using the BLAST program with all other parameters as default. The FASTA formats of all sequences were processed, and mature miRNA sequences were aligned against the unique ESTs using the ClustalW multiple sequence alignment tool[11,12].
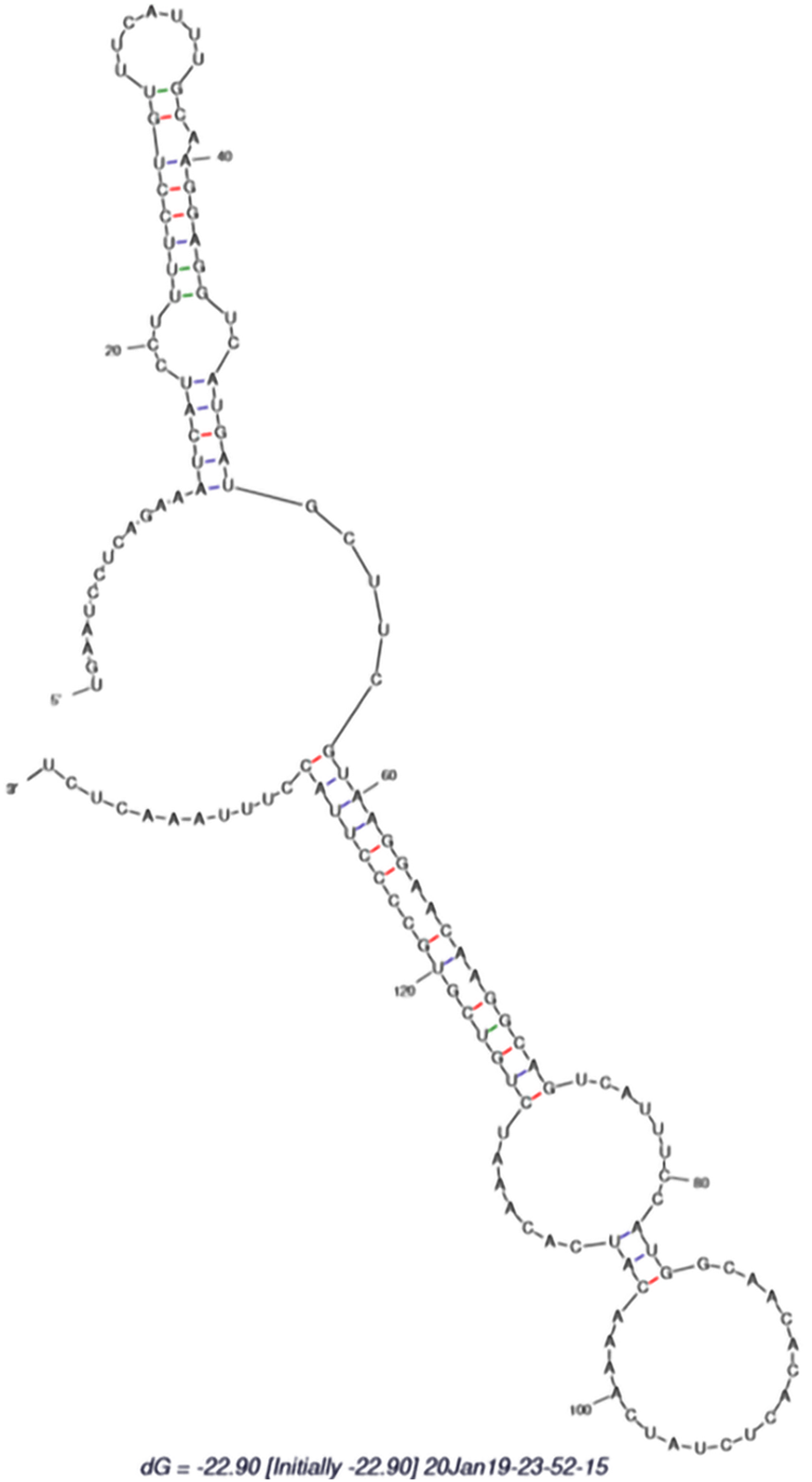
Selected EST sequences with not more than five mismatches were valid for this nonprotein encoding phenomenon using BLAST against the protein database at the National Center for Biotechnology Information using BLASTx with a default parameter[11,12] EST sequences were aligned to reference pre-miRNA sequences. Then the aligned portion was expressed as candidate pre-miRNA sequence[11]. Figure 1 shows the secondary structure of identified hsa-miR-802-5p. The incorporated pre-miRNAs were confirmed for secondary structure using mFold (http://www.mfold.rna.albary.edu/).
