INTRODUCTION
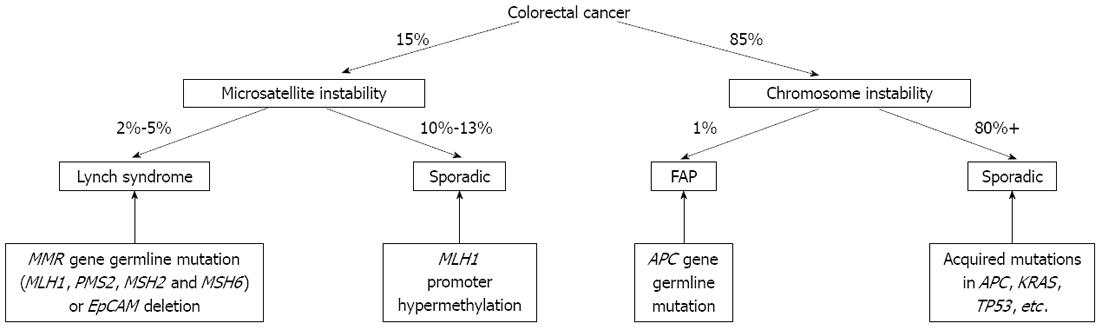
Colorectal cancer (CRC) is the third most prevalent cancer and the third most common cause of cancer death in both males and females in the United States[1]. However, widespread screening for CRC and progress in its treatment have both contributed to a recent decline in the incidence of mortality of the disease. In parallel, much progress has been made in the understanding of the molecular and genetic basis of CRC[2-4]. Chromosomal instability (CIN) and microsatellite instability (MSI) constitute the predominant tumorigenic pathways in CRC (Figure 1)[5-8]. CIN is associated with high mutation rates in genes tightly linked to the development of CRC, such as APC, KRAS, SMAD4, PI3KCA SOX9, ARID1A, FAM123B and TP53, which lead to the development of CIN tumors[5]. MSI is a form of genetic instability caused by alterations in the DNA mismatch repair (MMR) system. Although the majority of CRCs develop through the CIN pathway, approximately 15% of CRCs display MSI due to germline mutations, epigenetic silencing of MMR gene or a combination of these factors[9].
Figure 1 Molecular classification of colorectal cancer.
EpCAM: Epithelial cell adhesion molecule; MMR: DNA mismatch repair; FAP: Familial adenomatous polyposis; APC: Adenomatous polyposis coli.
Germline mutations in MMR genes cause a cancer susceptibility syndrome called Lynch syndrome, previously referred to as hereditary nonpolyposis CRC. These individuals are predisposed to CRC and multiple other cancers including endometrial, gastric, ovarian, urothelial, hepatobiliary tract, brain, small intestine, pancreatic, and skin (specifically sebaceous adenomas or carcinomas and benign keratoacanthomas) cancers[10-12]. Approximately 90% of CRCs occurring in Lynch syndrome patients exhibit MSI. The exact prevalence of Lynch syndrome among CRCs is unclear. A prospective, multicenter, nationwide study (the EPICOLON study), consisting of patients newly diagnosed with CRC in 20 community hospitals in Spain, showed the prevalence was only 0.9% compared with 2.9%-3.5% in other studies[13,14]. A most recent study showed that the prevalence of Lynch syndrome is 3.1% in all CRCs[15]. Many studies have found that some CRCs occurred in non-Lynch syndrome patients also showed MSI (sporadic CRCs with MSI) and CRCs with MSI showed different clinical-pathological features, prognosis and response to chemotherapeutic agents comparing to microsatellite-stable CRCs[9,16]. Increased emphasis has been placed on the importance of MSI testing for all newly diagnosed individuals with CRCs.
