Published online Jul 15, 2016. doi: 10.4251/wjgo.v8.i7.532
Peer-review started: February 17, 2016
First decision: March 9, 2016
Revised: March 19, 2016
Accepted: April 7, 2016
Article in press: April 12, 2016
Published online: July 15, 2016
Processing time: 143 Days and 17.9 Hours
AIM: To investigate the microRNA (miRNA) expression during histological progression from colorectal normal mucosa through adenoma to carcinoma within a lesion.
METHODS: Using microarray, the sequential changes in miRNA expression profiles were compared in colonic lesions from matched samples; histologically, non-neoplastic mucosa, adenoma, and submucosal invasive carcinoma were microdissected from a tissue sample. Cell proliferation assay was performed to observe the effect of miRNA, and its target genes were predicted using bioinformatics approaches and the expression profile of SW480 transfected with the miRNA mimics. mRNA and protein levels of the target gene in colon cancer cell lines with a mimic control or miRNA mimics were measured using qRT-PCR and Western blotting. The expression levels of miRNA and target gene in colorectal tissue samples were also measured.
RESULTS: Microarray analysis identified that the miR-320 family, including miR-320a, miR-320b, miR-320c, miR-320d and miR-320e, were differentially expressed in adenoma and submucosal invasive carcinoma. The miR-320 family, which inhibits cell proliferation, is frequently downregulated in colorectal adenoma and submucosal invasive carcinoma tissues. Seven genes including CDK6 were identified to be common in the results of gene expression array and bioinformatics analyses performed to find the target gene of the miR-320 family. We confirmed that mRNA and protein levels of CDK6 were significantly suppressed in colon cancer cell lines with miR-320 family mimics. CDK6 expression was found to increase from non-neoplastic mucosa through adenoma to submucosal invasive carcinoma tissues and showed an inverse correlation with miR-320 family expression.
CONCLUSION: MiR-320 family affects colorectal tumor proliferation by targeting CDK6, plays important role in its growth, and is considered to be a biomarker for its early detection.
Core tip: We investigated for the first time the sequential changes of miRNA expression profiles in colonic lesions from matched samples; histologically, non-neoplastic mucosa, adenoma, and submucosal invasive carcinoma were microdissected from a tissue sample. We have shown that the miR-320a, miR-320b, miR-320c, miR-320d are downregulated from colorectal adenoma and miR-320e is downregulated from colorectal submucosal carcinoma tissue and the miR-320 family suppresses tumor proliferation by targeting CDK6. The miR-320 family may play an important role in the growth of colorectal tumors and can be considered as a biomarker for the early detection of colorectal tumors.
- Citation: Tadano T, Kakuta Y, Hamada S, Shimodaira Y, Kuroha M, Kawakami Y, Kimura T, Shiga H, Endo K, Masamune A, Takahashi S, Kinouchi Y, Shimosegawa T. MicroRNA-320 family is downregulated in colorectal adenoma and affects tumor proliferation by targeting CDK6. World J Gastrointest Oncol 2016; 8(7): 532-542
- URL: https://www.wjgnet.com/1948-5204/full/v8/i7/532.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v8.i7.532
Colorectal cancer (CRC) is one of the most common malignancies resulting in cancer-related deaths in the world[1]. The adenoma-carcinoma sequence, which involves a series of changes from normal colorectal epithelium through an adenoma to an invasive and metastatic tumor, has been widely recognized as an important developmental mechanism in CRC[2]. According to this theory, the colorectal adenoma (CRA) is considered to be a precursor lesion of CRC; the removal of CRAs by polypectomy has been shown to reduce the incidence and mortality due to CRCs[3]. Endoscopic mucosal resection and endoscopic submucosal dissection (ESD) enable resection of almost all intramucosal neoplasia with submucosal invasion of less than 1000 μm in the colon[4]. Thus, detection of colorectal tumors in the early stages has become increasingly important in treatment and prognosis.
The adenoma-carcinoma sequence is accompanied by several genetic and epigenetic alterations, such as mutations of cancer-associated genes and epigenetic modifications, including changes in DNA methylation, histone modifications, and microRNAs (miRNAs)[5,6]. However, the involvement of miRNAs in the mechanism of this sequence remains undetermined. miRNAs are small (19-23 nucleotide) endogenous non-coding RNAs that regulate gene expression by targeting the 3′ untranslated region (UTR) of mRNA. miRNAs play fundamental roles in various biological processes[7]. Accumulating evidence indicates that miRNAs are frequently dysregulated in human cancers[8], and alterations of miRNA expression in colorectal tumors have been well documented. For example, miRNA profiles of CRC compared with those of normal mucosa[9] and adenoma[6,10] have been reported. However, limited reports exist on the sequential changes in miRNA expression during histological progression from normal colonic mucosa through colorectal adenoma to early carcinoma in a lesion from a patient.
