Published online Jul 15, 2012. doi: 10.4251/wjgo.v4.i7.170
Revised: May 22, 2012
Accepted: May 27, 2012
Published online: July 15, 2012
AIM: To investigate into the diversity of UGT1A1 polymorphism across three different districts in Japan and highlight genetic differences among the population in Japan.
METHODS: We enrolled 50 healthy volunteers from each of the Yamaguchi (western part of Japan), Kochi (southern part of Japan) and Akita (northern part of Japan) prefectures. Blood samples (7 mL) were collected from each participant and stored in EDTA for subsequent genotyping by fragment size analysis, direct sequencing and TaqMan assay of UGT1A1*28, UGT1A7*3/UGT1A9*22 and UGT1A1*93/UGT1A1*6/UGT1A1*27/UGT1A1*60/UGT1A7 (-57), respectively.
RESULTS: The only statistically significant differences in allele polymorphisms among the group examined were for UGT1A1*6. The Akita population showed more UGT1A1*6 heterozygosity (P = 0.0496).
CONCLUSION: Our study revealed no regional diversity among UGT1A1, UGT1A7 or UGT1A9 polymorphisms in Japan.
-
Citation: Kobayashi M, Hazama S, Takahashi K, Oba K, Okayama N, Nishioka M, Hinoda Y, Oka M, Okamoto K, Maeda H, Nakamura D, Sakamoto J, Mishima H. Is there diversity among
UGT1A1 polymorphism in Japan. World J Gastrointest Oncol 2012; 4(7): 170-175 - URL: https://www.wjgnet.com/1948-5204/full/v4/i7/170.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v4.i7.170
Irinotecan with fluoropyrimidine is approved worldwide as a first-line chemotherapeutic agent for metastatic colorectal cancer[1-5]. Although prolonged survival has been reported with the use of this drug, severe diarrhea and neutropenia have also been reported as dose-limiting toxicities in 20%-35% of patients treated by the agent. Recent studies revealed that the risk of such severe toxicities might be associated with genetic variation in irinotecan metabolism, indicating a possible predictive factor.
Irinotecan is activated by hydrolysis to SN-38, a potent topoisomerase I inhibitor[6] that is primarily inactivated through biotransformation into SN-38 glucuronide (SN-38G) by the enzyme uridine diphosphate glucuronosyltransferase isoform 1A1 (UGT1A1)[7]. In addition, the toxicity of irinotecan has been correlated with polymorphisms in the number of TA repeats in one of the promoter regions of the UGT1A1 gene (UGT1A1 *28), which affects transcriptional efficiency[8]. Because of the clinical importance of the glucuronidation pathway in irinotecan treatment, UGT1A1 *28 was proposed as a potent predictor for severe toxicity[9-11]. Recently, a novel prospective dose-finding study of irinotecan alone based on UGT1A1*6 and *28 genotyping was reported[4,12]. These results showed that the UGT1A1 *6 or *28 genotype status could be used to determine RD (recommended doses) of irinotecan. We conducted a prospective phase II study of FOLFIRI for metastatic colorectal cancer in Japan, analyzed the UGT1A1*28 and *6 polymorphisms and demonstrated that the combination of the UGT1A1*28 and *6 polymorphism is important to predict the adverse event of the CPT-11[5].
The role of UGT1A1*28 alleles in the toxicity and pharmacokinetics of irinotecan is considerably different between Asians and Caucasians. Only homozygotes of *28 have been associated with neutropenia in Caucasians[11,13-15], whereas both homozygote and heterozygote *28 patients have shown severe toxicity with irinotecan in Japan[4,9]. Other results revealed that SN-38 glucuronidation was highly impaired in heterozygotes, as previously reported[9,16]. Such ethnic differences may be associated with other genetic variants of UGT1A family polymorphisms, such as UGT1A1*60, *6, UGT1A7*3 and UGT1A9*22, which were demonstrated in linkage disequilibrium experiments with UGT1A1*28[17-22]. Such genotype variation could affect SN-38 glucuronidation and also the severe irinotecan-related toxicity. This study aimed to clarify the regional differences in UGT enzyme polymorphisms among three different districts in Japan that are widely different, both geographically and culturally.
The 50 volunteers from Akita, Kochi and Yamaguchi prefectures comprised of 8 males and 42 females, 6 males and 44 females, and 11 males and 39 females, respectively, with an average age of 37.5, 43.8 and 38.4 years, respectively. The examinee demographics are shown in Table 1.
| Akita | Kochi | Yamaguchi | |
| Sex | |||
| Male | 8 | 6 | 11 |
| Female | 42 | 44 | 39 |
| Age (yr) | 37.4 (23-55) | 43.8 (24-66) | 38.4 (18-67) |
Blood samples (7 mL) were collected from each participant and stored in EDTA for subsequent analysis. Examinees were limited to those whose parents and grandparents came from the same region.
Written informed consent was obtained from all participants.
Genomic DNA was extracted from peripheral blood anti-coagulated with EDTA-2Na, using a conventional NaI method[23]. UGT1A1*28, UGT1A7*3/UGT1A9*22 and UGT1A1*93/UGT1A1*6/UGT1A1*27/UGT1A1*60/UGT1A7 (-57) were genotyped by fragment size analysis, direct sequencing and TaqMan assay, respectively. Primers and probes used in this study are shown in Table 2.
| Gene | Variant | Primers and probes1 | |
| UGT1A1*28 | -53 TA6/TA7 | F-FAM | 5'-gtgacacagtcaaacattaacttgt-3' |
| R | 5'-gcctttgctcctgccagaggtt-3' | ||
| UGT1A7*3 | N129K | F | 5'-tacactctggaggatcagga-3' |
| W208R | R | 5'-tattgggcatcacgggtttg-3' | |
| UGT1A9*22 | -118 T10/T9 | F | 5'-acttaacattgcagcacagg-3' |
| R | 5'-atgggcaaaagccttgaact-3' | ||
| UGT1A1*93 | -3156 G/A | F | 5'-cagaagggctagagaggaggaa-3' |
| R | 5'-cttgctctcaaaactctgggataga-3' | ||
| FAM | 5'-cctgtccaagctca-3' | ||
| VIC | 5'-cacctgtctaagctca-3' | ||
| UGT1A1*6 | 211 G/A | C 559715 20 | |
| UGT1A1*27 | 686 C/A | C 2307598 20 | |
| UGT1A1*60 | -3279 T/G | C 1432134 10 | |
| UGT1A7 (-57) | -57 T/G | C 287265 10 | |
For fragment size analysis, PCR reactions were performed in a total volume of 10 μL containing template DNA (80 ng/μL) according to the manufacturer’s instructions (Ex Taq; Takara, Tokyo, Japan). The amplification was carried out with a Gene Amp PCR System PC808 (ASTEC, Tokyo, Japan), with an initial denaturation at 95 °C for 2 min followed by 27 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 20 s, and extension at 72 °C for 30 s. The PCR products of TA6 and TA7, whose sizes were 94 bp and 96 bp, respectively, were mixed with Hi-Di formamide, including the internal size standard (GeneScan 500, Applied Biosystems, CA, USA) at a 1:10 (vol/vol) ratio. Then, samples were run in the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Fragment sizes were determined by comparison with the internal size standard (GeneScan LIZ-500) using the local Southern algorithm and the data were analyzed by GeneMapper™ software version 3.5 (Applied Biosystems).
For direct sequencing, PCR amplifications were performed using the Gene Amp PCR System PC808 (ASTEC, Tokyo, Japan) with Ex Taq polymerase. Amplification conditions were 30 cycles of 95 °C for 30 s, each annealing temperature for 20 s, and 72 °C for 30 s. PCR products were purified using ExoSAP-IT (Amersham Bioscience, Tokyo, Japan) for 20 min at 37 °C and then for 20 min at 80 °C. Sequencing reactions were carried out using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Tokyo, Japan). After purification with ethanol, the reaction products were analyzed using an ABI 3100-Avant Genetic Analyzer (Applied Biosystems).
TaqMan assays of PCR products were performed according to the manufacturer’s protocol. Specific forward/reverse PCR primers and TaqMan probes for UGT1A1*93 were custom-synthesized by Applied Biosystems. Primers and probes for UGT1A1*6, UGT1A1*27, UGT1A1*60, UGT1A7 (-57) were purchased from Applied Biosystems (TaqMan SNP Genotyping Assays). Reaction mixtures were loaded into 384 well plates and placed in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). PCR amplifications were performed as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles of PCR with a denaturation at 95 °C for 15 s, and one step annealing/extension for 1 min at 60 °C.
Proportions of wild-type, hetero-type and homo-type were calculated with 95% Agresti-Coull confidence intervals (95% CI)[24]. Fisher’s exact test with a two-sided significance level of 0.05 was used for comparing the areas. For a two-sided 95% CI for a binomial proportion whose true value is varied from 0.5 to 0.1, a sample size of 50 yields a half-width of, at most, 14% in any situations of the true value.
Tables 3 and 4 list the polymorphisms of UGT1A1 allele *28, *6, *60, *27 and *93 (-3156), UGT1A7 *3 (N129K, W208R, -57) and UGT1A9*22. The incidence of wild-type UGT1A1*28 in the Akita, Kochi and Yamaguchi cohorts was 82% (95% CI: 69 to 90), 74% (95% CI: 60 to 84) and 74% (95% CI: 60 to 84), respectively (P-value = 0.663). The incidence of homozygous UGT1A1*28 across the three districts was only 1.3% (95% CI: 0.0 to 5.0).
| UGT1A1*28 (P = 0.663) | UGT1A1*6 (P = 0.0496) | UGT1A1*27 (P = 1.000) | UGT1A1*60 (P = 0.766) | UGT1A1-3156 | |||||||||||
| 6/6 | 6/7 | 7/7 | A/A | G/A | G/G | A/A | C/A | C/C | G/G | T/G | T/T | A/A | G/A | G/G | |
| A | 41 (82) | 8 (16) | 1 (2) | 1 (2) | 20 (40) | 29 (58) | 0 (0) | 0 (0) | 50 (100) | 2 (4) | 19 (38) | 29 (58) | 1 (2) | 8 (16) | 41 (82) |
| K | 37 (74) | 13 (26) | 0 (0) | 0 (0) | 14 (28) | 36 (72) | 0 (0) | 1 (2) | 49 (98) | 1 (2) | 25 (50) | 24 (48) | 0 (0) | 13 (26) | 37 (74) |
| Y | 37 (74) | 12 (24) | 1 (2) | 3 (6) | 9 (18) | 38 (76) | 0 (0) | 0 (0) | 50 (100) | 2 (4) | 22 (44) | 26 (52) | 1 (2) | 12 (24) | 37 (74) |
| UGT1A7 N129K (P = 0.853) | UGT1A7 W208R (P = 0.409) | UGT1A7 -57 (P = 0.409) | UGT1A9*22 (P = 0.993) | |||||||||
| G/G | T/G | T/T | C/C | T/C | T/T | G/G | T/G | T/T | 9/9 | 9/10 | 10/10 | |
| A | 7 (14) | 24 (48) | 19 (38) | 2 (4) | 23 (46) | 25 (50) | 2 (4) | 23 (46) | 25 (50) | 5 (10) | 24 (48) | 21 (42) |
| K | 8 (16) | 20 (40) | 22 (44) | 4 (8) | 17 (34) | 29 (58) | 4 (8) | 17 (34) | 29 (58) | 6 (12) | 22 (44) | 22 (44) |
| Y | 5 (10) | 23 (46) | 22 (44) | 4 (8) | 14 (28) | 32 (64) | 4 (8) | 14 (28) | 32 (64) | 5 (10) | 23 (46) | 22 (44) |
The only statistical difference in allele polymorphisms examined among the three groups was in UGT1A1*6. The incidence of wild-type UGT1A1*6 across the Akita, Kochi and Yamaguchi populations was 58% (95% CI: 44 to 71), 72% (95% CI: 58 to 83) and 76% (95% CI: 62 to 86), respectively, while the incidence of heterozygous-type UGT1A1*6 was 40%, 28% and 18%, respectively. Volunteers from Akita showed the most heterozygosity in UGT1A1*6, although the P-value was 0.0496.
The participants in this study were mostly nurses and other medical staff from hospitals in the three Japanese prefectures. Around 95% of the nurses in Japan are women; thus the predominance of female subjects in this study.
There are several reports about the distribution of UGT1A1 polymorphisms worldwide. However, these studies were limited to the promoter region, UGT1A1*28[8,25-27], and demonstrated that UGT1A1*28 homozygosity is frequent in Europe (5.0%-14.8%), Africa (5.9%-17.9%) and the Indian subcontinent (19.2%-24.0%), compared to East Asia, which comprises mainly of the Chinese (1.2%-5.0%)[25,26]. Hall et al[25] showed that sub-Saharan Africa, especially Cameroon, was 33% homozygous for UGT1A1*28, which is a fairly high frequency even compared to Caucasians and Indians.
The incidence of homozygous UGT1A1*28 across the three districts of our data in Japan was only 1.3%, which is comparable to the 1.0% reported by Hall et al[25]. Premawardhena et al[26] also reported a wider diversity of repeat numbers among individuals from North and Central America with varying degrees of African ancestry. Our data demonstrated that the repeat number of (TA) was 6/6, 6/7 and 7/7, which is the same as those reported for Europeans and other Asians. Hitherto, no studies have investigated the regional diversity in UGT1A1-family polymorphism within one country, although our study now indicates that there is no diversity of UGT1A1*28 polymorphism in Japan.
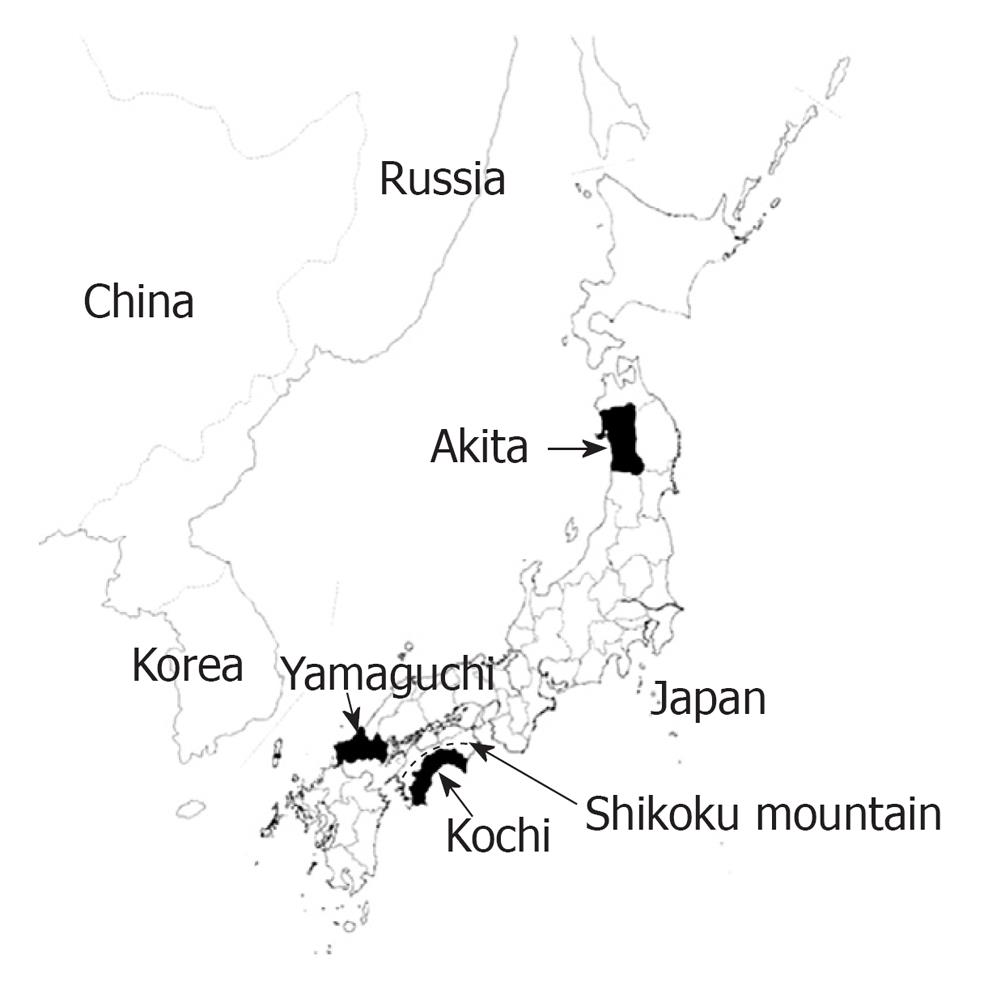
In this study, we selected the Akita, Kochi and Yamaguchi prefectures (Figure 1). Akita represents the northern part of Japan, while the Kochi prefecture on Shikoku Island was obstructed from communication with other prefectures by the Shikoku mountain range in ancient times. Thus, both prefectures have developed a unique dialect and less communication with each other historically. On the other hand, Yamaguchi is one of the nearest prefectures to the Korean Peninsula in Japan. All the prefectures chosen have also developed a unique culture.
Our study revealed no regional diversity of UGT1A1, UGT1A7 and UGT1A9 polymorphisms in Japan. Only UGT1A1*6 showed a statistically significant difference among these three regions in Japan, with more G/A type in the Akita prefecture compared to the other two regions. However, the p-value for the UGT1A1*6 polymorphism was marginal (P-value = 0.0496) and the statistical significance is easily changeable due to the selection of the sampling population. The number of UGT1A1*6 homozygotes was not different among the three districts, with allele frequencies for Akita, Kochi and Yamaguchi of 2.2%, 1.4% and 1.5%, respectively.
Our study is an exploratory research about the diversity of UGT1A1 in Japan. Before the study, we speculated that Akita may have the same tendency of UGT1A1 polymorphism as Caucasians, i.e. Akita may have more polymorphism in UGT1A1*28 and less polymorphism in UGT1A1*6. However, our study revealed that UGT1A1*28 showed no diversity and UGT1A1*6 did not show less polymorphism, although this was not random sampling and generalizability of our population could not be guaranteed.
As described, heterozygotes of UGT1A1*28 are extremely rare in the Japanese population compared to Caucasians and the incidence of heterozygotes and homozygotes of UGT1A1*28 across the three districts combined was 22.0% and 0.013%, respectively.
Our study also demonstrated that the UGT1A1*6 polymorphisms, G/A and A/A, occurred at a rate of 28.7% and 2.7%, respectively, in Japan. Kaniwa et al[28] examined the variants of UGT1A1*6 in Caucasian and African-American populations. Caucasians showed only two heterozygotes among 150 blood samples, while none were found among the African-Americans. Our study confirmed the Japanese standard data for UGT1A1 polymorphism frequencies, which shows more variants for UGT1A1*6 compared to Caucasian and African-American samples.
Jinno et al[29] examined the glucuronidation of SN-38, a potent inhibitor of topoisomerase 1, by human UGT1A1 variants in Cos-1 cells. The variant 211G<A (G71R) (UGT1A1*6) reduced the glucuronidation activity more than 686C>A (P229Q) (UGT1A1*27). Moreover, hyperbilirubinemia observed in Japanese and Taiwanese patients with the P229Q variant is mainly attributable to the TA7 variation. Thus, UGT1A1*6 plays an important role during chemotherapy with irinotecan in East Asian populations[28,30].
Finally, the variant sequences in exon 1, UGT1A1*6 and UGT1A1*27, have been identified only in the Japanese. Thus, Japanese studies could focus more on these two genotypes, which might be more closely associated with drug sensitivity in Japanese patients than in Caucasians[31-33].
Our ongoing studies will compare UGT1A gene polymorphism worldwide, starting in Asian populations and gradually spreading to Europeans. Such investigations may also clarify the movement of people throughout history.
Irinotecan with fluoropyrimidine is approved worldwide as a first-line chemotherapeutic agent for metastatic colorectal cancer. Although prolonged survival has been reported with the use of this drug, severe diarrhea and neutropenia have also been reported as dose-limiting toxicities in 20%-35% of patients treated by the agent. Recent studies revealed that the risk of such severe toxicities might be associated with genetic variation in irinotecan metabolism, indicating a possible predictive factor.
This study aimed to clarify the regional differences in UGT enzyme polymorphisms among three different districts in Japan that are widely distant, both geographically and culturally.
The authors enrolled 50 healthy volunteers from each of the Yamaguchi (western part of Japan), Kochi (southern part of Japan), and Akita (northern part of Japan) prefectures. Blood samples were collected from each participant and stored in EDTA for subsequent genotyping by fragment size analysis, direct sequencing, and TaqMan assay of UGT1A1*28, UGT1A7*3/UGT1A9*22, and UGT1A1*93/UGT1A1*6/UGT1A1*27/UGT1A1*60/UGT1A7 (-57), respectively.
The authors found that the only statistically significant differences in allele polymorphisms among the group examined were for UGT1A1*6. The Akita population showed more UGT1A1*6 heterozygosity. This study revealed no regional diversity among UGT1A1, UGT1A7 or UGT1A9 polymorphisms in Japan.
Kobayashi et al aimed to clarify the regional differences in UGT enzyme polymorphisms among three different districts in Japan that are widely distant, both geographically and culturally. The study seems interesting, but the sample size is somewhat small.
Peer reviewer: Kwang-Huei Lin, PhD, Professor, Associate Dean, College of Medicine, Chang-Gung University, 259 Wen-Hwa 1st Road, Taoyuan 333, Taiwan, China
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
| 1. | Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2407] [Cited by in RCA: 2381] [Article Influence: 95.2] [Reference Citation Analysis (1)] |
| 2. | Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343:905-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2273] [Cited by in RCA: 2221] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 3. | Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2282] [Cited by in RCA: 2200] [Article Influence: 104.8] [Reference Citation Analysis (1)] |
| 4. | Hazama S, Nagashima A, Kondo H, Yoshida S, Shimizu R, Araki A, Yoshino S, Okayama N, Hinoda Y, Oka M. Phase I study of irinotecan and doxifluridine for metastatic colorectal cancer focusing on the UGT1A1*28 polymorphism. Cancer Sci. 2010;101:722-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K, Kato T, Nagata N, Kimura H, Oba K. Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol. 2011;41:477-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991;51:4187-4191. [PubMed] |
| 7. | Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 495] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 8. | Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism. Proc Natl Acad Sci USA. 1998;95:8170-8174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 636] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 9. | Ando Y, Saka H, Ando M, Sawa T, Muro K, Ueoka H, Yokoyama A, Saitoh S, Shimokata K, Hasegawa Y. Polymorphisms of UDP-glucuronosyltransferase gene and irinotecan toxicity: a pharmacogenetic analysis. Cancer Res. 2000;60:6921-6926. [PubMed] |
| 10. | Massacesi C, Terrazzino S, Marcucci F, Rocchi MB, Lippe P, Bisonni R, Lombardo M, Pilone A, Mattioli R, Leon A. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106:1007-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, Karrison T, Janisch L, Ramírez J, Rudin CM. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 723] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 12. | Ura T, Satoh T, Tsujinaka T, Sasaki Y, Yamazaki K, Munakata M, Okamura S, Yamada Y, Hyodo I, Sakata Y. A genotype-directed dose-finding study of irinotecan based on UGT1A1 *28 and *6 polymorphisms in Japanese patients with gastrointestinal cancer (UGT0601). Ann Oncol. 2008;19 Suppl 8:abstr 406P. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D'Andrea M, Pasetto LM, Pessa S, Errante D, De Pangher V. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061-3068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Mathijssen RH, Marsh S, Karlsson MO, Xie R, Baker SD, Verweij J, Sparreboom A, McLeod HL. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9:3246-3253. [PubMed] |
| 15. | Stewart CF, Panetta JC, O'Shaughnessy MA, Throm SL, Fraga CH, Owens T, Liu T, Billups C, Rodriguez-Galindo C, Gajjar A. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol. 2007;25:2594-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 16. | Araki K, Fujita K, Ando Y, Nagashima F, Yamamoto W, Endo H, Miya T, Kodama K, Narabayashi M, Sasaki Y. Pharmacogenetic impact of polymorphisms in the coding region of the UGT1A1 gene on SN-38 glucuronidation in Japanese patients with cancer. Cancer Sci. 2006;97:1255-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 17. | Han JY, Lim HS, Shin ES, Yoo YK, Park YH, Lee JE, Jang IJ, Lee DH, Lee JS. Comprehensive analysis of UGT1A polymorphisms predictive for pharmacokinetics and treatment outcome in patients with non-small-cell lung cancer treated with irinotecan and cisplatin. J Clin Oncol. 2006;24:2237-2244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 238] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 18. | Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 19. | Saito Y, Sai K, Maekawa K, Kaniwa N, Shirao K, Hamaguchi T, Yamamoto N, Kunitoh H, Ohe Y, Yamada Y. Close association of UGT1A9 IVS1+399C& gt; T with UGT1A1*28, *6, or *60 haplotype and its apparent influence on 7-ethyl-10-hydroxycamptothecin (SN-38) glucuronidation in Japanese. Drug Metab Dispos. 2009;37:272-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Yamamoto N, Takahashi T, Kunikane H, Masuda N, Eguchi K, Shibuya M, Takeda Y, Isobe H, Ogura T, Yokoyama A. Phase I/II pharmacokinetic and pharmacogenomic study of UGT1A1 polymorphism in elderly patients with advanced non-small cell lung cancer treated with irinotecan. Clin Pharmacol Ther. 2009;85:149-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Lankisch TO, Schulz C, Zwingers T, Erichsen TJ, Manns MP, Heinemann V, Strassburg CP. Gilbert's Syndrome and irinotecan toxicity: combination with UDP-glucuronosyltransferase 1A7 variants increases risk. Cancer Epidemiol Biomarkers Prev. 2008;17:695-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 22. | Fujita K, Ando Y, Nagashima F, Yamamoto W, Eodo H, Araki K, Kodama K, Miya T, Narabayashi M, Sasaki Y. Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is associated with reduced activity for SN-38 in Japanese patients with cancer. Cancer Chemother Pharmacol. 2007;60:515-22. doi: 10.1007/s00280-006-0396-1. |
| 23. | Wang L, Hirayasu K, Ishizawa M, Kobayashi Y. Purification of genomic DNA from human whole blood by isopropanol-fractionation with concentrated Nal and SDS. Nucleic Acids Res. 1994;22:1774-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 148] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 24. | Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52:119-126. [RCA] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 249] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 25. | Hall D, Ybazeta G, Destro-Bisol G, Petzl-Erler ML, Di Rienzo A. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics. 1999;9:591-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Premawardhena A, Fisher CA, Liu YT, Verma IC, de Silva S, Arambepola M, Clegg JB, Weatherall DJ. The global distribution of length polymorphisms of the promoters of the glucuronosyltransferase 1 gene (UGT1A1): hematologic and evolutionary implications. Blood Cells Mol Dis. 2003;31:98-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 102] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 27. | Mercke Odeberg J, Andrade J, Holmberg K, Hoglund P, Malmqvist U, Odeberg J. UGT1A polymorphisms in a Swedish cohort and a human diversity panel, and the relation to bilirubin plasma levels in males and females. Eur J Clin Pharmacol. 2006;62:829-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Kaniwa N, Kurose K, Jinno H, Tanaka-Kagawa T, Saito Y, Saeki M, Sawada J, Tohkin M, Hasegawa R. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33:458-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Jinno H, Tanaka-Kagawa T, Hanioka N, Saeki M, Ishida S, Nishimura T, Ando M, Saito Y, Ozawa S, Sawada J. Glucuronidation of 7-ethyl-10-hydroxycamptothecin (SN-38), an active metabolite of irinotecan (CPT-11), by human UGT1A1 variants, G71R, P229Q, and Y486D. Drug Metab Dispos. 2003;31:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 85] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Huang CS, Luo GA, Huang ML, Yu SC, Yang SS. Variations of the bilirubin uridine-diphosphoglucuronosyl transferase 1A1 gene in healthy Taiwanese. Pharmacogenetics. 2000;10:539-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 84] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995;333:1171-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1077] [Cited by in RCA: 1004] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 32. | Akaba K, Kimura T, Sasaki A, Tanabe S, Ikegami T, Hashimoto M, Umeda H, Yoshida H, Umetsu K, Chiba H. Neonatal hyperbilirubinemia and mutation of the bilirubin uridine diphosphate-glucuronosyltransferase gene: a common missense mutation among Japanese, Koreans and Chinese. Biochem Mol Biol Int. 1998;46:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 52] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Maruo Y, Nishizawa K, Sato H, Doida Y, Shimada M. Association of neonatal hyperbilirubinemia with bilirubin UDP-glucuronosyltransferase polymorphism. Pediatrics. 1999;103:1224-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 125] [Article Influence: 4.8] [Reference Citation Analysis (0)] |