MOLECULAR BASIS OF THE MMR SYSTEM
The human genome is dynamic. It is estimated that each cell undergoes > 20 000 DNA damaging events and > 10 000 replication errors per cell per day[17]. One of the mechanisms to repair replication errors is the MMR system. The MMR system, a DNA repair pathway which is conserved from bacteria to humans, targets base-base mismatches and insertion-deletion mismatches that arise as a result of replication errors[18]. A proficient MMR system enhances replication accuracy 1000-10 000-fold[16]. A hallmark of MMR-deficient cells is instability (replication errors) at microsatellite regions. Microsatellites are mono-, di-nucleotide or higher-order nucleotide repeats such as (A)n or (CA)n that are distributed throughout the entire genome, and due to their repetitive pattern, they are prone to errors during DNA replication. The terminology used for the MMR system in eukaryotes is based on the analogous system in prokaryotes, best characterized in Escherichia coli (E. coli)[16]. The major E. coli MMR proteins include MutS and MutL[19]. Eukaryotic MutS homologs include MSH2, MSH3 and MSH6, and are primarily responsible for recognizing mismatches and recruiting MutL to the mismatch location. MutL homologs include MLH1, PMS1 and PMS2. Eukaryotic cells possess two MutS activities that function as heterodimers and share MSH2 as a common subunit: MutSα (MSH2-MSH6 heterodimer) and MutSβ (MSH2-MSH3 heterodimer). Eukaryotic MutL activities also function as heterodimeric complexes with MLH1 serving as a common subunit including MutLα (MLH1-PMS2 heterdimer), MutLβ (MLH1-PMS1 heterodimer) and MutLγ (MLH1-MLH3 complex)[20]. Specifically, when a mismatch exists, MSH2 will form a MutSα or MutSβ complex. Both MutSα and MutSβ can then recruit either MutLα, MutLβ, or the MutLγ complex, which in turn will mediate the processes of mismatch recognition and enzymatic repair[9,16,19,20]. Mutations in the MMR system lead to the accumulation of errors in DNA, which results in MSI.
GENETIC BASIS FOR LYNCH SYNDROME AND SPORADIC CRCS WITH MSI
Lynch syndrome is a genetically heterogeneous disorder which is caused by autosomal dominant germline mutations in MMR genes. The overall risk of CRCs in individuals with this syndrome is 75% by the age of 70 years and cancers occur predominantly in the right side of the colon. The mean age at diagnosis of CRC in individuals with Lynch syndrome is younger (approximately 42-61 years) than that in the general population (approximately 65 years). Mutations in MLH1, MSH2, MSH6 and PMS2 are found in 32%, 38%, 14% and 15% of Lynch syndrome cases, respectively[11,21]. Individuals with mutations in the MSH6 and PMS2 genes have a somewhat lower risk of CRC and a later age of onset of CRC compared with individuals with mutations in the MLH1 and MSH2[10,22,23]. Endometrial carcinoma is the most common extra-colonic carcinoma in Lynch syndrome and occurs in 28%-60% of women with an average age at diagnosis of 47-55 years, compared to a sporadic rate of 2%-3% in women with an average age at diagnosis in the mid 60s in the general population[10]. Endometrial cancers are more frequently associated with mutations in the MSH2 and MSH6 genes than the MLH1 or PMS2 genes.
Recently, deletions of the epithelial cell adhesion molecule (EpCAM) gene (previously known as TACSTD1, tumor-associated calcium signal transducer 1), which is located upstream of MSH2, have been described in a subset of families with Lynch syndrome[24]. Deletions affecting the 3’ exons of the EpCAM gene lead to a transcriptional read-through and mediate epigenetic silencing of the MSH2 allele in a mosaic pattern. Therefore CRCs in individuals with heterozygous germline EpCAM deletions will be MSH2-negative MSI cancers[25]. Though frequency of EpCAM deletions have been reported in different populations[24-26], further research is needed to confirm the prevalence and clinical phenotype of EpCAM deletions.
MSI CRCs that are not associated with germline mutations in the MMR system and Lynch syndrome are commonly referred to as “sporadic CRCs with MSI”. Sporadic CRCs with MSI account for about 10%-13% of CRCs with MSI. The most frequent cause of sporadic MSI is acquired promoter hypermethylation of MLH1. Hypermethylation of CpG islands in the promoter regions of both copies of MLH1 gene leads to inactivation of the gene and loss of expression of the MLH1 gene product in a manner analogous to the germline mutations of DNA MMR genes seen in Lynch syndrome. These sporadic CRCs with MSI are similar histologically to Lynch syndrome CRCs and, like Lynch syndrome, CRCs are more likely to be located in the right colon and they tend to have a better overall prognosis. In contrast to Lynch syndrome, however, these MSI CRCs do not present with a strong hereditary background nor occur at a young age, but tend to be more common in older population. Testing for mutations in the gene for the B-type Raf kinase (BRAF) can help distinguish sporadic CRCs with MSI from Lynch syndrome-associated CRCs. BRAF, a serine/threonine protein kinase, is an immediate downstream effector of KRAS in the MAP kinase signaling pathway. An activating mutation of BRAF is often present when the promoter region of the MLH1 gene is methylated. About 90% of the mutations in the BRAF gene in CRCs are transversion (1799 T>A), identified as V600E. Recently, reviewing the BRAF V600E mutation in 4562 tumors from 35 studies and MLH1 promoter methylation in 2975 tumors from 43 studies, Parsons et al[27] demonstrated that the BRAF V600E mutation occurred in 63.5% of CRCs displaying MLH1 promoter hypermethylation or MLH1/PMS2 protein loss. The frequency of BRAF V600E mutation in MSS CRCs was only 5.0%. More importantly, BRAF mutations are virtually absent in Lynch syndrome-associated tumors, and this is a very useful feature for distinguishing Lynch syndrome from sporadic CRCs with MSI. Evidence of MLH1 promoter hypermethylation or a BRAF V600E mutation is highly predictive of a sporadic CRC with MSI. Individuals with unmethylated MLH1 promoter and wild type BRAF should undergo further testing for Lynch syndrome. However, there are rare case reports of hypermethylation of the MLH1 promoter as the “second-hit” in a patient with a germline mutation[22,23].
DETECTION OF MSI
Currently, MSI is detected indirectly by demonstrating absence of expression of MMR proteins by immunohistochemical staining (IHC), or more directly by polymerase chain reaction (PCR)-based amplification of specific microsatellite repeats.
IHC of MMR proteins
The principle of using IHC of MMR proteins to indirectly indicate the presence of MSI is that the absence of one or more of the MMR proteins can cause MSI. Antibodies against MMR proteins such as MLH1, PMS2, MSH2 and MSH6 are commercially available and can be used to provide information of functionality of the MMR system. Loss of expression and the pattern of loss of expression of one or more of these proteins suggest deficient MMR, and indicate which gene harbors a germline mutation or has been inactivated by hypermethylation. As mentioned earlier, eukaryotic MMR proteins form functional heterodimers. MSH2 dimerizes with either MSH6 or MSH3, and then recruits heterodimers of MLH1 and PMS2 or MLH1 and PMS1 to excise the mismatched nucleotides. MSH2 and MLH1 proteins are the common subunits of their respective heterodimeric complexes, and when mutated, a loss of both the common subunits and their associated partner proteins by IHC is typically observed. However, the opposite is generally not true, since other proteins, such as MSH3, MLH3 and PMS1, may bind to the common subunits to stabilize them. Loss of staining of MSH6 or PMS2 alone is typically observed with germline mutations in each of these respective genes but with retained positive staining of corresponding MSH2 or MLH1. Understanding the expression patterns of MMR proteins and genetic basis of Lynch syndrome and sporadic CRCs with MSI are crucial to the interpretation of the IHC results and for guiding the further molecular analysis. For example, a CRC that fails to stain for both MLH1 and PMS2, but retains expression of MSH2 and MSH6, is due to an alteration in the MLH1 gene. However, determining whether the deficiency of MLH1 is due to a germline mutation or promoter hypermethylation requires further investigation (MLH1 hypermethylation test and/or BRAF mutation test). A CRC that shows loss of expression of both MSH2 and MSH6 is most often consistent with defective MMR through MSH2 germline mutations (Lynch syndrome), and this finding should be followed by genetic testing of MSH2. As mentioned earlier, a subset of Lynch syndrome is due to deletion of EpCAM, a gene upstream of MSH2. The deletion of EpCAM will lead to somatic hypermethylation of MSH2 and finally loss of expression of MSH2. A recent study showed that a lack of EpCAM immunostaining in MSH2-negative CRCs is indicative of EpCAM gene alterations with a 100% specificity[28], and also EpCAM negative immunostaining can be detected even at a precancerous stage[29]. Therefore, performance of EpCAM IHC before molecular analysis is suggested to be included in the algorithm approach to Lynch syndrome identification in MSH2-negative CRC cases.
PCR-based MSI testing
The principle of using PCR-based MSI testing is to detect the presence of different lengths of specific microsatellite repeats in tumor cells comparing to normal tissues caused by mismatches due to the absence of one or more of the MMR proteins. In 1997, National Cancer Institute (NCI) workshop established a reference panel of microsatellites for clinical and research testing, and also defined the criteria for diagnosing MSI. The core panel consists of two mononucleotide repeats (BAT25, BAT26) and three dinucleotide repeats (D5S346, D2S123, D17S250). Nineteen ‘‘alternative loci’’ are also suggested. Three categories of MSI have been established based on the following criteria: MSI-high (MSI-H), indicating instability at two or more loci (or > 30% of loci if a larger panel of markers is used); MSI-low (MSI-L), indicating instability at one locus (or in 10%-30% of loci in larger panels); and MSS, indicating no loci with instability (or < 10% of loci in larger panels)[30]. MSI-L CRCs do not appear to differ clinically or pathologically from MSS CRCs, and generally MSI-L CRCs are categorized as group of MSS CRCs[31]. MSI-L cases usually only show instability for dinucleotide markers, so the assessment of dinucleotides alone could lead to the misclassification of MSI-L as MSI-H. By contrast, mononucleotides BAT25 and BAT26 are nearly monomorphic. In 2002, NCI workshop (the revised Bethesda guidelines) addended new guidelines with recommendations of testing additional mononucleotide markers in tumors with instability at only dinucleotide loci, as mononucleotide markers are more reliable in the identification of MSI-H tumors[31]. Recent years, the uses of panels containing more mononucleotide markers and the availability of commercial kits including predominant mononucleotide markers have been improving the sensitivity and specificity[32-34].
Comparison of IHC and PCR-based MSI testing
The results of MMR IHC and PCR-based MSI testing have been shown to be largely concordant (97.80% concordance, exact 95%CI: 96.27-98.82)[35]. Studies have shown that IHC for the MMR proteins MLH1, PMS2, MSH2 and MHS6 provides a rapid, cost-effective, sensitive, and highly specific technique for screening CRC for MSI. Reviewing the IHC results of 16 series representing 3494 cases, Rigau et al[36] demonstrated that the following performances of IHC in assessing MSI: sensitivity, 92.4%; specificity, 99.6%; positive predictive value, 98.5%; and negative predictive value, 97.8%, which are comparable to PCR-based molecular MSI testing. In one previous large study, IHC in CRCs for MLH1 and MSH2 provided a rapid, cost-effective, sensitive (92.3%), and extremely specific (100%) method for screening for DNA MMR defects. The predictive value of normal IHC for an MSS/MSI-L phenotype was 96.7%, and the predictive value of abnormal IHC was 100% for an MSI-H phenotype[37]. The major advantage of IHC is that it is widely available in general pathology laboratories. Another advantage of IHC is that tumors with MSH6 germline mutations sometimes lack MSI in PCR-based testing owing to a functional redundancy in the MMR system, but demonstrate loss of MSH6 staining by IHC[16]. Furthermore, a key advantage to the use of IHC is its ability to guide and direct genetic testing. However, rare missense mutations, which are reported usually in MLH1 and MSH6 genes, affect protein function other than protein translation and antigenicity. IHC will still show positive staining despite MSI[16,22]. In these cases, PCR-based MSI testing can help to determine whether there are true functional MMR proteins through these mutations.
ERA OF UNIVERSAL MSI TESTING
The diagnosis of Lynch syndrome and recognition of sporadic CRCs with MSI have important implications regarding cancer prevention, surveillance and management. Studies have shown that MSI-H CRCs carry a better prognosis compared to those with MSS CRCs[38]. In addition, stage II MSI-H CRCs achieved similar progression free survival and overall survival with or without 5-flurouracil (5-FU)-based neoadjuvant chemotherapy[39]. Therefore, patients with stage II MSI-H CRC are not recommended to receive 5-FU based adjuvant chemotherapy. As mentioned earlier, individuals with Lynch syndrome have significantly higher risks of developing extra-colonic malignancies besides early onset of CRC. Intensive cancer surveillance has shown to substantially reduce cancer-related death in this group of patients[40]. Most recently, it also has shown that aspirin can be used as a chemopreventive agent in carriers of Lynch syndrome to prevent the development of CRCs and extra-colonic carcinomas[41].
Historically, diagnosis of Lynch syndrome relied on clinical characteristics of personal and family history of cancer. The Amsterdam criteria[42], later revised to Amsterdam II criteria[43] are now well-recognized to be too stringent and insufficiently sensitive because of small family sizes, unfamiliarity with Lynch syndrome by clinicians, lack of documentation of tumors in the family, and/or reduced penetrance of the tumors in the family. With the availability of molecular diagnostic testing, the Bethesda guidelines[44], and then the revised Bethesda guidelines[31], were developed to select patients who should undergo MSI analysis. These guidelines incorporated tumor histopathology features into their criteria, including the presence of tumor infiltrating lymphocytes, Crohn’s-like lymphocytic reaction, mucinous/signet-ring differentiation, and/or a medullary growth pattern. However, data suggests that the clinical guidelines and histopathology features are neither sensitive nor specific in determining the presence or absence of MSI. For example, up to 50% of mutation carriers do not meet the Amsterdam criteria and 40%-45% of families who fulfill the Amsterdam criteria do not demonstrate MSI on tumor testing or MMR gene germline mutations[16,23]. In an effort to improve the detection rate of Lynch syndrome individuals and sporadic CRCs with MSI, it has been suggested that all CRCs (universal testing) should be tested for MSI using either a PCR-based or an IHC approach[45]. Julié et al[46] compared the performance of the revised Bethesda guidelines with universal molecular testing in 214 newly diagnosed CRC patients. The revised Bethesda guidelines identified 42.1% of patients for MSI testing. Of these 4.2% were MSI positive and 6 were MMR mutation-positive. However, using a universal MSI testing strategy in these patients, 9.8% were found to be MSI positive and 5.1% of the MSI positive patients were MMR mutation-positive. Thus, the authors concluded that the revised Bethesda guidelines does not adequately identify mutation carriers and CRCs with MSI[46]. Morrison et al[47] compared the MSI detection rate in 445 primary CRCs resected between November 2006 and March 2009, when MSI testing was based on histopathology features and age, with the rate in 145 CRCs resected between July 2009 and July 2010 when a universal testing paradigm was used. The overall Lynch syndrome screening rate between November 2006 and March 2009 was 34.8%, and the extrapolated MSI-H rate was 8.5% (38/445). Strict adherence to the revised Bethesda guidelines, that is, without testing CRC diagnosed in patients over 60 years, would have missed 26 (68.4%) MSI CRCs. The overall Lynch syndrome screening rate between July 2009 and July 2010 was 76.3% and the MSI rate was 20.6% (30/145). These data indicated that the revised Bethesda guidelines is inadequate for Lynch syndrome screening when personal and family cancer history is not available to the pathologist, a universal screening paradigm greatly increased the rate of MSI testing and MSI CRC detection[47]. Most recently, Pérez-Carbonell et al[48] investigated 2093 patients with CRC from the EPICOLON I and II cohorts and found the revised Bethesda guidelines strategy failed to detect 14.3% cases with Lynch syndrome and 57.1% cases with probable non-sporadic MSI-H tumors. The authors concluded that routine screening of patients with CRC for Lynch syndrome using immunohistochemistry or PCR-based MSI testing has better sensitivity for detecting mutation carriers than the Bethesda guidelines alone[48]. Many studies have identified other histopathologic features, which are included in the revised Bethesda guidelines, such as right-sided location, lack of “dirty necrosis”, a circumscribed/expansile growth pattern, histologic heterogeneity, lack of intratumoral budding, and carcinoma associated with sessile serrated adenoma/polyp (serrated pathway) are all suggestive of MSI-H[49-52]. However, our experience and that of others have shown that around 3%-6% of CRCs with feature of “dirty necrosis” and a portion of left-sided tumors do show MSI-H, especially with MSH6 loss[22].
Recent data have shown that testing for MMR expression can be performed on the diagnostic CRC biopsy samples prior to definitive surgery[53], with results comparable to those obtained on the surgical resection specimens[54,55]. Using this approach the diagnosis of Lynch syndrome can be made preoperatively, and this information can help the surgeon in planning the operative approach (extended colectomy, subtotal colectomy, or total colectomy) and in recommending screening for cancers in other organs. Another argument for early testing for MMR expression is the fact that neoadjuvant chemotherapy and radiation can cause aberrant or loss of immunoexpression of MMR proteins. Diminished MMR staining in treated tumors should prompt IHC evaluation of pretreatment biopsy samples before genetic testing is pursued for Lynch syndrome[56].
ALGORISM FOR MSI TESTING
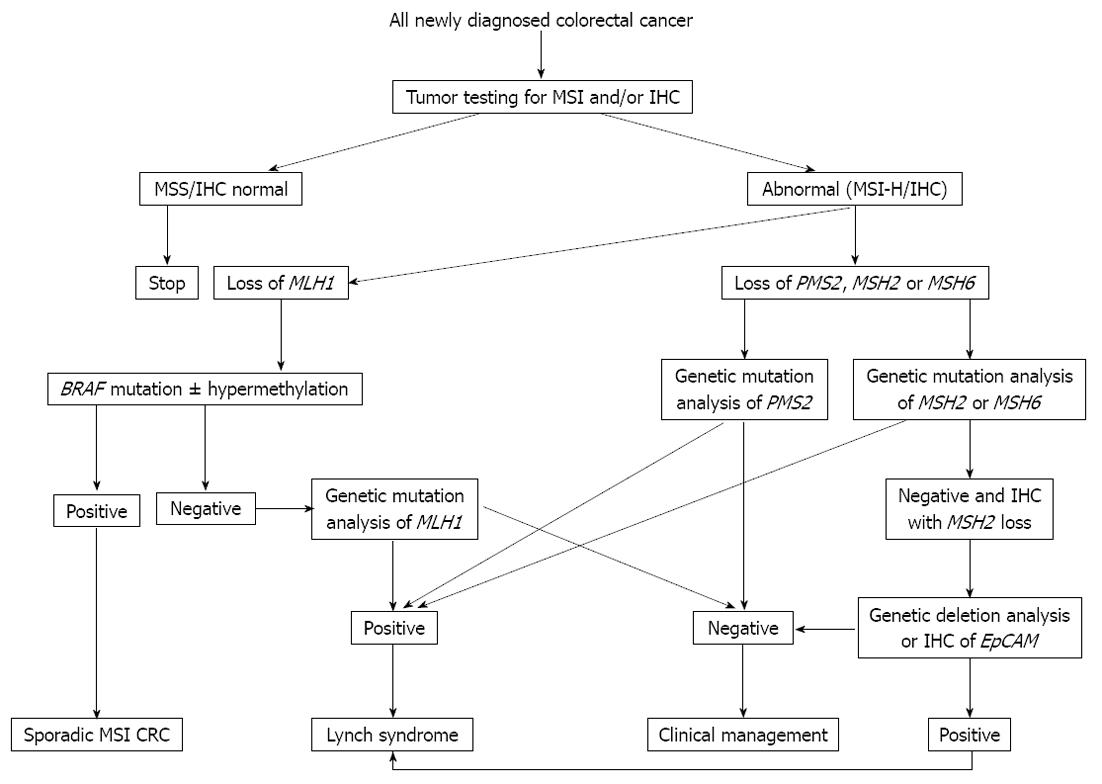
Even though the increased emphasis has been placed on the importance of MSI testing and recommendations have been proposed to identify individuals at risk for Lynch syndrome[10], and both PCR-based and IHC MSI detection are highly sensitive methods for the identification of individuals with defective MMR[21], the approach of universal MSI testing for all newly diagnosed CRCs has not been widely accepted and understood. A recent survey of Canadian hospitals demonstrated that up to 21.2%, 42.1% and 38.2% of respondents either do not have access or are uncertain whether they have access to MMR-IHC, PCR-based MSI testing, and genetic counseling services respectively[57]. It has been demonstrated that the highest detection rate of Lynch syndrome in CRC is achieved through integrated efforts of pathologists, clinicians (surgeons, gastroenterologists, and family doctors) and genetic counselors[58]. However, only 13.1% of respondents have an integrated multidisciplinary approach to Lynch syndrome detection. A recent survey of United States hospitals reported that routine tumor testing with IHC, PCR-based MSI testing, or both is currently performed at 71% of NCI comprehensive cancer centers, 36% of American College of Surgeons-accredited community hospital comprehensive cancer programs, but only 15% of community hospital cancer programs[59]. Awareness of the importance of MSI testing and an appropriate algorithmic approach (Figure 2), starting with PCR-based MSI testing or IHC analysis on all newly diagnosed CRC specimens (universal testing) will help recognize Lynch syndrome and distinguish sporadic CRCs with MSI and Lynch syndrome effectively.
Figure 2 Testing algorithm for Lynch syndrome and sporadic microsatellite instability colorectal cancer.
MSI: Microsatellite instability; IHC: Immunohistochemical staining; CRC: Colorectal cancer; EpCAM: Epithelial cell adhesion molecule.
CONCLUSION
Many studies have shown the importance of MSI testing in diagnosing Lynch syndrome and predicting prognosis and response to chemotherapeutic agents. Increased emphasis has been placed on the importance of MSI testing for all newly diagnosed individuals with CRCs. Both IHC and PCR-based MSI testing show close concordance and high sensitivity and specificity in detecting MSI. The current clinical guidelines and histopathology features are indicative of, but not sensitive and specific in diagnosing Lynch syndrome and CRCs with MSI. Currently, there are evidences that universal testing for MSI starting with either IHC or PCR-based MSI testing is cost effective, sensitive, specific and is getting widely accepted.
ACKNOWLEDGMENTS
We are grateful to Dr. Michal G Rose, MD, Cancer Center and Department of Medicine, Yale University School of Medicine and VA Medical Center, for critical revising and commenting on the manuscript.
P- Reviewer Russo A S- Editor Song XX L- Editor A E- Editor Xiong L