Furthermore, according to the Paris and Japanese classification[11,12], a flat colorectal lesion exceeding 10 mm in diameter is classified as a “laterally spreading type (LST)” and subclassified into a granular (G) or a non-granular (NG) type. The percentage of gene mutation in the protruded tumor is hypothesized to be different from that of the LST[13]. When we focused on miRNA, expression changes of some miRNAs in exophytic and flat elevated tumors were reported[14]; however, there is no analysis comparing miRNA expressions of the LST with those of the protruded tumor.
Therefore, we analyzed miRNA expressions of both LSTs and protruded tumors as a specific feature of the stepwise progression from adjacent non-neoplastic mucosa to adenoma and submucosal invasive carcinoma using matched samples to compare accurate miRNA expression in each phase.
Formalin-fixed, paraffin-embedded (FFPE) colorectal tissue samples that included carcinoma, adenoma, and adjacent non-neoplastic mucosa were obtained from patients who underwent ESD at Tohoku University between January 2011 and December 2014 and provided written informed consent for study participation. All the cancers were limited to submucosal carcinomas, which were classified as T1M0N0 (American Joint Committee on Cancer Staging Manual) in this study. The 18 colorectal samples were LST (15 carcinoma with adenoma and 3 carcinoma without adenoma) and the 3 colorectal samples were protruded-type carcinoma with adenoma.
A senior pathologist reviewed the histopathological grades and types of all cases. Regions of colorectal carcinoma, pre-existing adenoma, and adjacent non-neoplastic mucosa in a resection specimen were microdissected using a laser microdissection (LMD) system (Leica Microsystems, Wetzlar, Germany). This study was reviewed and approved by the ethics committee and the institutional review board at the Tohoku University Hospital.
Human colon cancer cell lines HT29 and SW480 were obtained from the American Type Culture Collection (Manassas, VA) and then grown in Dulbecco’s Modified Eagle’s Medium supplemented with 10% (v/v) fetal bovine serum at 37 °C in a humidified atmosphere of 5% CO2. The human intestinal epithelial cells (InEpC) were obtained from Lonza, Japan.
We used mirVana™ miRNA 320a, b, c, d, and e mimic; mimic control; inhibitor; and inhibitor control (Life Technologies, Carlsbad, CA). These RNA oligonucleotides were transiently transfected into SW480 and HT29 cells using the Lipofectamine RNAiMAX Reagent (Invitrogen, United States) according to the manufacturer’s instructions.
Total RNA from microdissected FFPE samples was purified using a miRNAeasy FFPE kit (QIAGEN, Hilden, Germany). Total RNA isolated from cultured cells using TRIzol reagent (Invitrogen) was purified with an RNeasy Mini kit (QIAGEN).
The RNA from FFPE samples was labeled with a miRNA complete labeling and Hyb kit (Agilent Technologies, Santa Clara, CA, United States), and the labeled probes were hybridized onto Agilent Human miRNA Microarray Rel 19.0 according to the manufacturer’s instructions. The arrays were scanned and the data were extracted and analyzed using the Agilent Feature Extraction software and Agilent GeneSpring software.
RNA samples were reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit and miScript II RT Kit (QIAGEN). Each cDNA sample was analyzed in triplicate using the QuantiFast SYBR Green PCR kit (QIAGEN) with the StepOnePlus real-time PCR system (Life Technologies). A comparative ΔCt method was used for the quantification of the miR-320 family and CDK6 expression; expression levels of the hsa-miR-320 family and CDK6 were normalized by those of U6 and GAPDH, respectively. A miScript Primer Assay (QIAGEN) was used for the miR-320 family and U6. The following primer sets were used for other quantitative reverse transcription (qRT)-PCR assays: CDK6 Forward: 5′-GGATAAAGTTCCAGAGCCTGGAG-3′; CDK6 Reverse: 5′-GCGATGCACTACTCGGTGTGAA-3′ and GAPGH Forward: 5′-ATCAGCAATGCCTCCTGCAC-3′; Reverse: 5′-ATGGCATGGACTGTGGTCAT-3′.
Human colon cancer cell lines SW480 were seeded in 96-well plates, and cell proliferation was measured 24, 48, 72, 96, and 120 h later using the CellTiter96 Aqueous One Solution Cell Proliferation Assay (MTS assay; Promega, Madison, WI, United States) according to manufacturer’s instructions.
Gene expression profiling of SW480 transfected with a mimic control or miR-320a mimics was performed using the SurePrint G 3Human Gene Expression 8x60K v2 Microarray Kit (Agilent Technologies) according to the manufacturer’s instructions. Candidates of miRNA target genes were selected according to the results of these mRNA expression analysis and two different bioinformatics algorithms-TargetScan (http://www.targetscan.org) and Pic tar (http://pictar.mdc-berlin.de/).
Total cell lysates were prepared using a mammalian cell extraction kit (BioVision, Mountain View, CA, United States). Protein concentrations in the lysates were measured using the BCA Protein Assay kit (Pierce Chemical Co., Rockford, IL, United States). Equal amounts of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. After incubation with Tris-buffered saline and Tween-20 containing an ECL blocking agent, the membranes were incubated with primary antibodies against CDK6 (Cell Signaling Technology, Inc., Danvers, MA, United States) or α-tubulin (B512, Sigma) at 4 °C overnight and further incubated with secondary antibodies for 1 h at room temperature. Reactive bands were detected using the ECL Prime Western Blotting Detection Reagent (GE Healthcare, Bucks, United Kingdom).