While selecting the RNA sequence from the EST resource as a candidate miRNA, the following criteria were referred as per Priyanka et al[11]: (1) RNA sequence must fold into an appropriate stem-loop hairpin 2D structure; (2) Mature miRNA sequence site in one arm of the hairpin structure; (3) miRNAs should have less than seven mismatches with the opposite miRNAs* sequence in the other arm; and (4) Predicted 2D structures have higher negative energy minimal free energy (≤ -18 kcal/mol). The prediction of miR-802-5p targets was determined using Target Scan. Table 1 represents the characteristics of a mature miR-802-5p.
| Source miRNA | Source organism | PL | MFE ∆ G | MS | Strand | A + U, % |
| hsa-miR-802 | Homo sapiens | 94 | -22.90 | CAGUAACAAAGAUUCAUCCUUGU | 3’ | 70 |
To validate this research paper, the available human T2DM ESTs were selected from the National Center for Biotechnology Information EST database for miR-802-5p and evaluated through the bioinformatics approach. The methodology for the identification of miR-802-5p was carried out as described by Priyanka et al[11] and Bai et al[12]. The source sequences, length of the precursor sequences, minimum folding energy and A + U content of the predicted miRNA are shown in Table 1. Secondary structural analysis of the pre-miRNA related sequence of the noncoding ESTs revealed the presence of miR-802-5p as shown in Figure 1. The minimum folding free energy was -37.90. It contained 61% A + U. From the above findings, it is clearly evident that miR-802-5p is present in T2DM ESTs, suggesting that it might have clinical relevance with disease progression. In addition, miRNA target analysis has been analyzed by the Target Scan online computational tool (http://www.targetscan.org/vert_72/) to identify miR-802-5p targets. Table 2 represents the identified targets for miR-802-5p.
| SI. No. | Target gene | Representative transcript | Gene name | Representative miRNA |
| 1 | TMED9 | ENST00000332598.6 | Transmembrane emp24 transport domain containing 9 | hsa-miR-802 |
| 2 | PCNP | ENST00000296024.5 | PEST proteolytic signal containing nuclear protein | hsa-miR-802 |
| 3 | C3orf58 | ENST00000441925.2 | Chromosome 3 open reading frame 58 | hsa-miR-802 |
| 4 | NUS1 | ENST00000368494.3 | Nuclear undecaprenyl pyrophosphate synthase 1 homolog | hsa-miR-802 |
| 5 | ZNF597 | ENST00000301744.4 | Zinc finger protein 597 | hsa-miR-802 |
In conclusion, miR-802-5p, a novel miRNA has been identified from human T2DM through a computational approach. However, further studies about miR-802-5p are required to prove how it is involved in the suppression and progression of T2DM. This computational approach proves the role of miRNAs and creates the platform for further research studies both in vitro and in vivo.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhang LL S-Editor: Gao CC L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, Xiao X, Wang Y. Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer. 2017;8:4106-4116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 2. | O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1160] [Cited by in RCA: 1272] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 3. | Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 367] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 4. | Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res. 2012;22:1163-1172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 5. | Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44:839-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1257] [Cited by in RCA: 1371] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 6. | Thanabalasingham G, Pal A, Selwood MP, Dudley C, Fisher K, Bingley PJ, Ellard S, Farmer AJ, McCarthy MI, Owen KR. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes Care. 2012;35:1206-1212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 7. | Nanditha A, Ma RC, Ramachandran A, Snehalatha C, Chan JC, Chia KS, Shaw JE, Zimmet PZ. Diabetes in Asia and the Pacific: Implications for the Global Epidemic. Diabetes Care. 2016;39:472-485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 321] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 8. | Chawla A, Chawla R, Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J Endocrinol Metab. 2016;20:546-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 539] [Cited by in RCA: 606] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 9. | Veazie S, Winchell K, Gilbert J, Paynter R, Ivlev I, Eden KB, Nussbaum K, Weiskopf N, Guise JM, Helfand M. Rapid Evidence Review of Mobile Applications for Self-management of Diabetes. J Gen Intern Med. 2018;33:1167-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 10. | Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Chen Q, Xia T, Wan XC. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 314] [Cited by in RCA: 277] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 11. | Priyanka P, Panagal M, Sivakumar P, Gopinath V, Ananthavalli R, Karthigeyan M, Paramasivam S, SR SK, Sekar D. Identification, expression, and methylation of miR-7110 and its involvement in type 1 diabetes mellitus. Gene Rep. 2018;11:229-234. [RCA] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Bai L, Li J, Panagal M, M B, Sekar D. Methylation dependent microRNA 1285-5p and sterol carrier proteins 2 in type 2 diabetes mellitus. Artif Cells Nanomed Biotechnol. 2019;47:3417-3422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |