Published online Nov 15, 2015. doi: 10.4251/wjgo.v7.i11.271
Peer-review started: May 15, 2015
First decision: June 2, 2015
Revised: June 20, 2015
Accepted: September 30, 2015
Article in press: October 9, 2015
Published online: November 15, 2015
Processing time: 187 Days and 2.9 Hours
Colorectal cancer (CRC) remains a leading cause of cancer death in both men and women worldwide. Among the factors and mechanisms that are involved in the multifactorial etiology of CRC, autophagy is an important transformational switch that occurs when a cell shifts from normal to malignant. In recent years, multiple hypotheses have been considered regarding the autophagy mechanisms that are involved in cancer. The currently accepted hypothesis is that autophagy has dual and contradictory roles in carcinogenesis, but the precise mechanisms leading to autophagy in cancer are not yet fully defined and seem to be context dependent. Autophagy is a surveillance mechanism used by normal cells that protects them from the transformation to malignancy by removing damaged organelles and aggregated proteins and by reducing reactive oxygen species, mitochondrial abnormalities and DNA damage. However, autophagy also supports tumor formation by promoting access to nutrients that are critical to the metabolism and growth of tumor cells and by inhibiting cellular death and increasing drug resistance. Autophagy studies in CRC have focused on several molecules, mainly microtubule-associated protein 1 light chain 3, beclin 1, and autophagy related 5, with conflicting results. Beneficial effects were observed for some agents that modulate autophagy in CRC either alone or, more often, in combination with other agents. More extensive studies are needed in the future to clarify the roles of autophagy-related genes and modulators in colorectal carcinogenesis, and to develop potential beneficial agents for the prognosis and treatment of CRC.
Core tip: This review describes the role of autophagy in cancer, focusing on the involvement of autophagy in colorectal cancer (CRC). Initially, we describe the steps and components of autophagy, and we then further highlight the dual role of autophagy in cancer, where it can potentially act as both a promoter and an inhibitor during the transformation from normal to malignant cell. In particular, we emphasize the major autophagy genes involved in CRC pathogenesis along with autophagy-modulating agents and their modes of action in the context of CRC therapy.
- Citation: Burada F, Nicoli ER, Ciurea ME, Uscatu DC, Ioana M, Gheonea DI. Autophagy in colorectal cancer: An important switch from physiology to pathology. World J Gastrointest Oncol 2015; 7(11): 271-284
- URL: https://www.wjgnet.com/1948-5204/full/v7/i11/271.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v7.i11.271
Despite advances in diagnosis and treatment, colorectal cancer (CRC) remains one of the major causes of cancer death in both sexes worldwide: It is the third most common diagnosed cancer in males and the second most common in females[1]. It is well known that many risk factors, including multiple genes and environmental influences, are involved in malignant transformation. Recent research provides new data regarding the complex mechanisms involved in colorectal carcinogenesis. Among these mechanisms, autophagy is important in the switch from normal to malignant colorectal cells. The involvement of autophagy in cancer appears to be context specific, with evidence suggesting that it can have a dual role in both tumor suppressing and tumor promoting activities. Moreover, autophagy performs important functions in different processes that are connected to carcinogenesis, including inflammation, immune response and genome stability.
Here, we describe the involvement of autophagy in carcinogenesis, with a particular emphasis on CRC. We summarize the components and steps of macroautophagy (herein referred to as autophagy), and we emphasize the conflicting roles of autophagy in cancer, indicating that it has both promoter and suppressor mechanisms during malignant transformations. The second part of this study is focused on the autophagy genes and proteins that are associated with CRC. Finally, the effects of autophagy-based drugs in CRC treatment are discussed.
Autophagy is an evolutionarily conserved catabolic process that is characterized by cellular self-digestion and the removal of excessive, long-lived or dysfunctional organelles and proteins[2]. Autophagy occurs as a physiological process in normal cells at a basal level to assure cellular homeostasis, or as a strategic survival mechanism that recycles energy and nutrients under special conditions. Hypoxia, stress and nutrient deprivation trigger autophagy as a critical adaptive response during starvation[3]. Three morphologically distinct forms of autophagy can be distinguished: macroautophagy, microautophagy and chaperone-mediated autophagy[4]. Macroautophagy is identified by the presence of double membrane vesicles known as an autophagosomes, which engulf cytoplasmic components that include damaged organelles and deliver them to lysosomes for degradation. The other two forms, microautophagy and chaperone-mediated autophagy, involve a direct membrane invagination to engulf damaged proteins and the translocation of soluble cytosolic proteins by chaperone-dependent selection across the lysosomal membrane, respectively[5,6].
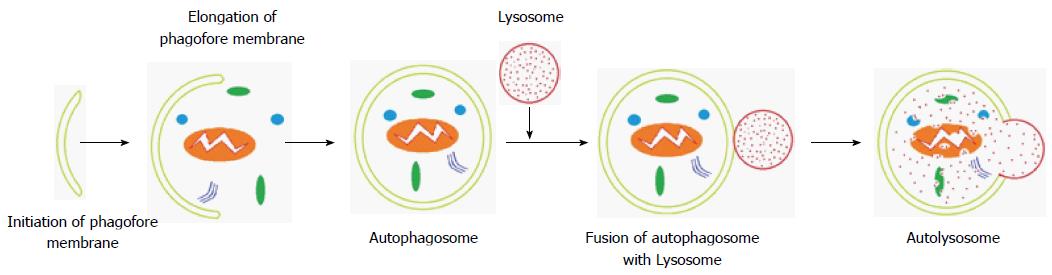
Autophagy-related genes (ATGs) play a critical role in facilitating the regulation of well-orchestrated autophagy. To date, thirty-six ATGs have been identified[7]. Autophagosome formation is initiated by unc-51-like kinase (ULK) and class III phosphatidylinositol 3-kinase (PI3K) complexes. The ULK complex consists of ATG13, ATG101, ULK1/2 and family-interacting protein FIP200[8,9]. Under normal growth conditions, the mammalian target of rapamycin (mTOR) complex inhibits the formation of the ULK complex, in effect blocking autophagy, and the ULK components are dissociated. Various stimuli (e.g., hypoxia, starvation) inhibit mTOR, allowing the ULK kinase complex to be activated, which initiates the formation of an isolation membrane (Figure 1) called a phagophore[10,11]. The origin of phagophores has not been explained, but the plasma membrane, endoplasmic reticulum, Golgi apparatus and mitochondria are all possible sources[12]. The completion of this critical step is driven by vacuolar sorting protein 34, a class III PI3K that is bound to beclin-1, and other ATG proteins (e.g., ATG14), which generate PI3K, the second complex, that catalyzes the production of phosphatidylinositol-3-phosphate[10,13].
Autophagosome elongation and closure steps and the further conversion to a nascent closed autophagosome are controlled by two ubiquitin-like conjugates. First, ATG12 forms a conjugate with ATG5 under the control of ATG7 and ATG10, which have E1 and E2-like enzyme activity, respectively. The resulting ATG12-ATG5 complex interacts with ATG16L1 to form a multimeric ATG12-ATG5-ATG16L1 conjugate that is located on the outer surface of the autophagosomal membrane. It will dissociate from the membrane upon completion of the autophagosome[14,15]. The second ubiquitin-like pathway involves the conjugation of the microtubule-associated protein 1-light chain 3 (LC3-I) to the lipid phosphatidylethanolamine (PE) by ATG7 and ATG3, which is an E2-like enzyme, to form the membrane-bound LC3-II. LC3 is initially synthesized as a precursor protein, proLC3, and is immediately processed to LC3-I by ATG4 through cleavage of its C-terminal amino acid. The membrane-bound form of LC-3, LC-3II, is recruited to both sides of the autophagosomal membrane[16,17]. After fusion with lysosomes, LC3-II on the cytoplasmic face of the autolysosome can be delipidated by ATG4 and recycled, whereas proteins located on internal surface of the autophagosome are processed for degradation by lysosomal enzymes in autolysosomes. During the maturation process, lysosomal-associated membrane protein 2 and the Ras-related protein Rab-7a facilitate autophagosome fusion with endocytic and lysosomal compartments to form an autolysosome. Autophagic cargo is then degraded through the activity of lysosomal proteases[18-21].
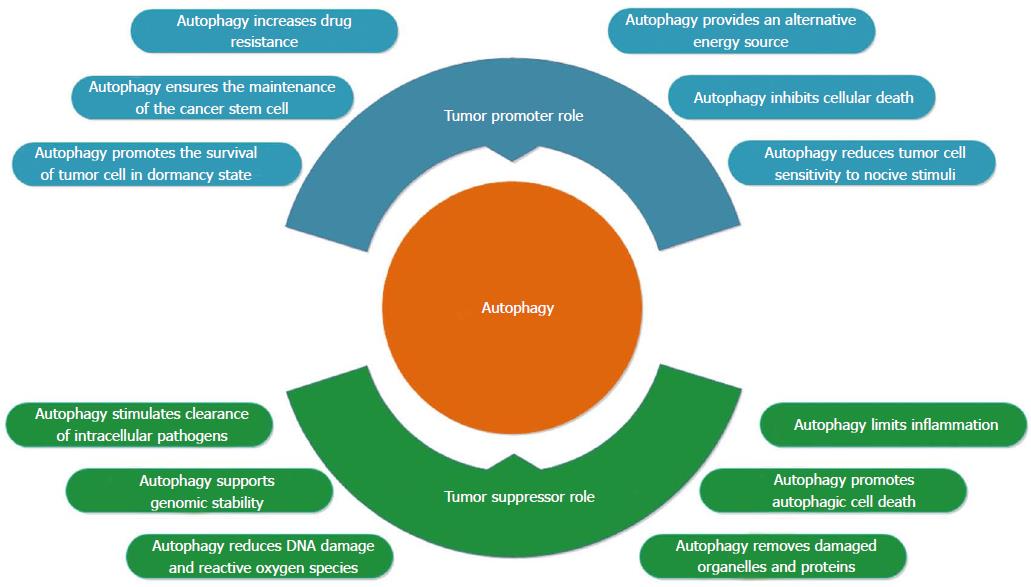
Autophagy plays crucial roles in the pathogenesis of various human diseases, including cancer, neurodegenerative diseases, infection, and cardiovascular, metabolic, and pulmonary diseases, and aging[22]. The currently accepted hypothesis is that autophagy has dual, contradictory roles in carcinogenesis (Figure 2). First, autophagy is a surveillance mechanism in normal cells, where it acts to protect cells from malignant transformations by removing damaged organelles and aggregated proteins and reducing DNA damage, reactive oxygen species (ROS) and mitochondrial abnormalities. However, autophagy also supports tumor formation by providing access to nutrients that are critical to the metabolism and growth of tumor cells, and by inhibiting cellular death and increasing drug resistance[7,23]. The response of cells to autophagy during cancer metastasis is stage dependent. Autophagy may help to reduce cancer metastasis in the early steps of tumor cell dissemination by promoting inflammatory responses against tumors. Furthermore, autophagy limits tumor necrosis and the expansion of dormant cancer cells into micrometastases, in tandem with impairing oncogene-induced senescence[24]. Autophagy seems to support metastasis during advanced stages of cancer by increasing the survival of detached metastatic cells in the absence of extracellular matrix, and by supporting the dissemination of cancer cells to distant organ sites by triggering tumor cells that lack a connection with the extracellular matrix in the new environment to shift to a dormant state until appropriate conditions occur[24,25].
Autophagy can prevent the transformation from normal to malignant through several suppressive mechanisms. An appropriate autophagic response is necessary for genome stability and for the clearance of mutagens because it acts to prevent the accumulation of the genetic defects that accompany malignant transformations. Damaged mitochondria and the redox-active aggregates of ubiquitinated proteins are removed by autophagy, resulting in avoidance of the overproduction of highly genotoxic ROS[26]. Inhibition of autophagy switches off this protection and can expose cells to ROS cytotoxicity, which promotes the activation of oncogenes[27,28]. In addition to mitophagy, autophagy supports genomic stability by enabling the discarding of micronuclei that are produced by cell cycle anomalies[29], and it may also promote autophagic cell death, known as type II programmed cell death, under certain conditions[30,31].
The impact of autophagy on tumor progression exhibits a significant degree of context dependence[23]. BECN1 gene studies in hormone-related cancers unmasked, for the first time, the possible tumor suppressing role of autophagy[32,33]. There remains significant debate regarding the role of BECN1 as a tumor suppressor due to the proximity of BECN1 to BRCA1, a well-known tumor suppressor gene. Both of these genes are located on human chromosome 17q21[34]. The role of autophagy as an important tumor suppressive process that has been demonstrated in murine experiments. Lack of BECN1 gene in embryoid bodies leads to embryonic death[35], and mice with a heterozygotic deletion of BECN1 demonstrate increased susceptibility to tumorigenesis in multiple tissues[36,37]. Similarly, mice deficient for ATG5 and ATG7 died after birth[38,39], while mice with mosaic deletion of ATG5 and liver-specific ATG7-deficient mice developed only benign liver adenomas[40]. Mice lacking autophagy genes ATG5 or ATG7 acquired premalignant pancreatic cancer, while the progression to pancreatic cancer driven by KRasG12D was blocked[41]. ATG7 deletion in a murine model (BrafV600E-induced lung cancer) initially accelerated the proliferation of tumor cells, but at later stages of tumorigenesis it reduced tumor burden, blocked conversion to a more malignant phenotype and increased the life spans of experimental mice[42]. In the absence of autophagy, the advance to cancer can be arrested, resulting in protection from conversion into malignant cells. Progression to a malignant phenotype may require additional genetic alterations[43].
In addition, autophagy is involved in both innate and adaptive immune responses, by which it prevents the establishment and proliferation of malignant cells[44]. Malignant transformation can be stimulated by an inflammatory microenvironment, which contains high amounts of potentially genotoxic ROS as well as various mitogenic cytokines[45]. Autophagy limits inflammation by efficiently disposing of inflammasomes, thereby inhibiting the pro-inflammatory signals that are delivered by some pattern recognition receptors, such as RIG-I-like receptors[46], and limiting the abundance of B-cell CLL/lymphoma 10, a protein that is involved in pro-inflammatory NF-κB signaling[47]. Autophagy ensures a well-coordinated and appropriate response, enabling crucial cells in the immune system to develop properly and to produce interferon, secrete antimicrobial peptides or present antigens to stimulate adaptive immunity. Dying malignant cells may determine innate and/or adaptive antitumor immune responses by recruiting antigen-presenting cells and other cellular components of the immune system. Thus, defects in autophagy may prevent the host immune system from properly recognizing and eliminating premalignant and malignant cells. Moreover, autophagy mediates potent anti-inflammatory effects[48,49].
Autophagy plays a key role in the first line of defense against pathogens and thus has anticarcinogenic effects that combat viral and bacterial infections. A xenophagic response is required for the stimulation of pathogen-specific immune responses and for the rapid clearance of intracellular pathogens[48]. Some of these processes are associated with digestive cancers (e.g., Helicobacter pylori, which is associated with gastric carcinoma, or Streptococcus bovis, which may cause colorectal carcinoma)[50,51].
Autophagy seems to promote malignant progression and resistance to therapy following the initiation of tumor growth[2,27]. As a conserved cellular survival mechanism, tumor cells can use autophagy to provide a backup energy source for survival and expansion[52]. During the progression of tumors, malignant cells are under metabolic stress as a result of a high proliferation rate and exposure to hypoxia, and nutrient deprivation due to inadequate blood supply or selective pressure from therapeutic intervention[53]. Tumor cells usually have a high proliferation rate, which demands more energy and resources than normal cells, and both ATP and metabolites can be obtained by increasing autophagy[54]. Although angiogenesis does occur in tumors, the availability of glucose and glutamine is reduced in some tumor regions due to the leakiness of tumor-associated vessels and continued hypovascularization[55].
Autophagy is activated in the hypoxic areas of tumors, and the inhibition of autophagy by AKT activation or by monoallelic disruption of BECN1 promotes cell death specifically in those regions. These results support hypothesis that tumor cells can use autophagy as a surveillance mechanism under metabolic stress conditions, to provide an alternative energy source for the survival and proliferation of malignant cells[52].
The pro-malignant role of autophagy has been demonstrated in tumor studies in which the inhibition of autophagy was linked to reduced tumor processes. Moreover, down-regulating the expression of essential autophagy proteins impaired tumor growth and led to the accumulation of abnormal mitochondria and reduced oxygen consumption, and autophagy was necessary to support the growth of Ras-driven tumors[56]. However, increased autophagy has also been associated with poor outcomes and short disease-free periods in human pancreatic cancers[57]. In vitro studies have shown that the survival of Ras-driven cancer cells requires autophagy and that gaining autophagy results in a marked increase in the survival of malignant cells under conditions of metabolic stress[28]. Inhibiting autophagy by deleting ATG5 prevents the progression of premalignant lesions to cancer in either a p53-independent or p53-dependent manner[41,58]. Furthermore, deletion of ATG7 decreases the tumor growth rate and induces nonmalignant tumor formation. In addition, non-Ras-driven tumoral cell types also need autophagy for survival, and the loss of autophagy has been shown to inhibit malignant tumor development. For example, FIP200 deletion significantly reduced proliferation and suppressed mammary tumor initiation and progression in a mouse model of breast cancer driven by the PyMT oncogene[59]. In a Palb2 knockout mouse model, heterozygous deletion of the autophagy gene BECN1 reduced Palb2-associated mammary tumorigenesis in a p53-dependent manner, indicating that in the presence of DNA damage and oxidative stress, autophagy can support tumor development by suppressing p53[60].
Autophagy can improve the resistance of cancer cells to detachment from the basal membrane, resulting in transformed cells that are less sensitive to therapy-induced cell death. Moreover, this activity sustains the survival of cancer cells that enter a state of dormancy or senescence in response to therapy and ensures the maintenance of the cancer stem cell compartment[23].
Autophagic responses favor the growth and progression of established tumors by reducing their sensitivity to different stimuli that would normally promote their death[61]. KRasG12D-driven pancreatic adenocarcinoma cells that enter a state of dormancy in response to oncogene ablation have recently been shown to activate autophagy to efficiently counteract metabolic stress[62], demonstrating the functional and phenotypic features of cancer stem cells. In addition, mammary cancer stem cells are often characterized by elevated autophagic flux, and their ability to efficiently form tumors in vivo appears to rely on autophagy, as tumor formation can be abolished through the genetic inhibition of BECN1 or ATG4A[63,64]. Thus, autophagy may also sustain tumor progression by preserving the viability of the cancer stem cell compartment and/or by promoting the persistence of dormant cancer cells.
Moreover, autophagy is required not only for the emission of immunostimulatory signals by malignant cells succumbing to specific anticancer agents but also for the activation of tumor-targeting innate and adaptive immune responses[49]. Cancer cells that have been isolated from established tumors where autophagy was inhibited were less resistant to exogenous stimuli than their wild-type counterparts[61]. In line with these data, autophagy-deficient tumors are often more sensitive to several chemotherapeutic agents and radiation therapy than their autophagy-proficient counterparts[65,66]. Cancer cells that are exposed to therapeutic interventions can also undergo senescence. Although senescent cells do not proliferate, they may support disease relapse by releasing a wide panel of pro-inflammatory and mitogenic cytokines into the microenvironment[67].
The autophagy machinery involves multiple genes and proteins that have critical functions in complex autophagic pathways, and these genes may be involved in the important switch from normal to colorectal pathology under specific conditions (Table 1).
| Gene/protein | Expression level in colorectal cancer |
| LC3/LC3-II | Higher expression, especially in advanced stages[20] |
| Higher expression associated with aggressiveness[71] | |
| Higher perinuclear expression associated with positive prognosis[77] | |
| Higher levels in DLD-1 and SW480 CRC lines treated with autophagy inhibitors[72] | |
| Higher levels in CRC cell lines treated with 5-FU[73] | |
| Higher levels in CRC cell lines treated with 5-FU and radiotreated[74] | |
| Lower levels associated with good outcome and treatment response[75,76] | |
| Negative expression associated with poor clinical outcome and survival[87] | |
| BECN1/ | Higher expression, negatively linked to metastasis[82] |
| Beclin-1 | Higher expression associated with favorable outcome[83] |
| Higher expression associated with longer survival in patients treated with 5-FU[84] | |
| Higher expression associated with a worse survival in patients treated with 5-FU[85] | |
| Higher expression associated with metastasis and worse prognosis[86] | |
| Lower levels associated with increased survival in advanced CRC patients treated with cetuximab[75,76] | |
| Lower levels associated with poor clinical outcome and survival[87] | |
| Lower levels associated with a good response after chemoradiation in patients with rectal cancer[88] | |
| ATG5 | Higher levels associated with lymphovascular invasion[92] |
| Lower levels[91] | |
| Lower expression associated with poor clinical outcome survival[87] | |
| Lower expression enhanced sensitivity to oxaliplatin[93] | |
| ATG10 | Higher expression associated with tumor lymph node metastasis and poor survival[95] |
| ATG16L1 | ATG16L1T300A polymorphism improved overall survival in human CRC patients[116] |
| BCL2/Bcl-2 | Higher levels associated with migration and invasion[105] |
| Higher levels associated with resistance to paclitaxel[106] | |
| Bif-1 | Lower levels[109] |
The LC3 gene family encodes three isoforms (LC3A, LC3B, and LC3C) and is the mammalian homologue of yeast ATG8[68]. The isoform LC3B is cleaved into the soluble form LC3B-I, which is conjugated with PE to generate the lipidated form (LC3B-II). LC3B-II accumulates specifically on nascent autophagosomes and is one of the most widely and reliably used markers for autophagy[69]. LC3 was the first autophagy marker proposed to be involved in human CRC[70]. LC3-II is overexpressed in CRC compared to normal tissue, especially in advanced stages[20]. Zheng et al[71] reported that LC3B-II was overexpressed in cancer cells and that autophagy enhanced the aggressiveness of CRC. LC3B expression in the peripheral areas of CRC tissues was correlated with tumor differentiation, growth pattern at the tumor margin, pN and pStage, as well as vessel and nerve plexus invasion. An increased level of LC3-II protein was found in DLD-1 and SW480 CRC-derived cell lines that were treated with a combination of autolysosome inhibitors. Association with 3-methyl adenine (3-MA), an inhibitor of PI3K, blocks autophagosome formation and led to increased apoptosis in treated CRC cell lines[72]. The treatment of CRC cell lines with 5-fluorouracil (5-FU) activated the autophagic process as a protective mechanism in cancerous cells, increased LC3-II levels and reduced the rate of apoptosis compared with untreated cell lines, and an increase in the apoptotic rate was induced by adding 3-MA to 5-FU[73]. Similar results were reported by Schonewolf et al[74], who reported that both 5-FU treated and radiotreated CRC cell lines showed an increase in autophagy. After adding chloroquine (CQ) to the treatment, these authors reported an increase in the sensitivity of malignant cells to apoptosis. However, in early stages, LC3-II expression levels were decreased compared with normal tissue[20]. A low LC3 value has been associated with a good response to treatment and a good survival prognosis, especially in patients with advanced CRC[75,76]. Perinuclear LC3A expression has been shown to be a positive predictor in patients with stage IIA-III colorectal adenocarcinomas who were treated with only surgery, whereas an increased autophagic response was linked to metastasis and a worse prognosis[77].
BECN1, the mammalian orthologue of yeast ATG6, encodes the beclin-1 protein, which exerts its biological activities through three identified structural domains: A Bcl-2 homology domain, a central coiled-coiled domain and an evolutionarily conserved domain[78]. Beclin-1 plays a pivotal role in autophagy as a component of the autophagy class III PI3K complex. By interacting with different factors, it regulates autophagy pathways, resulting in the gain (e.g., AMBRA 1, UVRAG) or loss (e.g., Bcl-2) of autophagy. Moreover, beclin-1 dysfunction has been linked to immune disorders, neurodegenerative diseases and cancer[79].
BECN1 plays a controversial role in colorectal carcinomas in that it supports tumorigenesis[80] but may also inhibit CRC cell growth[81]. Higher expression levels of BECN1 have been reported in malignant colorectal tissue than in normal colorectal mucosa[82], with overexpression being especially associated with advanced stages of CRC[75,83-85]. Using immunohistochemistry, Ahn et al[80]. showed increased BECN1 expression in 95% of colorectal carcinoma samples compared to normal mucosal epithelial tissue, but they found no significant association with invasion, metastasis or stage. High BECN1 expression has been linked to a good prognosis and longer survival in patients with stage IIIB colorectal carcinoma[83]. Consistent with these findings, an increased level of BECN1 expression was strongly associated with longer 5-year survival in patients with locally advanced colon carcinomas who were treated with 5-FU chemotherapy for six months after surgery[84]. Overexpression of BECN1 in patients with resected stage II and III colon carcinomas who were treated with 5-FU-based adjuvant therapy was associated with worse overall survival, supporting a role for autophagy in drug resistance[85]. Moreover, in a meta-analysis, overexpression of BECN1 was associated with a poor prognosis and metastasis in patients with CRC[86]. Furthermore, low levels of BECN1 were correlated with a longer survival in advanced CRC patients who were treated with cetuximab-containing chemotherapy[75,76]. Supporting this hypothesis, a lack of the expression of the autophagy-related proteins LC3B, ATG5 and beclin-1 is associated with poor clinical outcomes and poor survival in CRC patients[87]. Rectal adenocarcinoma patients exhibiting low expression levels of BECN1 were more likely to experience a good response to chemoradiation than patients with increased expression levels of BECN1[88]. Moreover, the expression levels of BECN1 were reduced in a panel of human neoplasms, including brain tumors and gastric and colorectal carcinomas[89].
ATG5 protein is encoded by the ATG5 gene and forms a complex with ATG12 that participates in autophagosome membrane elongation[22]. Mutations in the ATG2B, ATG5, ATG9B, and ATG12 genes have been associated with CRC and gastric cancer[90]. An association between mutations in the ATG5 gene and reduced levels of ATG5 protein expression has been shown in gastrointestinal cancers, including CRC[91]. ATG5 expression was down-regulated in 95% of CRC patients and, interestingly, increased ATG5 expression was associated with lymphovascular invasion[92]. Other research showed that ATG5 is down-regulated in colorectal carcinoma, in both tissue samples and cell lines, and that down-regulation of ATG5 in CRC enhanced sensitivity to oxaliplatin[93]. Heterozygous deletion of ATG5 predisposed mice to intestinal adenoma growth and enhanced the antitumor effect of interferon gamma. In CRC mouse models, treatment with ursolic acid promoted autophagic cell death through a path mediated by ATG5[94].
The ATG10 gene has been mapped to chromosome 5 and encodes an E2 ubiquitin ligase-like enzyme that has essential functions in vesicle elongation, where it catalyzes the conjugation of ATG5 and ATG12[22]. ATG10 was found to be upregulated in CRC tissues and high protein expression of ATG10 was associated with tumor lymph node metastasis and invasion. Moreover, the presence of ATG10 was correlated with poor survival, indicating that ATG10 may be a potential prognostic marker for CRC[95].
The AMBRA1 gene encodes the activating molecule in beclin-1-regulated autophagy (Ambra1) protein, which has roles in autophagy, cell growth, cell death, embryonic development and carcinogenesis[96]. AMBRA1 is mutated in a subset of colorectal neoplasms[97].
The UV radiation resistance-associated gene (UVRAG) encodes a tumor suppressor protein that induces autophagy by interacting with BECN1. In addition to its function in autophagy, UVRAG is also involved in endocytic trafficking, DNA damage repair and apoptosis[98]. UVRAG, in association with BECN1, supports the maintenance of genomic stability by protecting established CRC cells against radiation-induced DNA damage[99]. UVRAG is heterozygous mutated in a high proportion of gastric and colonic tumors[100,101].
The BCL2 gene encodes the antiapoptotic B-cell lymphoma 2 (Bcl-2) protein, which inhibits autophagy by directly binding to the BH3 domain of beclin-1 and blocking its activity[102]. A recent report suggested that the prosurvival Bcl-2 protein modulates autophagy only indirectly, by inhibiting the apoptosis mediators Bax and Bak[103]. Bcl-2 has been associated with migration and invasion of malignant cells and with the prevention of apoptosis in pT3 CRC patients[104,105]. In addition, the overexpression of Bcl-2 in CRC was correlated with resistance to paclitaxel[106]. Furthermore, the role of Bcl-2 in modulating autophagy has been investigated in different cancer cell lines, including colon carcinoma, where the deletion of the BH4 domain in the Bcl-2 protein in HT29 colon carcinomas was not found to affect tumorigenicity[107].
The Bif-1 gene encodes Bax-interacting factor (Bif-1), also known as endophilin B1, which is involved in the control of membrane dynamics in cytosolic organelles, such as the Golgi complex and mitochondria, as well as in autophagosomes. Bif-1 induces the formation of autophagosomes and modulates autophagy-enhancing PI3K lipid kinase activity by interaction with beclin-1 through UVRAG[108]. The expression of Bif-1 was found to be reduced in colorectal carcinomas and the loss of Bif-1 suppressed programmed cell death and promoted colon adenocarcinomas. Bif-1 null mice developed normally, with the exception of an enlarged spleen, but they had an increased incidence of spontaneous tumor formation: 82.8% of Bif-1 null mice developed lymphoma compared with 14.3% of their wild-type counterparts[109].
Autophagy has also been linked to CRC through inflammatory bowel disease (IBD). In the complex pathogenesis leading to colitis-associated cancer, the severity of inflammation is a risk factor for CRC[110]. Cytokines released by epithelial and immune cells play an important role, and autophagy can affect the regulation of both inflammation and immune system functions[22]. Autophagy contributes to intestinal homeostasis by ensuring intracellular defenses against microbes, by maintaining the integrity of secretory granules in Paneth cells, and by regulating the inflammasome or mediating antigen presentation[111]. Genome-wide association studies provided the first link between autophagy and IBD by showing that the ATG16L1 T300A polymorphism is associated with an increased risk of Crohn’s disease (CD)[112-114]. In addition, IRGM, NOD2, and LRRK2 have been identified as additional markers of CD risk, and autophagy and DAP1 were associated with ulcerative colitis[115]. Recently, the ATG16L1T300A polymorphism was found to improve overall survival in human CRC patients and to enhance the production of type I interferon[116].
Recent data indicate that only tumors that utilize excessive levels of autophagy, even in nutrient-rich conditions and in the absence of stressful stimuli, respond to autophagy inhibitors in vivo[117]. This suggests that only a fraction of cancer patients may benefit from the administration of autophagy inhibitors. Along similar lines, autophagy has been shown to underlie, at least in part, the therapeutic activity of some anticancer regimens[118,119].
Autophagy promotes cancer cell survival under stressful conditions or nutrient deprivation and thus may contribute to chemoresistance. The drugs targeting various autophagy pathways can either induce gain or loss of autophagy. The exaggerated and sustained autophagy that is trigged by anticancer therapies can lead to type II cell death in various cancers, including CRC. Increased autophagy in the early stages of cancers can induce protection by suppressing tumorigenesis, necrosis, and chronic inflammation[13]. On the contrary, inhibition of autophagic influx may accelerate the initial steps of tumorigenesis and reduce protein degradation, and as a consequence, the reduced protein turnover might induce the early tumor progression.
In advanced stages, tumor cells use autophagy to survive cellular metabolic stress and to provide essential nutrients to tumor cells that are experiencing ischemia. Therefore, inhibiting autophagy in late-stage cancers can suppress tumor progression by blocking this prosurvival mechanism in nutrient-deprived tumor cells and by preventing protein recycling and cellular growth[120]. On the other hand, inhibition of autophagy can also lead to a decrease in the antitumorigenic activity achieved by promoting non-apoptotic cell death.
This prosurvival autophagy mechanism can be overcome by inhibition. Autophagy-inhibiting compounds include lysosomotropic agents[121]. These agents target acidic compartments, such as lysosomes, but are not specific to tumor cells and therefore have a range of effects on other cells. Lysosomotropic agents cross the lysosomal membrane and are then protonated within the acidic vesicle[122]. This results in an increased pH, which prevents cellular degradation and indirectly inhibits autophagy. Preclinical studies have demonstrated the effects of lysosomotropic agents, including CQ, which include the indirect modulation of late-stage autophagy[123]. Furthermore, CQ inhibits phospholipase A2 and lysophospholipid acylhydrolase, enzymes that are required for the acidification of lysosomes[124].
Treating human colon carcinoma HT29 cells with CQ sensitized mouse colon cancers to antiangiogenic and cytotoxic therapy[93]. Moreover, the combination of CQ and 5-FU displayed a significant advantage over treatment with 5-FU alone in inhibiting tumor growth in colon 26 cells, which are a CRC cell line[125]. A combination of the autophagy inhibitor CQ and vorinostat, a histone deacetylase inhibitor, was shown to significantly reduce tumor growth and induce apoptosis in a colon cancer xenograft model[126]. Notably, the combination of CQ with saracatinib, an inhibitor of Src nonreceptor tyrosine kinase, enhanced apoptotic cell death and resulted in 64% tumor growth inhibition compared with saracatinib alone[127]. Autophagy inhibitors shown synergy with proteasome inhibitors; for example, the simultaneous use of bortezomib and CQ in a colon cancer xenograft model decreased tumor growth to a greater extent than the use of either of these drugs alone[128].
Interestingly, treatment of human HCT-15 colon adenocarcinoma culture cells with B-group soyasaponins induced autophagy and suppressed proliferation through a marked increase in autophagic cell death[129]. In addition to its effects on cell viability and anchorage-independent growth inhibition, the flavonoid quercetin induced autophagic processes in Ha-Ras transformed human colon cells and has been proposed to have anticancer properties[130]. Vitamin D can trigger autophagy by enhancing BECN1 expression and inducing PI3KC3 expression[131]. Cetuximab (an antibody for EGFR) generates autophagy and it is currently used to treat K-Ras mutation-negative, EGFR-expressing, metastatic CRC[121]. Moreover, MS-275, a synthetic benzamide derivative of HDAC, promoted Atg7 protein expression and induced autophagy to switch to apoptosis through the modulation of p38 in human colon cancer cells[132].
Curcumin is a natural polyphenolic compound that is isolated from the plant Curcuma longa. In addition to apoptosis, curcumin also promotes autophagic cell death type II[133] by inhibiting the Akt/mTOR/p70S6K pathway or by activating the ERK1/2 pathway[134]. The proliferation of HT-29 and HCT-15 human colon cancer cell lines was inhibited by curcumin treatment, which arrested the cell cycle in the G2/M phase with no detected apoptosis[135]. Curcumin administered in combination with 5-FU plus oxaliplatin resulted in increased inhibition of growth and enhanced apoptosis in HCT-116 and HT-29 colon cancer cells compared to each of these drugs alone, and these effects were attained mainly through the attenuation of the EGFR and IGF-1R signaling pathways[136]. The induction of autophagy activation and ROS production was observed in HCT116 human colon cancer cells that were treated with curcumin, and they showed higher mRNA and protein LC3 levels[137].
Autophagy facilitates cancer cell resistance to chemotherapy treatments, and the inhibition of autophagy may resensitize resistant tumor cells to anticancer therapy, thus enhancing the efficacy of the treatment. For example, imatinib induces nonapoptotic autophagic cell death, while the inhibition of autophagy enhances its cytotoxicity, but only at a late stage[138]. Autophagy activation was observed in colon cancer stem cells by analysis of the expression of the intestine-specific transcription factor Cdx1, which plays a crucial role in chemoresistance to paclitaxel[106]. Similarly, autophagy increased resistance to photodynamic therapy-induced apoptosis in CRC stem-like cells[139]. However, this report did not address whether the protective autophagy that was induced in cancer stem cells was due to a drug-mediated response to stress or to the inherent ability of cancer stem cells to maintain a high threshold for autophagy. Suppression of protective autophagy by 3-MA was reported to enhance the therapeutic efficacy of cisplatin and 5-FU in digestive cancers, including colon cancer[140].
Many mTOR inhibitors with effective antitumor activity have been developed. However, they also have downstream effects that include the activation of autophagy, which is linked to prosurvival mechanisms in tumor cells through the recycling of damaged cellular contents. The addition of an autophagy inhibitor could solve this complication by excluding this alternate recovery pathway and sensitizing malignant cells to anticancer therapies[141,142].
Taken together, these observations suggest that autophagy supports the progression of established neoplasms through several mechanisms and that pharmacological inhibitors of autophagy may exert robust antineoplastic effects, at least in some settings.
Future research aimed at exploring the context specific role of autophagy in particular cancer types can provide new opportunities to develop personalized therapeutic strategies based on the regulation of autophagy, and autophagy modulators may become a targetable option for enhancing the efficacy of anticancer therapies used alone or, more likely, in combination with other chemotherapeutic drugs[120].
Multiple genes and proteins are involved in the complex steps of autophagy. Recent evidence has suggested that autophagy plays an important role in all stages of carcinogenesis, by influencing initiation, progression and metastatic capacity in tumors. The precise mechanisms that involve autophagy in cancer are not yet defined, and they seem to be context dependent, having both promoting and inhibiting roles. During the first steps of cancer, autophagy may have a suppressive effect, whereas it may alternatively act as tumor promoter during advanced cancer stages. It is necessary to determine how these dual roles of autophagy in CRC are regulated and identify the signals, molecules, and mechanisms that enable autophagy to play a dominant pro-malignant role in one situation and the opposite role in another. The most important research on CRC has been focused on several molecules, mainly LC3, BECN1, ATG5, and these studies have produced conflicting results. Several therapeutic agents that modulate autophagy in CRC have been developed and show promising results supporting their use either alone or, more likely, in combination with other drugs. Further research is required to better understand the relationship between CRC and autophagy, and to produce potentially beneficial agents for the prognosis and therapy of CRC.
P- Reviewer: Haerian BS, Yin PH S- Editor: Yu J L- Editor: A E- Editor: Jiao XK
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21371] [Article Influence: 2137.1] [Reference Citation Analysis (3)] |
| 2. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5748] [Article Influence: 338.1] [Reference Citation Analysis (1)] |
| 3. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5293] [Article Influence: 311.4] [Reference Citation Analysis (0)] |
| 4. | Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2430] [Cited by in RCA: 2818] [Article Influence: 156.6] [Reference Citation Analysis (0)] |
| 5. | Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3678] [Cited by in RCA: 4869] [Article Influence: 347.8] [Reference Citation Analysis (0)] |
| 6. | Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 679] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 7. | Panda PK, Mukhopadhyay S, Das DN, Sinha N, Naik PP, Bhutia SK. Mechanism of autophagic regulation in carcinogenesis and cancer therapeutics. Semin Cell Dev Biol. 2015;39:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 8. | Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992-2003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1369] [Cited by in RCA: 1579] [Article Influence: 98.7] [Reference Citation Analysis (0)] |
| 9. | Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 836] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 10. | Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 403] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 11. | Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 162] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 12. | Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1033] [Cited by in RCA: 1043] [Article Influence: 86.9] [Reference Citation Analysis (0)] |
| 13. | Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 830] [Cited by in RCA: 982] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 14. | Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84-91. [PubMed] |
| 15. | Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 583] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 16. | Ren F, Shu G, Liu G, Liu D, Zhou J, Yuan L, Zhou J. Knockdown of p62/sequestosome 1 attenuates autophagy and inhibits colorectal cancer cell growth. Mol Cell Biochem. 2014;385:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720-5728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4962] [Cited by in RCA: 5534] [Article Influence: 221.4] [Reference Citation Analysis (0)] |
| 18. | Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094-19104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 597] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 19. | Bhutia SK, Mukhopadhyay S, Sinha N, Das DN, Panda PK, Patra SK, Maiti TK, Mandal M, Dent P, Wang XY. Autophagy: cancer’s friend or foe? Adv Cancer Res. 2013;118:61-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 171] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 20. | Chen Z, Li Y, Zhang C, Yi H, Wu C, Wang J, Liu Y, Tan J, Wen J. Downregulation of Beclin 1 and impairment of autophagy in a small population of colorectal cancer. Dig Dis Sci. 2013;58:2887-2894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109-122. [PubMed] |
| 22. | Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1792] [Cited by in RCA: 1914] [Article Influence: 159.5] [Reference Citation Analysis (0)] |
| 23. | Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 905] [Cited by in RCA: 953] [Article Influence: 95.3] [Reference Citation Analysis (0)] |
| 24. | Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: another double-edged sword. Curr Opin Cell Biol. 2010;22:241-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 255] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 25. | Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 430] [Cited by in RCA: 738] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 26. | Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 908] [Cited by in RCA: 945] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 27. | Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 667] [Cited by in RCA: 735] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 28. | Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 29. | Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Mariño G, Vitale I, Jemaá M, Harper F, Pierron G. Autophagic removal of micronuclei. Cell Cycle. 2012;11:170-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 30. | Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679-2688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1271] [Cited by in RCA: 1328] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 31. | Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004-1010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1204] [Cited by in RCA: 1154] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 32. | Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679-1688. [PubMed] |
| 33. | Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2446] [Cited by in RCA: 2700] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 34. | Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12:485-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 150] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 35. | Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 492] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 36. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1534] [Cited by in RCA: 1722] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 37. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1836] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 38. | Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2154] [Cited by in RCA: 2294] [Article Influence: 109.2] [Reference Citation Analysis (0)] |
| 39. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1755] [Cited by in RCA: 1926] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 40. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 41. | Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 580] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 42. | Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 353] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 43. | Zhi X, Zhong Q. Autophagy in cancer. F1000Prime Rep. 2015;7:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 44. | Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 364] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 45. | Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 849] [Cited by in RCA: 866] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 46. | Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050-14055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 476] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 47. | Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 168] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 48. | Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1278] [Cited by in RCA: 1521] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 49. | Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1808] [Cited by in RCA: 1896] [Article Influence: 158.0] [Reference Citation Analysis (0)] |
| 50. | Zhang L, Sung JJ, Yu J, Ng SC, Wong SH, Cho CH, Ng SS, Chan FK, Wu WK. Xenophagy in Helicobacter pylori- and Epstein-Barr virus-induced gastric cancer. J Pathol. 2014;233:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 831] [Cited by in RCA: 892] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 52. | Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1605] [Cited by in RCA: 1587] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 53. | Naumov GN, Folkman J, Straume O. Tumor dormancy due to failure of angiogenesis: role of the microenvironment. Clin Exp Metastasis. 2009;26:51-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 54. | White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1226] [Cited by in RCA: 1380] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 55. | Jiang X, Overholtzer M, Thompson CB. Autophagy in cellular metabolism and cancer. J Clin Invest. 2015;125:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 171] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 56. | Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460-470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1090] [Cited by in RCA: 1055] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 57. | Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 133] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 58. | Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 398] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 59. | Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510-1527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 324] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 60. | Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, Barnard N, Ganesan S, Karantza V, White E. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Discov. 2013;3:894-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 61. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2886] [Cited by in RCA: 2760] [Article Influence: 184.0] [Reference Citation Analysis (0)] |
| 62. | Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 983] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 63. | Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261-2272, 2261-2272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 281] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 64. | Wolf J, Dewi DL, Fredebohm J, Müller-Decker K, Flechtenmacher C, Hoheisel JD, Boettcher M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013;15:R109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 65. | Ko A, Kanehisa A, Martins I, Senovilla L, Chargari C, Dugue D, Mariño G, Kepp O, Michaud M, Perfettini JL. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ. 2014;21:92-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 66. | Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman NK. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 191] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 67. | López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10570] [Cited by in RCA: 10407] [Article Influence: 867.3] [Reference Citation Analysis (0)] |
| 68. | Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. 2014;127:3-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 198] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 69. | Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181-206. [PubMed] |
| 70. | Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503-2518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 929] [Cited by in RCA: 1146] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 71. | Zheng HY, Zhang XY, Wang XF, Sun BC. Autophagy enhances the aggressiveness of human colorectal cancer cells and their ability to adapt to apoptotic stimulus. Cancer Biol Med. 2012;9:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 72. | Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677-9684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 287] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 73. | Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900-1909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 74. | Schonewolf CA, Mehta M, Schiff D, Wu H, Haffty BG, Karantza V, Jabbour SK. Autophagy inhibition by chloroquine sensitizes HT-29 colorectal cancer cells to concurrent chemoradiation. World J Gastrointest Oncol. 2014;6:74-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 75. | Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH, Chen XX, Wang F, Xia LP. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol. 2011;17:4779-4786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 76. | Yang M, Zhao H, Guo L, Zhang Q, Zhao L, Bai S, Zhang M, Xu S, Wang F, Wang X. Autophagy-based survival prognosis in human colorectal carcinoma. Oncotarget. 2015;6:7084-7103. [PubMed] |
| 77. | Giatromanolaki A, Koukourakis MI, Harris AL, Polychronidis A, Gatter KC, Sivridis E. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol. 2010;63:867-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 78. | Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1794] [Cited by in RCA: 1947] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 79. | Sahni S, Merlot AM, Krishan S, Jansson PJ, Richardson DR. Gene of the month: BECN1. J Clin Pathol. 2014;67:656-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 80. | Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 81. | Koneri K, Goi T, Hirono Y, Katayama K, Yamaguchi A. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer Res. 2007;27:1453-1457. [PubMed] |
| 82. | Zhang MY, Gou WF, Zhao S, Mao XY, Zheng ZH, Takano Y, Zheng HC. Beclin 1 expression is closely linked to colorectal carcinogenesis and distant metastasis of colorectal carcinoma. Int J Mol Sci. 2014;15:14372-14385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 83. | Yang Z, Ghoorun RA, Fan X, Wu P, Bai Y, Li J, Chen H, Wang L, Wang J. High expression of Beclin-1 predicts favorable prognosis for patients with colorectal cancer. Clin Res Hepatol Gastroenterol. 2015;39:98-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 84. | Li BX, Li CY, Peng RQ, Wu XJ, Wang HY, Wan DS, Zhu XF, Zhang XS. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009;5:303-306. [PubMed] |
| 85. | Park JM, Huang S, Wu TT, Foster NR, Sinicrope FA. Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther. 2013;14:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 86. | Han Y, Xue XF, Shen HG, Guo XB, Wang X, Yuan B, Guo XP, Kuang YT, Zhi QM, Zhao H. Prognostic significance of Beclin-1 expression in colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:4583-4587. [PubMed] |
| 87. | Choi JH, Cho YS, Ko YH, Hong SU, Park JH, Lee MA. Absence of autophagy-related proteins expression is associated with poor prognosis in patients with colorectal adenocarcinoma. Gastroenterol Res Pract. 2014;2014:179586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 88. | Zaanan A, Park JM, Tougeron D, Huang S, Wu TT, Foster NR, Sinicrope FA. Association of beclin 1 expression with response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal carcinoma. Int J Cancer. 2015;137:1498-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709-730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1195] [Cited by in RCA: 1195] [Article Influence: 91.9] [Reference Citation Analysis (0)] |
| 90. | Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol. 2009;217:702-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 224] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 91. | An CH, Kim MS, Yoo NJ, Park SW, Lee SH. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol Res Pract. 2011;207:433-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 92. | Cho DH, Jo YK, Kim SC, Park IJ, Kim JC. Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 2012;32:4091-4096. [PubMed] |
| 93. | Selvakumaran M, Amaravadi RK, Vasilevskaya IA, O’Dwyer PJ. Autophagy inhibition sensitizes colon cancer cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res. 2013;19:2995-3007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 181] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 94. | Wang L, Wang Y, Lu Y, Zhang Q, Qu X. Heterozygous deletion of ATG5 in Apc(Min/+) mice promotes intestinal adenoma growth and enhances the antitumor efficacy of interferon-gamma. Cancer Biol Ther. 2015;16:383-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 95. | Jo YK, Kim SC, Park IJ, Park SJ, Jin DH, Hong SW, Cho DH, Kim JC. Increased expression of ATG10 in colorectal cancer is associated with lymphovascular invasion and lymph node metastasis. PLoS One. 2012;7:e52705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 96. | Cianfanelli V, De Zio D, Di Bartolomeo S, Nazio F, Strappazzon F, Cecconi F. Ambra1 at a glance. J Cell Sci. 2015;128:2003-2008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 97. | Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D’Orazio M. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17:20-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 178] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 98. | Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 643] [Cited by in RCA: 619] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 99. | Park JM, Tougeron D, Huang S, Okamoto K, Sinicrope FA. Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PLoS One. 2014;9:e100819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 100. | Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39:1059-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 125] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 101. | Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 795] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 102. | Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 556] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 103. | Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci USA. 2014;111:8512-8517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 104. | Sadowska A, Car H, Pryczynicz A, Guzińska-Ustymowicz K, Kowal KW, Cepowicz D, Kędra B. Expression of apoptotic proteins in human colorectal cancer and metastatic lymph nodes. Pathol Res Pract. 2014;210:576-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 105. | Koehler BC, Scherr AL, Lorenz S, Urbanik T, Kautz N, Elssner C, Welte S, Bermejo JL, Jäger D, Schulze-Bergkamen H. Beyond cell death - antiapoptotic Bcl-2 proteins regulate migration and invasion of colorectal cancer cells in vitro. PLoS One. 2013;8:e76446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 106. | Wu S, Wang X, Chen J, Chen Y. Autophagy of cancer stem cells is involved with chemoresistance of colon cancer cells. Biochem Biophys Res Commun. 2013;434:898-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 107. | Trisciuoglio D, De Luca T, Desideri M, Passeri D, Gabellini C, Scarpino S, Liang C, Orlandi A, Del Bufalo D. Removal of the BH4 domain from Bcl-2 protein triggers an autophagic process that impairs tumor growth. Neoplasia. 2013;15:315-327. [PubMed] |
| 108. | Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142-1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 724] [Cited by in RCA: 705] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 109. | Coppola D, Khalil F, Eschrich SA, Boulware D, Yeatman T, Wang HG. Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer. 2008;113:2665-2670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 110. | Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, Williams C, Price A, Talbot I, Forbes A. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126:451-459. [PubMed] |
| 111. | Gardet A, Xavier RJ. Common alleles that influence autophagy and the risk for inflammatory bowel disease. Curr Opin Immunol. 2012;24:522-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 112. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1519] [Cited by in RCA: 1458] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 113. | Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1435] [Cited by in RCA: 1389] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 114. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2114] [Cited by in RCA: 2047] [Article Influence: 120.4] [Reference Citation Analysis (0)] |
| 115. | Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1095] [Cited by in RCA: 1048] [Article Influence: 74.9] [Reference Citation Analysis (1)] |
| 116. | Grimm WA, Messer JS, Murphy SF, Nero T, Lodolce JP, Weber CR, Logsdon MF, Bartulis S, Sylvester BE, Springer A. The Thr300Ala variant in ATG16L1 is associated with improved survival in human colorectal cancer and enhanced production of type I interferon. Gut. 2015;Feb 2; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 117. | Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8:200-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 327] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 118. | Salazar M, Carracedo A, Salanueva IJ, Hernández-Tiedra S, Lorente M, Egia A, Vázquez P, Blázquez C, Torres S, García S. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359-1372. [PubMed] |
| 119. | Vara D, Salazar M, Olea-Herrero N, Guzmán M, Velasco G, Díaz-Laviada I. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: role of AMPK-dependent activation of autophagy. Cell Death Differ. 2011;18:1099-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 209] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 120. | Radogna F, Dicato M, Diederich M. Cancer-type-specific crosstalk between autophagy, necroptosis and apoptosis as a pharmacological target. Biochem Pharmacol. 2015;94:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 131] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 121. | Duffy A, Le J, Sausville E, Emadi A. Autophagy modulation: a target for cancer treatment development. Cancer Chemother Pharmacol. 2015;75:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 122. | Kaufmann AM, Krise JP. Lysosomal sequestration of amine-containing drugs: analysis and therapeutic implications. J Pharm Sci. 2007;96:729-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 210] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 123. | Rosich L, Xargay-Torrent S, López-Guerra M, Campo E, Colomer D, Roué G. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin Cancer Res. 2012;18:5278-5289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 124. | Sotelo J, Briceño E, López-González MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337-343. [PubMed] |
| 125. | Sasaki K, Tsuno NH, Sunami E, Kawai K, Hongo K, Hiyoshi M, Kaneko M, Murono K, Tada N, Nirei T. Resistance of colon cancer to 5-fluorouracil may be overcome by combination with chloroquine, an in vivo study. Anticancer Drugs. 2012;23:675-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 126. | Carew JS, Medina EC, Esquivel JA, Mahalingam D, Swords R, Kelly K, Zhang H, Huang P, Mita AC, Mita MM. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14:2448-2459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 127. | Wu Z, Chang PC, Yang JC, Chu CY, Wang LY, Chen NT, Ma AH, Desai SJ, Lo SH, Evans CP. Autophagy Blockade Sensitizes Prostate Cancer Cells towards Src Family Kinase Inhibitors. Genes Cancer. 2010;1:40-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 128. | Ding WX, Ni HM, Gao W, Chen X, Kang JH, Stolz DB, Liu J, Yin XM. Oncogenic transformation confers a selective susceptibility to the combined suppression of the proteasome and autophagy. Mol Cancer Ther. 2009;8:2036-2045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 129. | Ellington AA, Berhow M, Singletary KW. Induction of macroautophagy in human colon cancer cells by soybean B-group triterpenoid saponins. Carcinogenesis. 2005;26:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 130. | Psahoulia FH, Moumtzi S, Roberts ML, Sasazuki T, Shirasawa S, Pintzas A. Quercetin mediates preferential degradation of oncogenic Ras and causes autophagy in Ha-RAS-transformed human colon cells. Carcinogenesis. 2007;28:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 93] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 131. | Wang J, Lian H, Zhao Y, Kauss MA, Spindel S. Vitamin D3 induces autophagy of human myeloid leukemia cells. J Biol Chem. 2008;283:25596-25605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 132. | Zhan Y, Gong K, Chen C, Wang H, Li W. P38 MAP kinase functions as a switch in MS-275-induced reactive oxygen species-dependent autophagy and apoptosis in human colon cancer cells. Free Radic Biol Med. 2012;53:532-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 133. | Reuter S, Eifes S, Dicato M, Aggarwal BB, Diederich M. Modulation of anti-apoptotic and survival pathways by curcumin as a strategy to induce apoptosis in cancer cells. Biochem Pharmacol. 2008;76:1340-1351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 238] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 134. | Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 408] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 135. | Hanif R, Qiao L, Shiff SJ, Rigas B. Curcumin, a natural plant phenolic food additive, inhibits cell proliferation and induces cell cycle changes in colon adenocarcinoma cell lines by a prostaglandin-independent pathway. J Lab Clin Med. 1997;130:576-584. [PubMed] |
| 136. | Patel BB, Sengupta R, Qazi S, Vachhani H, Yu Y, Rishi AK, Majumdar AP. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122:267-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 155] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 137. | Lee YJ, Kim NY, Suh YA, Lee C. Involvement of ROS in Curcumin-induced Autophagic Cell Death. Korean J Physiol Pharmacol. 2011;15:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 138. | Shingu T, Fujiwara K, Bögler O, Akiyama Y, Moritake K, Shinojima N, Tamada Y, Yokoyama T, Kondo S. Inhibition of autophagy at a late stage enhances imatinib-induced cytotoxicity in human malignant glioma cells. Int J Cancer. 2009;124:1060-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 139. | Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, Hung SC, Hsiao M, Yao CJ, Shieh MJ. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy. 2014;10:1179-1192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 140. | Li J, Hou N, Faried A, Tsutsumi S, Takeuchi T, Kuwano H. Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann Surg Oncol. 2009;16:761-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 231] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 141. | Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 142. | Vignot S, Faivre S, Aguirre D, Raymond E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann Oncol. 2005;16:525-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 486] [Article Influence: 24.3] [Reference Citation Analysis (0)] |