Data from at least three independent experiments were analyzed. Statistical analysis was conducted using Excel (Microsoft). The difference between two groups was analyzed using the paired t-test for miRNA array data and the Student’s t-test for qRT-PCR data. The statistical significance of correlations between the expressions of the miR-320 family and CDK6 mRNA was evaluated using Pearson’s correlation analysis. P < 0.05 was considered statistically significant.
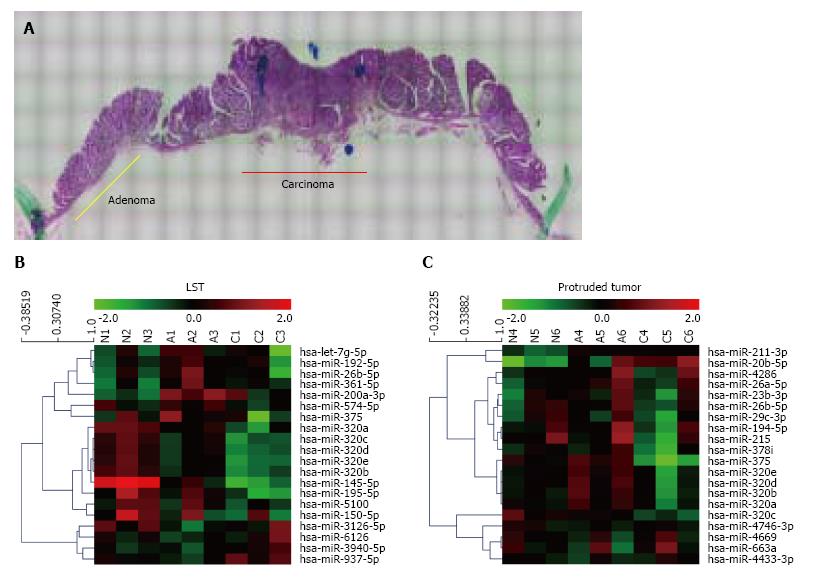
Sequential changes of miRNA expression profiles from matched samples, histologically non-neoplastic mucosa, adenoma, and submucosal invasive carcinoma microdissected by LMD from a tissue sample (Figure 1A), were assessed. To differentiate tumor forms, these analyses were conducted in each of the LSTs (n = 3) and protruded tumors (n = 3) (Figure 1B and C). All three LSTs were of the granular type. Seven miRNAs in LSTs and 23 miRNAs in protruded tumors showed significantly higher or lower expression in early carcinomas than that in adenomas, with expression in non-neoplastic mucosa as the baseline. The top 10 miRNAs in each form are summarized in Table 1. Comparing these results, six of the 10 miRNAs, including five belonging to the miR-320 family (320a, b, c, d, and e) were identical. In addition, we confirmed the downregulation of miR-195 (LST), miR-375, miR-378 (protruded tumor), and miR-26b (both forms) in early cancer, which has been previously reported in several studies of CRCs[9,15,16]. Decreased expression of miR-320a and 320b in CRC has also been reported previously[6,17,18]. It is interesting that these changes in miR-320 family expressions are common in early carcinomas both in LSTs and protruded tumors; therefore, we focused on the miR-320 family in the subsequent analysis.
| Systematic name | Expression levels vs normal (mean ± SD) | P value | ||
| Adenoma | Carcinoma | |||
| Laterally spreading types (n = 3) | ||||
| 1 | hsa-miR-320b | 0.649 ± 0.170 | 0.402 ± 0.146 | 0.00488 |
| 2 | hsa-miR-320e | 0.666 ± 0.159 | 0.376 ± 0.085 | 0.0213 |
| 3 | hsa-miR-937-5p | 0.842 ± 0.169 | 1.446 ± 0.253 | 0.0215 |
| 4 | hsa-miR-574-5p | 1.287 ± 0.477 | 0.881 ± 0.339 | 0.0373 |
| 5 | hsa-miR-320d | 0.705 ± 0.204 | 0.425 ± 0.108 | 0.0407 |
| 6 | hsa-miR-320a | 0.645 ± 0.140 | 0.412 ± 0.198 | 0.0417 |
| 7 | hsa-miR-26b-5p | 1.751 ± 0.307 | 1.003 ± 0.600 | 0.0477 |
| 8 | hsa-miR-320c | 0.676 ± 0.181 | 0.401 ± 0.072 | 0.0516 |
| 9 | hsa-miR-361-5p | 1.976 ± 0.124 | 1.469 ± 0.378 | 0.0762 |
| 10 | hsa-miR-195-5p | 0.729 ± 0.172 | 0.391 ± 0.338 | 0.0772 |
| Protruded tumors (n = 3) | ||||
| 1 | hsa-miR-320d | 1.450 ± 0.388 | 0.805 ± 0.369 | 0.000284 |
| 2 | hsa-miR-320b | 1.456 ± 0.386 | 0.834 ± 0.362 | 0.00203 |
| 3 | hsa-miR-375 | 1.312 ± 0.163 | 0.285 ± 0.174 | 0.00492 |
| 4 | hsa-miR-320e | 1.350 ± 0.278 | 0.772 ± 0.362 | 0.00724 |
| 5 | hsa-miR-320c | 0.879 ± 0.236 | 0.584 ± 0.219 | 0.0105 |
| 6 | hsa-miR-320a | 1.262 ± 0.208 | 0.810 ± 0.263 | 0.0110 |
| 7 | hsa-miR-663a | 0.985 ± 0.786 | 1.317 ± 0.798 | 0.0114 |
| 8 | hsa-miR-211-3p | 1.634 ± 0.193 | 1.533 ± 0.189 | 0.0129 |
| 9 | hsa-miR-378i | 1.110 ± 0.223 | 0.640 ± 0.333 | 0.0250 |
| 10 | hsa-miR-26b-5p | 1.261 ± 0.462 | 0.896 ± 0.360 | 0.0253 |
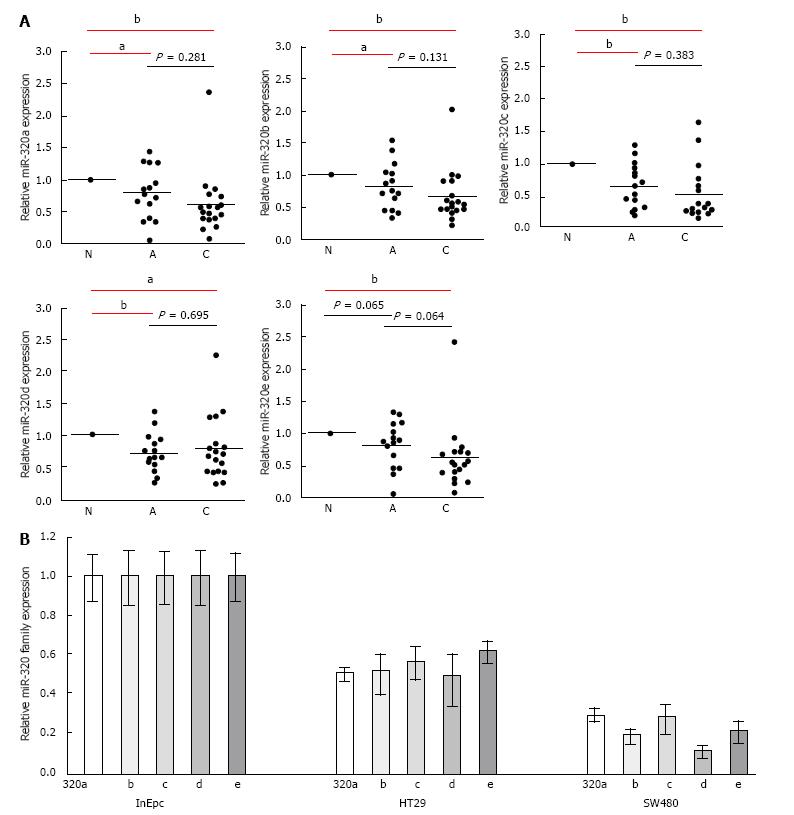
To confirm the downregulation of the miR-320 family in early carcinoma, we conducted qRT-PCR of the miR-320 family using 18 matched colorectal submucosal invasive carcinoma specimens in LSTs (Figure 2A). Expressions of all members of the miR-320 family except miR-320e in adenoma were significantly decreased not only in submucosal invasive carcinomas but also in adenomas compared with those in the non-neoplastic mucosa (P < 0.05 and P < 0.01, respectively). On comparing adenomas and carcinomas, expression levels of the miR-320 family, except for 320d, in carcinomas were lower than that in adenomas; however, differences were not statistically significant. These findings were confirmed in human colon cancers cell lines by qRT-PCR, and expression of the miR-320 family was found to be decreased in HT29 and SW480 compared with that in InEpC (Figure 2B).
These results indicated that the expression of the miR-320 family decreased from the early stages of the adenoma-carcinoma sequence.
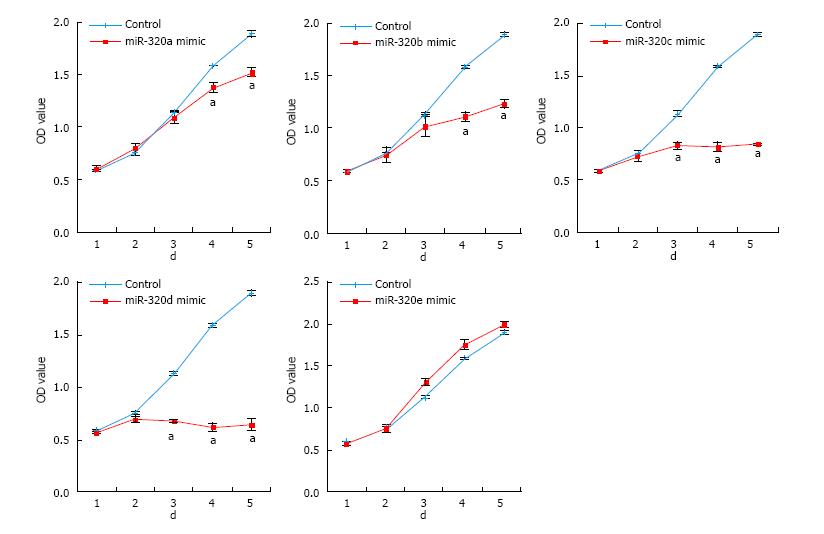
To reveal the role of the miR-320 family in CRC, cell proliferations in SW480 transfected with the miR-320 family were analyzed by MTS assay for 5 d. Overexpression of miR-320a, 320b, 320c, or 320d significantly inhibited the cell growth of SW480 (Figure 3), and inhibition of these miRNA significantly promoted proliferation of SW480 (data not shown). However, these findings were not observed with miR-320e.
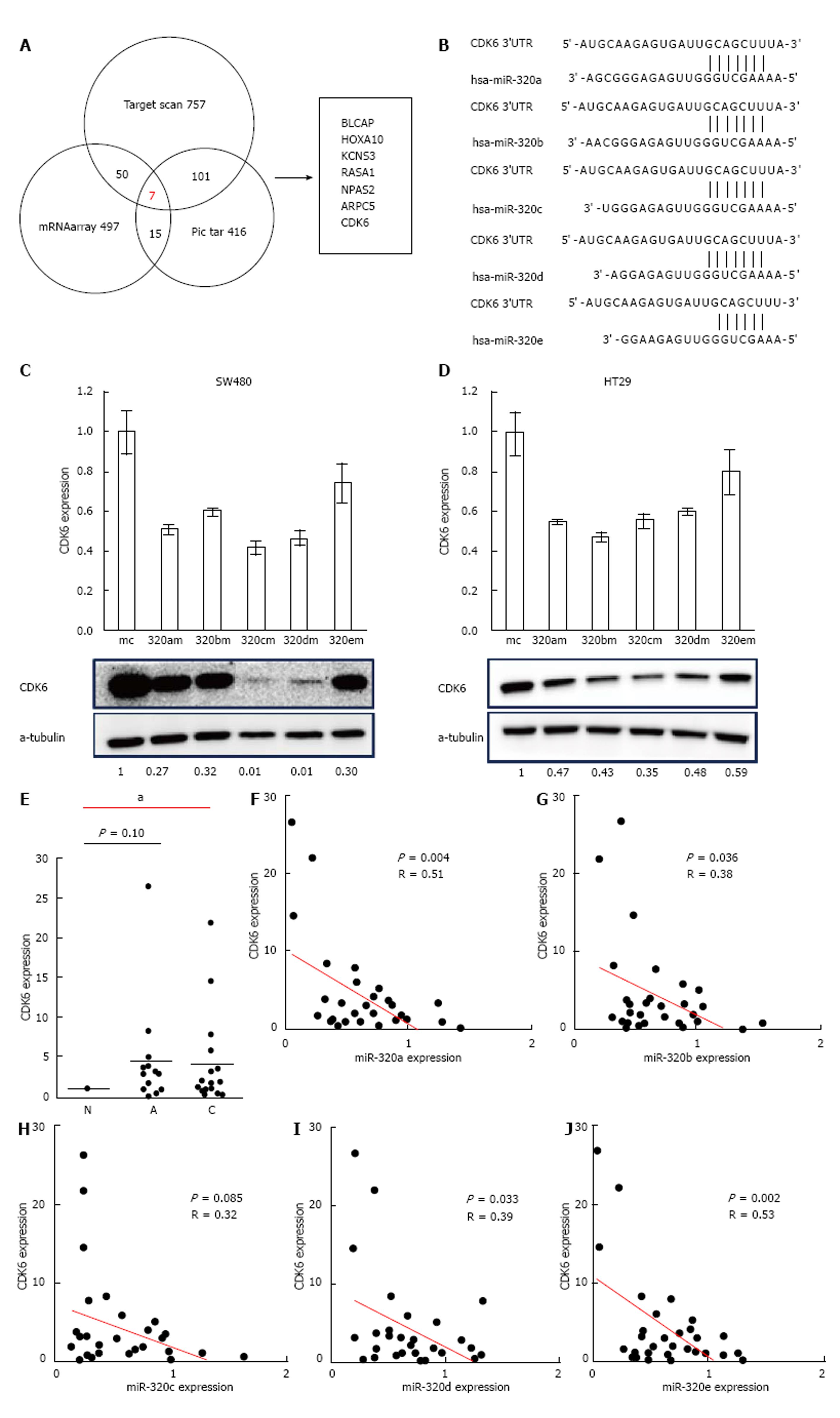
To find a target gene of the miR-320 family, we used miR-320a as a representative of the miR-320 family in microarray gene expression analysis and miRNA target prediction. Before the gene expression analysis, we confirmed that the expression of miR-320a in SW480 cells transfected with miR-320a mimics was increased by approximately 1600-fold compared with that in controls. Next, the gene expression analysis of SW480 with a mimic control or miR-320a mimics was conducted using microarray, and the expressions of 497 genes were found to be decreased in SW480 with miR-320a mimics (fold-change < 0.67). In addition to these tests, we conducted TargetScan and Pic tar analyses to detect potential targets of miR-320a; seven genes (BLCAP, HOXA10, KCNS3, RASA1, NPAS2, ARPC5, and CDK6) were identified to be common in the results of these three different analyses (Figure 4A). In seven candidate genes, CDK6 expression was shown to be associated with prognosis in patients with CRC[19]; moreover, the results of miRNA binding-site prediction analyses using bioinformatics tools (Target scan and microRNA.org) indicate that CDK6 has a putative miR-320 family’s binding site that is mapped to the 3′ UTR (Figure 4B). From these results, we selected CDK6 as a novel candidate target of the miR-320 family.
To examine whether the miR-320 family influences both mRNA and protein levels of CDK6, these were analyzed in colon cancer cell lines (HT29 and SW480) with a mimic control or miR-320 family mimics. Both mRNA and protein levels of CDK6 were significantly suppressed in cells with miR-320 family mimics (Figure 4C and D).
To confirm the changes of CDK6 expression in colorectal tissues, we investigated CDK6 expression in FFPE tissues of 18 matched samples. Two samples were excluded because of difficulty in detecting CDK6 expression. CDK6 expression in submucosal invasive carcinoma compared with non-neoplastic mucosa was significantly increased (P < 0.05, Figure 4E) and that in adenoma was also increased but without statistical significance (P = 0.10). Finally, we found that there is an inverse correlation between the expression levels of the miR-320 family, except miR-320c, and CDK6 in the 16 matched samples studied (P < 0.05, Figure 4F).
The main aim of this study was to investigate changes in miRNAs corresponding to the adenoma-carcinoma sequence and to clarify the functions of these miRNAs in carcinogenesis. We showed that the miR-320 family (a, b, c, d and e) were important factors in the progression of the early stages of colorectal tumors and found a novel target of the miR-320 family, CDK6. From the results of our comprehensive microarray miRNA analysis, miR-320a, b, c, d and e were downregulated from adenoma to submucosal invasive carcinoma in both LSTs and protruded tumors. It has been previously reported that miR-320a regulates tumor occurrence[20], progression[21], and metastasis[22] in CRC; therefore, we inferred that the miR-320 family may play an important role in colorectal carcinogenesis[23]. Moreover, 6 of the 10 miRNAs, including the miR-320 family, were identical between the LST-G type and protruded tumors; therefore, there is a possibility that the carcinogenic mechanisms of these two different forms share similar pathways. In contrast, the LST-NG type has been reported to show a significantly higher frequency of submucosal invasion than the LST-G type[24]; thus, their carcinogenic mechanisms might be different. Further verification of the carcinogenic mechanisms of the LST-NG type is warranted.
To investigate the expression of the miR-320 family in the colorectal tissue, we conducted expression analysis of the miR-320 family in 18 FFPE samples by RT-PCR. In addition to the results from our miRNA microarray analysis, which showed differences of the miR-320 expression between colorectal adenoma and carcinoma, qRT-PCR analyses showed progressively decreasing expression of the miR-320 family, except miR-320d, from non-neoplastic mucosa through adenoma to submucosal invasive carcinoma. There are some reports of a relationship between miRNA and colorectal adenoma[6,25], and miR-320a was downregulated from colorectal mucosa to low-grade dysplasia to high-grade dysplasia to adenocarcinomas in the same sample[6]. Our results support these previous findings, and this article presents the first report of the downregulation of miR-320b in CRA, miR-320c and d in CRA and CRC, and miR-320e in CRC. miR-320a has been shown to effectively regulate proliferation, cell cycle, invasion, migration, and epithelial-mesenchymal transition of CRC, particularly in advanced stages, and is a novel tumor and metastasis suppressor acting by directly targeting mRNAs of neuropilin 1, β-catenin, and other genes[17]. We confirmed that miR-320a inhibited cell proliferation in cancer cell lines but not how it affects tumor progression in earlier stages of the tumor. Further, the carcinogenic mechanisms of miR-320b, c, d, and e are poorly understood, particularly in CRC. Therefore, we first tried to identify novel targets of the miR-320 family to identify the carcinogenic mechanism in the early stages of the adenoma-carcinoma sequence. The most commonly used approach to find the target genes of miRNA is through bioinformatics algorithm. Recent reports have provided evidence that miRNAs may downregulate a greater number of transcripts than previously appreciated[26]; we adopted the results of the mRNA expression array analysis to narrow down the candidates. Finally, we selected seven target genes as common to two bioinformatics algorithms and our results of the mRNA array.
Carcinogenesis is believed to be caused by the dysregulation of the cell-cycle machinery. CDK6, a cyclin-D1-dependent kinase, plays an important role in G1/S phase transition of the cell cycle and sends signals modulating the control of cell development[27]. The function of CDK6 in CRC has been shown previously[19], and there are several reports on the relationships between CDK6 and miRNA in some types of cancers[28]. Moreover, it was reported that miR-320c inhibited tumor-like behaviors of bladder cancer by targeting CDK6[29]. However, no studies have examined the relationship between the expression of the miR-320 family and CDK6 expression in CRC.
We confirmed the decreased expression of mRNA and CDK6 in CRC cell lines transfected with the miR-320 family. In addition, we confirmed that CDK6 expression was downregulated in colorectal tumor tissues and that the expressions levels of the miR-320 family, except miR-320c, were negatively correlated with the mRNA expression levels of CDK6. These results suggested that the miR-320 family is targeted to CDK6 in CRC and has an influence on cell cycle. We report that the miR-320 family suppresses colorectal tumor progression by targeting CDK6.
In our study, the effects on tumor proliferation by overexpression of miR-320e were not observed, whereas overexpression of miR-320c and miR-320d resulted in a particularly strong reduction of tumor proliferation. CDK6 expression after introduction of miR-320e mimics in colon cancer cell lines indicated a similar tendency to be decreased at the mRNA and protein levels, but these were not as significant as the marked changes observed in other members of the miR-320 family. Targets of miRNAs can be predicted by requiring conserved Watson-Crick pairing to the 5′ region of the miRNA, known as the miRNA seed[30]. The seeds of miR-320e for CDK6 were maximum 6 mer. As the seeds of other members of the miR-320 family were maximum 8 mer, this difference in the structure of the miR-320 family might lead to differences in the effects on CDK6 expression. We believe that this results give support the relationship between miR-320 family and CDK6. Further validations of these are required.
In conclusion, we have shown that the miR-320 family, which suppresses tumor proliferation by targeting CDK6, is downregulated in colorectal adenoma and early colorectal carcinoma tissue. The miR-320 family may play an important role in the growth of colorectal tumors and can be considered a biomarker for the early detection of colorectal tumors.
According to the adenoma-carcinoma sequence, colorectal adenoma (CRA) is considered to be a precursor lesion of colorectal cancer (CRC); the removal of CRAs by polypectomy has been shown to reduce the incidence and mortality due to CRCs. Thus, detection of colorectal tumors in the early stages has become increasingly important in treatment and prognosis. The adenoma-carcinoma sequence is accompanied by several genetic and epigenetic alterations, such as mutations of cancer-associated genes and epigenetic modifications, including changes in microRNAs (miRNAs). However, the involvement of miRNAs in the mechanism of this sequence remains undetermined.
miRNAs are frequently dysregulated in human cancers, and alterations of miRNA expression in colorectal tumors have been well documented. For example, miRNA profiles of CRC compared with those of normal mucosa and adenoma. However, limited reports exist on the sequential changes in miRNA expression during histological progression from normal colonic mucosa through colorectal adenoma to early carcinoma in a lesion from a patient. miR-320a has been shown to effectively regulate proliferation, cell cycle, invasion, migration, and epithelial-mesenchymal transition of CRC. However, the carcinogenic mechanisms of miR-320b, c, d, and e are poorly understood, particularly in CRC, and needed further exploration.
The authors investigated for the first time the sequential changes of miRNA expression profiles in colonic lesions from matched samples; histologically, non-neoplastic mucosa, adenoma, and submucosal invasive carcinoma were microdissected from a tissue sample. They have shown for the first time that the miR-320b, miR-320c, miR320d are downregulated from colorectal adenoma and miR320e is downregulated from colorectal submucosal carcinoma tissue and the miR-320 family suppresses tumor proliferation by targeting CDK6.
This study suggests that the miR-320 family affects colorectal tumor proliferation by targeting CDK6, plays an important role in the growth of colorectal tumors. From these results, miR-320 family is considered as a biomarker for early detection of colorectal tumor.
The adenoma-carcinoma sequence is accompanied by several genetic and epigenetic alterations, such as mutations of cancer-associated genes and epigenetic modifications, including changes in DNA methylation, histone modifications, and miRNAs. miRNAs are small (19-23 nucleotide) endogenous non-coding RNAs that regulate gene expression by targeting the 3′ untranslated region of mRNA. miRNAs play fundamental roles in various biological processes.
The manuscript by Tadano et al investigated the expression of miRNA in colorectal adenoma and submucosal invasive carcinoma and revealed miR-320 family affects colorectal tumor proliferation by targeting CDK6. This article is overall interesting and gives new insight in the field of dysregulation of miRNAs and colorectal cancer.
P- Reviewer: Fang JY, Huang ZH, Wang YH S- Editor: Gong ZM L- Editor: A E- Editor: Lu YJ
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21363] [Article Influence: 2136.3] [Reference Citation Analysis (3)] |
| 2. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8003] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 3. | Zauber AG, Winawer SJ, O’Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2283] [Article Influence: 175.6] [Reference Citation Analysis (1)] |
| 4. | Tanaka S, Oka S, Kaneko I, Hirata M, Mouri R, Kanao H, Yoshida S, Chayama K. Endoscopic submucosal dissection for colorectal neoplasia: possibility of standardization. Gastrointest Endosc. 2007;66:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 349] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 5. | Leslie A, Carey FA, Pratt NR, Steele RJ. The colorectal adenoma-carcinoma sequence. Br J Surg. 2002;89:845-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 439] [Cited by in RCA: 462] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 6. | Gattolliat CH, Uguen A, Pesson M, Trillet K, Simon B, Doucet L, Robaszkiewicz M, Corcos L. MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur J Cancer. 2015;51:409-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16069] [Article Influence: 1004.3] [Reference Citation Analysis (2)] |
| 8. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 9. | Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamilton SR. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B, Meijer GA. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707-714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 194] [Cited by in RCA: 209] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 11. | The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3-43. [PubMed] |
| 12. | Kudo Se, Lambert R, Allen JI, Fujii H, Fujii T, Kashida H, Matsuda T, Mori M, Saito H, Shimoda T, Tanaka S, Watanabe H, Sung JJ, Feld AD, Inadomi JM, O’Brien MJ, Lieberman DA, Ransohoff DF, Soetikno RM, Triadafilopoulos G, Zauber A, Teixeira CR, Rey JF, Jaramillo E, Rubio CA, Van Gossum A, Jung M, Vieth M, Jass JR, Hurlstone PD. Nonpolypoid neoplastic lesions of the colorectal mucosa. Gastrointest Endosc. 2008;68:S3-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 363] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 13. | Takahashi T, Nosho K, Yamamoto H, Mikami M, Taniguchi H, Miyamoto N, Adachi Y, Itoh F, Imai K, Shinomura Y. Flat-type colorectal advanced adenomas (laterally spreading tumors) have different genetic and epigenetic alterations from protruded-type advanced adenomas. Mod Pathol. 2007;20:139-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Nakagawa Y, Akao Y, Taniguchi K, Kamatani A, Tahara T, Kamano T, Nakano N, Komura N, Ikuno H, Ohmori T. Relationship between expression of onco-related miRNAs and the endoscopic appearance of colorectal tumors. Int J Mol Sci. 2015;16:1526-1543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 16. | Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 17. | Zhang Y, He X, Liu Y, Ye Y, Zhang H, He P, Zhang Q, Dong L, Liu Y, Dong J. microRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep. 2012;27:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 18. | Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, Yan B, Bian YQ, Zhao J, Wang WZ. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun. 2012;420:787-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 128] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Meng LH, Zhang H, Hayward L, Takemura H, Shao RG, Pommier Y. Tetrandrine induces early G1 arrest in human colon carcinoma cells by down-regulating the activity and inducing the degradation of G1-S-specific cyclin-dependent kinases and by inducing p53 and p21Cip1. Cancer Res. 2004;64:9086-9092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Tsikitis VL, White I, Mori M, Potter A, Bhattcharyya A, Hamilton SR, Buckmeier J, Lance P, Thompson P. Differential expression of microRNA-320a, -145, and -192 along the continuum of normal mucosa to high-grade dysplastic adenomas of the colorectum. Am J Surg. 2014;207:717-722; discussion 722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Zhao H, Dong T, Zhou H, Wang L, Huang A, Feng B, Quan Y, Jin R, Zhang W, Sun J. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis. 2014;35:886-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 22. | Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, Goel A. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107:dju492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (1)] |
| 23. | Wan LY, Deng J, Xiang XJ, Zhang L, Yu F, Chen J, Sun Z, Feng M, Xiong JP. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun. 2015;457:125-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 24. | Uraoka T, Saito Y, Matsuda T, Ikehara H, Gotoda T, Saito D, Fujii T. Endoscopic indications for endoscopic mucosal resection of laterally spreading tumours in the colorectum. Gut. 2006;55:1592-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 307] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 25. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 376] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 26. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 3742] [Article Influence: 187.1] [Reference Citation Analysis (0)] |
| 27. | Sherr CJ. Cancer cell cycles. Science. 1996;274:1672-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3965] [Cited by in RCA: 4004] [Article Influence: 138.1] [Reference Citation Analysis (0)] |
| 28. | Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9:1809-1818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 29. | Wang X, Wu J, Lin Y, Zhu Y, Xu X, Xu X, Liang Z, Li S, Hu Z, Zheng X. MicroRNA-320c inhibits tumorous behaviors of bladder cancer by targeting Cyclin-dependent kinase 6. J Exp Clin Cancer Res. 2014;33:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 30. | Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3812] [Cited by in RCA: 3951] [Article Influence: 188.1] [Reference Citation Analysis (0)] |