Published online Jun 15, 2025. doi: 10.4251/wjgo.v17.i6.106161
Revised: March 24, 2025
Accepted: April 22, 2025
Published online: June 15, 2025
Processing time: 115 Days and 10 Hours
Colorectal cancer (CRC) is a leading cause of cancer-related morbidity and mor
To explore the specific roles and underlying mechanisms of exosomal miR-425-5p and miR-135b-3p in CRC progression.
Differentially expressed miRNAs were identified through microarray analysis of exosomes isolated from CRC tissues and adjacent normal mucosa. Functional roles of miR-425-5p and miR-135b-3p were evaluated in vitro using macrophage polarization, T cell differentiation, and vascular permeability assays, as well as in vivo tumor formation and metastasis experiments in nude mice. Validation expe
Exosomal miR-425-5p and miR-135b-3p were significantly upregulated in CRC compared to normal tissues. Functional studies revealed that miR-425-5p promo
Exosomal miR-425-5p and miR-135b-3p drive CRC progression by promoting immune suppression and vascular permeability. Their inhibition offers a promising strategy for modulating the tumor microenvironment and limiting CRC metastasis.
Core Tip: This study identifies exosomal miR-425-5p and miR-135b-3p as critical regulators of colorectal cancer (CRC) progression. These microRNAs (miRNAs) are significantly upregulated in CRC-derived exosomes and modulate the tumor microenvironment through distinct mechanisms. MiR-425-5p promotes immune suppression by inducing macrophage M2 polarization and skewing T cell differentiation toward pro-tumor subsets, while miR-135b-3p enhances vascular permeability and angiogenesis. Inhibition of these miRNAs in preclinical models significantly reduces tumor growth and metastasis, suggesting they represent promising therapeutic targets for CRC. The findings provide novel insights into exosomal miRNA-mediated immune modulation and vascular remodeling in CRC progression.
- Citation: Feng CZ, Zhong SQ, Ye SW, Zheng Z, Sun H, Zhou SH. Tumor-derived exosomal miR-425-5p and miR-135b-3p enhance colorectal cancer progression through immune suppression and vascular permeability promotion. World J Gastrointest Oncol 2025; 17(6): 106161
- URL: https://www.wjgnet.com/1948-5204/full/v17/i6/106161.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i6.106161
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide, with high rates of morbidity and mortality[1,2]. Despite advances in diagnosis and treatment, the mechanisms underlying CRC progression and metastasis remain incompletely understood[3,4]. Exosomes, small extracellular vesicles secreted by various cell types, have emerged as critical mediators of intercellular communication within the tumor microenvironment[5]. Notably, exosomal microRNAs (miRNAs) are increasingly recognized as pivotal regulators of cancer biology, influencing processes such as immune modulation, angiogenesis and metastasis[6-8]. Several studies have reported specific exosomal miRNAs linked to macro
Among these miRNAs, miR-425-5p and miR-135b-3p have been implicated in the progression of several cancers[17-22]. Previous studies have shown that miR-425-5p is involved in modulating immune responses and promoting tumor growth in hepatocellular carcinoma and pancreatic carcinoma[23,24]. In CRC, miR-425-5p is found upregulated in pa
In this study, we systematically address these gaps by investigating the roles of exosomal miR-425-5p and miR-135b-3p in CRC progression. Through microarray analysis of tumor tissue exosomes, we identified these miRNAs as highly upregulated in CRC. Functional enrichment analyses linked miR-425-5p to immune cell infiltration and miR-135b-3p to vascular permeability, highlighting novel regulatory functions in CRC progression. To further explore these roles, we examined their effects on immune modulation, angiogenesis and tumor growth using both in vitro and in vivo models.
Our findings demonstrate that exosomal miR-425-5p promotes macrophage polarization toward an immunosuppressive phenotype and skews T cell differentiation toward pro-tumor subsets, while miR-135b-3p enhances vascular permeability and angiogenesis. Furthermore, in vivo inhibition of these miRNAs suppresses tumor growth and metastasis, reprogramming the tumor microenvironment to a less aggressive state. By systematically elucidating the roles of these two miRNAs in immune regulation and vascular permeability - two underexplored aspects of exosomal miRNA function in CRC - our study provides novel insights into the tumor microenvironment and suggests potential therapeutic strategies targeting exosomal miRNAs in CRC.
CRC and adjacent normal tissue specimens were obtained from patients who underwent surgical procedures at Zhong
Human CRC cell lines (HCT116, SW620, SW480, HT-29), normal colon epithelial cell line (FHC), macrophage cell line (THP-1), primary naive CD4+ T cells, and human umbilical vein endothelial cells (HUVECs) were obtained from American Type Culture Collection (Rockville, MD, United States). Authentication and culture were conducted in specific media supplemented with 100 g/L foetal bovine serum (FBS) (Gibco, NY, United States). HCT116 and HT-29 were cultured in McCoy’s 5A medium, SW620 and SW480 in Leibovitz L-15, FHC in DMEM: F-12, and THP-1 in RPMI-1640 (Gibco, NY, United States). Primary naive CD4+ T cells were maintained in RPMI-1640 with 100 g/L HAS (Med Chem Express, NJ, United States), while HUVECs were grown in vascular cell basal medium with an endothelial cell growth kit-BBE (American Type Culture Collection, Rockville, MD, United States). All cells were incubated at 37 °C in a humidified 50 mL/L CO2 atmosphere.
Exosomes were isolated and characterized as previously described[30]. In brief, exosomes were extracted using an exo Easy Maxi Kit (Qiagen, Germany) according to the manufacturer’s instructions. Following isolation, exosomes were characterized using transmission electron microscopy to confirm morphology, nanoparticle tracking analysis for size distribution, and western blotting for exosomal markers, including tumor susceptibility gene 101 protein (TSG101) and CD81. The purity of exosome preparations was validated by assessing the absence of calnexin, an endoplasmic reticulum marker. The primary antibodies were anti-TSG101 (28283-1-AP), anti-CD81 (66866-1-Ig) and anti-calnexin (10427-2-AP) (all from Proteintech, IL, United States).
Total RNA from CRC patient-derived exosomes was extracted using the All-in-One microRNA extraction kit (Gene
Small RNA libraries were constructed using 100 ng total RNA input with the VAHTS Small RNA Library Prep Kit (Vazyme Biotech, Nanjing, China) compatible with Illumina platforms. The workflow involved sequential enzymatic processing, beginning with adapter ligation targeting small RNA species (miRNA, small interfering RNA, PIWI-interacting RNA). Reverse transcription was then performed to generate cDNA templates, followed by polymerase chain reaction (PCR) enrichment and purification of amplified libraries. Quality control was conducted via Agilent 2100 Bioanalyzer profiling using a DNA 1000 chip, with library quantification achieved through KAPA Biosystems’ library quantification assay (Kapa Biosystems, Wilmington, MA, United States).
Libraries were diluted, equimolarly pooled, and clustered before undergoing single-end sequencing (50 bp read length). Bioinformatics analysis, including differential expression and Kyoto Encyclopedia of Genes and Genomes enrichment analysis, was performed to identify significant molecular changes and related biological pathways.
Total RNA from exosomes was isolated as described in the microarray analysis section, while total RNA from cells was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. cDNA synthesis was carried out with either the miRNA first-strand cDNA Synthesis Kit (Vazyme, Nanjing, China) or MonScriptRTIII Super Mix (Monad, China).
PCRs were performed on a CFX96 Touch™ real-time PCR system (Bio-Rad, Hercules, CA, United States) using miRNA Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) or MonAmp ChemoHS qPCR Mix (Monad, China). U6 and GAPDH served as internal controls for exosomes and cells respectively. The relative gene abundance was calculated via the 2-ΔΔCT formula, and the primer sequences are listed in Table 1.
| Gene | Primer sequence (5’-3’) |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| MiR-425-5p | Forward: AAUGACACGAUCACUCCCGUUGA |
| Reverse: AGTGCAGGGTCCGAGGTATT | |
| MiR-135b-3p | Forward: AUGUAGGGCUAAAAGCCAUGGG |
| Reverse: AGTGCAGGGTCCGAGGTATT | |
| GAPDH | Forward: AATGACCCCTTCATTGAC |
| Reverse: TCCACGACGTACTCAGCGC | |
| IL-1β | Forward: AACCTCTTCGAGGCACAAGG |
| Reverse: AGATTCGTAGCTGGATGCCG | |
| IL-6 | Forward: CCTTCGGTCCAGTTGCCTTCT |
| Reverse: TCTGAGGTGCCCATGCTACA | |
| TNF-α | Forward: CTGAGACTCTCCTGTGCAGC |
| Reverse: ACTCCCTGAAGAGACGGTGA | |
| IL-4 | Forward: CTTTGCTGCCTCCAAGAACA |
| Reverse: GTTCCTGTCGAGCCGTTTCA |
Cells were lysed using RIPA buffer, and total protein concentration was measured via a BCA Protein Assay Kit (Pierce, Waltham, MA, United States). Equal amounts (20 μg) of protein were separated on a 120 g/L sodium-dodecyl sulfate gel electrophoresis gel (80 V, 40 minutes) and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 100 g/L nonfat milk, incubated overnight at 4 °C with primary antibodies, and then with secondary antibodies for 1 hour at room temperature. Signals were detected using the iBright 1500 system (Thermo Fisher, Waltham, MA, United States), and band intensities were quantified with ImageJ. Antibodies: Anti-zonula occludens-1 (anti-ZO-1) (21773-1-AP), anti-occludin (27260-1-AP), anti-claudin-5 (29767-1-AP) (all from Proteintech, IL, United States).
Transfection: MiR-425-5p andmiR-135b-3p inhibitors, along with negative control (NC) recombinant (Med Chem Express, NJ, United States), were transfected into CRC cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). Cells were cultured to 85% confluence, and exosomes were isolated using exosome concentration solution at 4 °C. Exosomes were then incubated with THP-1 cells, primary naïve T cells, or HUVECs for further analysis.
Flowcytometry: THP-1 cells were stained with CD86-FITC, inducible nitric oxide synthase (iNOS)-Alexa Fluor 488, CD163-PE, and CD206-APC. CD4+ T cells were labeled with CD4-FITC, interferon gamma (IFN-γ)-PE, interleukin 4 (IL4)-PE, IL17A-PE, CD25-FITC, forkhead box protein P3 (Foxp3)-PE, and Zombie NIR™ for live/dead discrimination. Tumor cells from mouse models were dissociated using Miltenyi Biotec’s dissociation kit and GentleMACS™, then stained with CD4-FITC, IFNγ-PE, IL4-PE, IL17A-PE, CD25-APC, Foxp3-PE, and 7-AAD. Flowcytometry was performed on a CytoFLEX S (Beckman Coulter, Brea, CA, United States), with data analyzed via Kaluza software. Antibodies are listed in Table 2.
| Antibody | Catalog number | Manufacture |
| THP-1 cells | ||
| Anti-CD86-FITC | 557343 | BD Biosciences, NJ, United States |
| Anti-iNOS | ab283655 | Abcam, United Kingdom |
| Alexa Fluor® 488 | ab236553 | Abcam, United Kingdom |
| Anti-CD163-PE | 560933 | BD Biosciences, NJ, United States |
| Anti-CD206-APC | 561763 | BD Biosciences, NJ, United States |
| CD4+ T cells | ||
| Anti-CD4-FITC | 561005 | BD Biosciences, NJ, United States |
| Anti-IFN-γ-PE | 561056 | BD Biosciences, NJ, United States |
| Anti-IL4-PE | 554485 | BD Biosciences, NJ, United States |
| Anti-IL17A-PE | 560438 | BD Biosciences, NJ, United States |
| Anti-CD25-FITC | 560990 | BD Biosciences, NJ, United States |
| Anti-Foxp3-PE | 560082 | BD Biosciences, NJ, United States |
| Mouse tumor cells | ||
| Anti-CD4-FITC | 553046 | BD Biosciences, NJ, United States |
| Anti-IFNγ-PE | 562020 | BD Biosciences, NJ, United States |
| Anti-IL4-PE | 562044 | BD Biosciences, NJ, United States |
| Anti-IL17A-PE | 561020 | BD Biosciences, NJ, United States |
| Anti-CD25-APC | 561048 | BD Biosciences, NJ, United States |
| Anti-Foxp3-PE | 563101 | BD Biosciences, NJ, United States |
Immunofluorescence staining: Cell slices or 4-μm tumor sections were blocked with 10 g/L BSA (Sigma - Aldrich, Saint Louis, MI, United States). They were then incubated overnight at 4 °C with primary antibodies: Anti-CD86 (13395-1-AP), anti-CD206 (18704-1-AP), anti-ZO-1 (21773-1-AP), anti-occludin (27260-1-AP), and anti-claudin-5 (29767-1-AP) (all from Proteintech, IL, United States). After washing with phosphate buffered saline, samples were incubated for 2 hours at room temperature with fluorescence-conjugated secondary antibodies (Abcam, United Kingdom). Nuclei were stained with DAPI (Beyotime, China), and images were taken using a ZEISS LSM 900 confocal microscope.
Tube formation assay: Matrigel (Corning, NY, United States) was polymerized in 24-well plates at 37 °C for 30 minutes. Treated HUVECs were seeded onto the matrigel and incubated at 37 °C in 50 mL/L CO2. Tube formation was observed at 12 hours under a microscope and quantified by counting the tube structures.
Transwell migration assay: Exosome-treated HUVECs in serum-free medium were seeded into transwell inserts (8-μm pore, BD Biosciences, NJ, United States). The lower chamber contained medium with 100 g/L FBS. After 12 hours, migrated cells on the membrane’s lower surface were stained with hematoxylin and counted under a light microscope in four random fields (200 ×).
Permeability assay: Rhodamine B isothiocyanate-dextran with a molecular weight around 70000 (Sigma, MI, United States) was introduced to into the upper compartment of transwell filters (0.4-μm pore, BD Biosciences, NJ, United States) seeded with HUVECs (105 cells/well). The HUVECs were co-cultured with CRC cells for 3 days. After incubation for 30 minutes, lower chamber fluorescence was quantified at 544 nm excitation/590 nm emission.
Aortic ring assay: Thoracic aortas from 8-12-week-old Sprague Dawley rats were sectioned (1 mm), placed on matrigel-coated wells, and incubated at 37 °C for 30 minutes. Vascular cell basal medium with 100 g/L FBS, vascular endothelial growth factor (VEGF) (10 ng/mL), and heparin (25 μg/mL) was added. After 24 hours, aortic rings were incubated with conditioned medium or exosomes from CRC cells transfected with miR-425-5p inhibitor or NC. Sprouts were quantified on day 5.
Female Balb/c-nude mice (4-6 weeks, Guangdong Zhiyuan Biomedical Technology Co., Ltd., China) were housed pathogen-free under a 12-hour light/dark cycle. Experimental procedures were approved by the Laboratory Animal Ethics Committee of Lai’an Technology (Approval No. G2024123).
Mice were subcutaneously injected with CRC cells (2 × 106/mouse) transfected with miR-425-5p inhibitor, miR-135b-3p inhibitor, both inhibitors, or NC. After 42 days, mice were exposure to excess CO2 to ensure unconsciousness, and tumor, liver, and lung tissues were collected immediately. Tumor volume, weight, and gross size were recorded, and metastatic nodules in the liver/lungs were counted.
Tumors were fixed informal in, embedded in paraffin, and sectioned (4 μm). Hematoxylin and eosin staining (Beyo
Data are presented as mean ± SD. Statistical analysis was performed using GraphPad Prism 8.2 (GraphPad Software, United States) with unpaired t test or one-way ANOVA, followed by Dunnett’s test for multiple comparisons. Statistical significance was considered as aP < 0.05, bP < 0.01, cP < 0.001, dP < 0.0001.
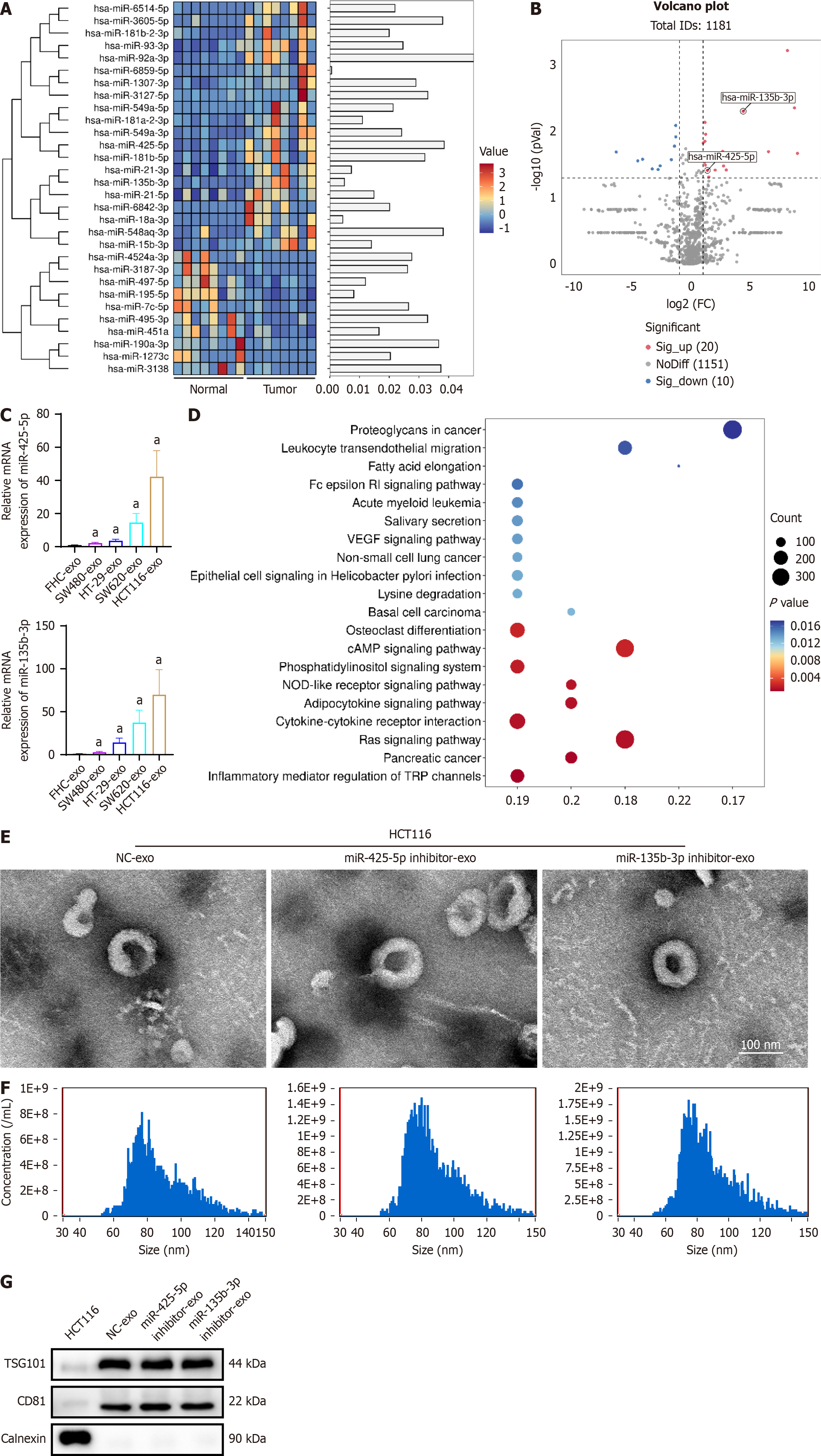
To explore the dysregulation of exosomal miRNAs, we extracted exosomes from tumor tissues and adjacent normal mu
To further investigate the effects of exosomal miR-425-5p on tumor cell-induced immune responses and exosomal miR-135b-3p on tumor cell-induced vascular permeability, we extracted exosomes from the most upregulated HCT116 cells transfected with miR-425-5p inhibitor or miR-135b-3p inhibitor. Characterization of HCT116-derived exosomes were carried out by transmission electron microscopy (Figure 1E) and nanoparticle tracking analysis (Figure 1F). In addition, western blot analysis confirmed the expression of specific exosome markers (TSG101 and CD81) and the absence of the endoplasmic reticulum resident protein calnexin, which serves as a negative control for exosome purity (Figure 1G).
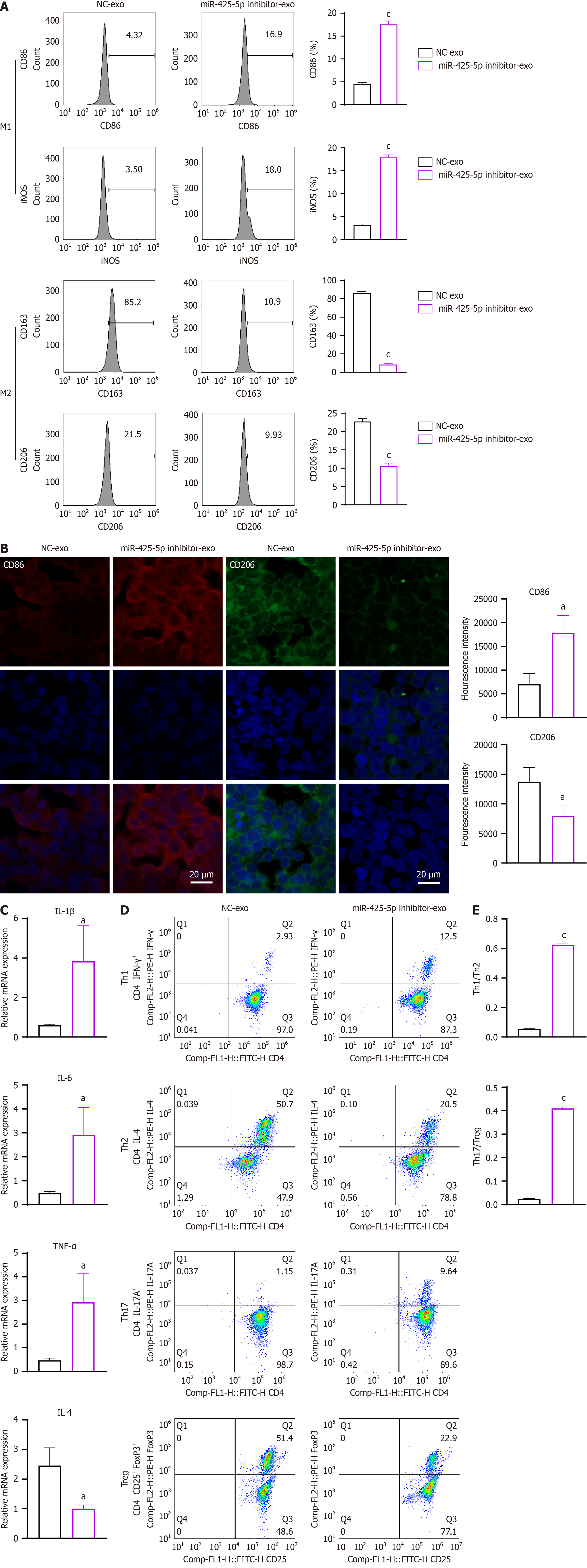
To investigate the effects of exosomal miR-425-5p on immune cells, exosomes isolated from HTC116 cells transfected with miR-425-5p inhibitor or NC were cultured with human macrophage cell line THP-1. Flow cytometry analysis showed an increased proportion of CD86+iNOS+ M1 macrophages [CD86+: P < 0.001, 95% confidence interval (CI): 10.66-15.26; iNOS+: P < 0.001, 95%CI: 13.65-16.09] and a decreased proportion of CD163+CD206+ M2 macrophages (CD163+: P < 0.001, 95%CI: -83.48 to -72.77; iNOS+: P < 0.001, 95%CI: -15.40 to -8.995) when THP-1 cultured with miR-425-5p inhibitor exosomes compared to NC (Figure 2A). Immunofluorescent results confirmed the changes, as evidenced by upregulation of M1 marker CD86 and downregulation of M2 marker CD206 when miR-425-5p was depleted (CD86: P < 0.05, 95%CI: 4036-17670; CD206: P < 0.05, 95%CI: -10519 to -1032) (Figure 2B). These results indicate that exosomal miR-425-5p from CRC cells affects macrophages polarization and depletion of it promotes toward a M2-like phenotype.
To further explore the role of exosomal miR-425-5p in T cell differentiation, primary naïve CD4+ T cells were cultured with exosomes isolated from HCT116 cells transfected with miR-425-5p inhibitor or NC. Real-time quantitative PCR analysis of T cell cytokines revealed significant changes in pro- and anti-inflammatory cytokine expression. In the miR-425-5p inhibitor group, proinflammatory cytokines IL-1β, IL-6, and TNF-α were significantly increased (IL-1β: P < 0.05, 95%CI: 0.3545-6.125; IL-6: P < 0.05, 95%CI: 0.5998-4.264; TNF-α: P < 0.05, 95%CI: 0.4767-4.433), while the anti-inflammatory cytokine IL-4 was markedly decreased (IL-4: P < 0.05, 95%CI: -2.439 to -0.4751) (Figure 2C). Flow cytometry analysis of CD4+ T cell subsets showed pronounced shifts in differentiation patterns. The proportion of CD4+IFNγ+ T-helper 1 (Th1) cells was significantly increased, whereas CD4+IL-4+ Th2 cells were decreased in the miR-425-5p inhibitor group (Th1/Th2: P < 0.001, 95%CI: 0.5486-0.5894) (Figure 2D and E). Additionally, CD4+IL-17A+ Th17 cells were elevated, and the proportion of immunosuppressive CD4+CD25+Foxp3+ regulatory T cells (Tregs) was reduced compared to the NC group (Th17/Treg: P < 0.001, 95%CI: 0.3680-0.4026) (Figure 2D and E). These results collectively indicate that exosomal miR-425-5p from CRC cells modulates T cell differentiation, and its depletion skews differentiation toward a proinflammatory Th1 and Th17 phenotype while reducing anti-inflammatory Th2 and immunosuppressive Treg subsets. To validate the effects of exosomal miR-425-5p on immune modulation, similar experiments were conducted using SW620 cells. The results, which corroborate the findings from HCT116 cells, are presented in Supplementary Figures 1 and 2, further supporting the role of miR-425-5p in regulating macrophage polarization and T cell differentiation.
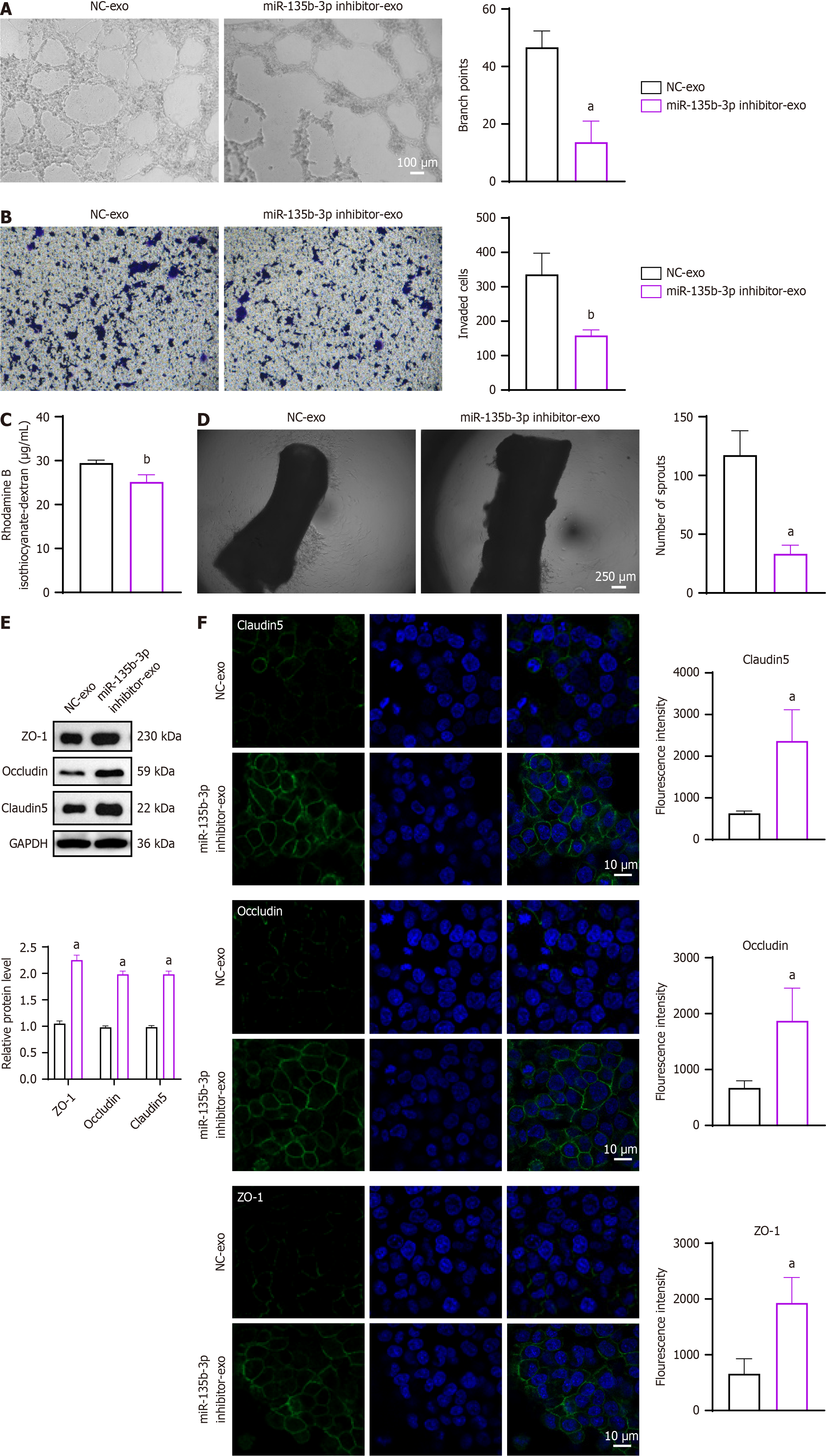
To explore the role of exosomal miR-135b-3p in vascular permeability, exosomes isolated from HCT116 cells transfected with miR-135b-3p inhibitor or NC were cultured with HUVECs. Microscopic observation revealed a reduction in angiogenesis in the inhibitor group compared to NC (branch points: P < 0.05, 95%CI: -47.92 to -18.08) (Figure 3A). Cell invasion assays showed a slight but consistent decrease in invasive capacity in the inhibitor group (invaded cells: P < 0.01, 95%CI: -279.8 to -75.51) (Figure 3B). Similarly, the rhodamine B permeability assay indicated a modest decrease in permeability in HUVECs treated with miR-135b-3p inhibitor exosomes (P < 0.01, 95%CI: -6.134 to -2.512) (Figure 3C). In the aortic ring assay, the number of microvascular sprouts was significantly reduced in the inhibitor group (P < 0.05, 95%CI: -119.3 to -48.71) (Figure 3D). ZO-1, occludin, and claudin-5 are tight junction proteins and lack of them leads to increased vascular permeability[31]. Western blot analysis demonstrated increased expression of them in the inhibitor group, indicating enhanced endothelial barrier integrity (ZO-1: P < 0.05, 95%CI: 1.206-1.374; occludin: P < 0.05, 95%CI: 0.9003-1.106; claudin-5: P < 0.05, 95%CI: 0.8941-1.106) (Figure 3E). Immunofluorescence analysis further confirmed the upregulation of ZO-1, occludin and claudin-5 in HUVECs treated with miR-135b-3p inhibitor exosomes (claudin-5: P < 0.05, 95%CI: 528.5-2945; occludin: P < 0.05, 95%CI: 239.5-2161; ZO-1: P < 0.05, 95%CI: 420.6-2121) (Figure 3F). These results suggest that exosomal miR-135b-3p from CRC cells contributes to increased vascular permeability and angiogenesis. Similar experiments conducted using SW620 cells yielded comparable results, further supporting the role of miR-135b-3p in vascular permeability. These findings are presented in Supplementary Figure 3.
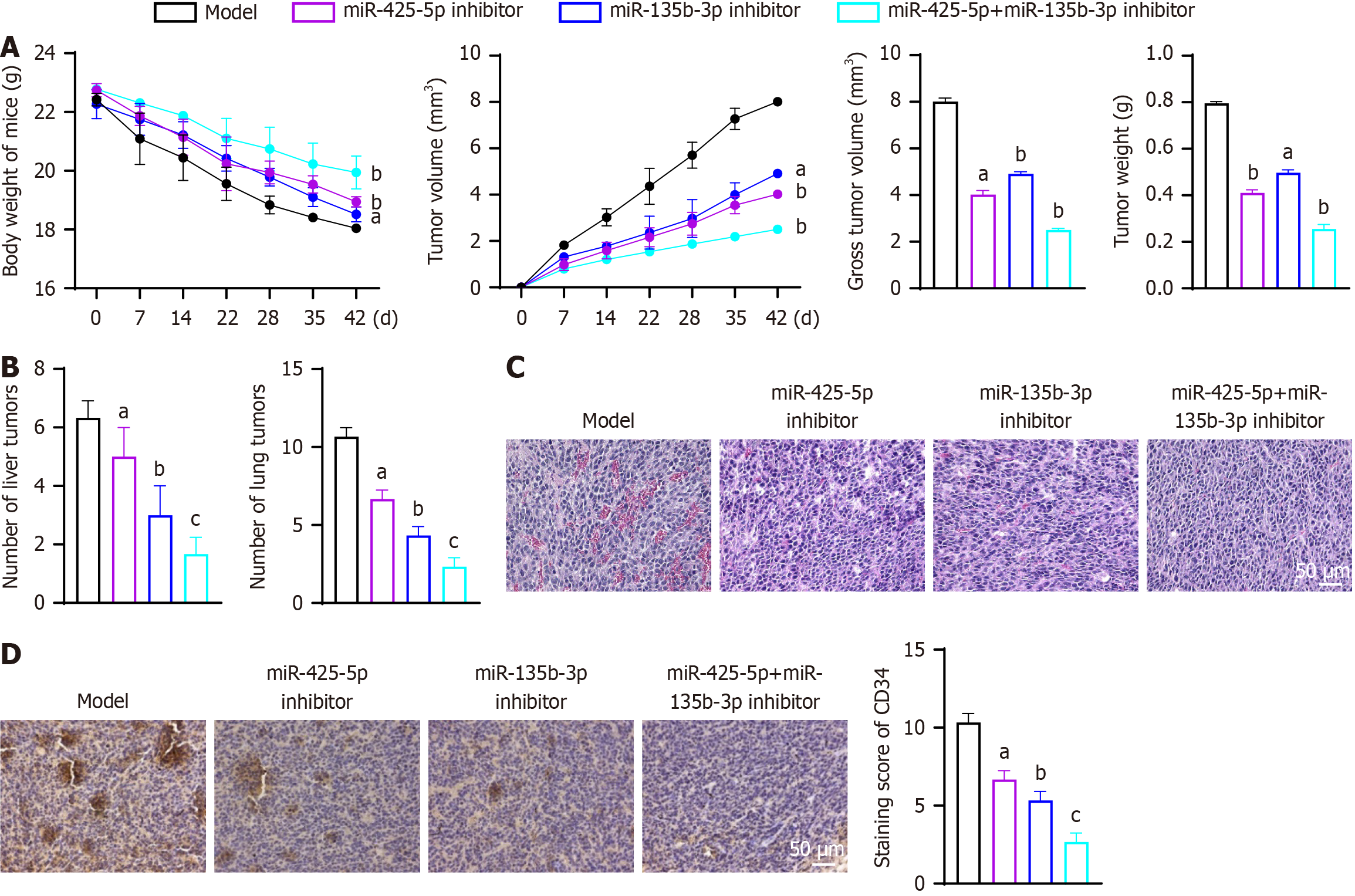
Inhibition of exosomal miR-425-5p and miR-135b-3p significantly suppressed tumor growth, metastasis, and altered the tumor microenvironment in vivo. Compared to the model group, tumor volume, weight, and gross size were markedly reduced in the miR-425-5p inhibitor group (gross tumor volume: P < 0.05, 95%CI: -4.327 to -3.651; tumor weight: P < 0.05, 95%CI: -0.4184 to -0.3523), the miR-135b-3p inhibitor group (gross tumor volume: P < 0.01, 95%CI: -3.431 to -2.755; tumor weight: P < 0.01, 95%CI: -0.3317 to -0.2656), and most notably in the combined inhibition group (gross tumor volume: P < 0.01, 95%CI: -5.846 to -5.170; tumor weight: P < 0.01, 95%CI: -0.5737 to -0.5076) (Figure 4A). Metastatic burden, as indicated by the number of liver and lung tumors, was also significantly decreased in the miR-425-5p inhibitor group (liver tumor: P < 0.05, 95%CI: -3.253 to 0.5865; lung tumor: P < 0.05, 95%CI: -5.358 to -2.642), the miR-135b-3p inhibitor group (liver tumor: P < 0.01, 95%CI: -5.253 to -1.414; lung tumor: P < 0.01, 95%CI: -7.691 to -4.976), with the combined inhibition group demonstrating the lowest metastasis rates (liver tumor: P < 0.001, 95%CI: -6.586 to -2.747; lung tumor: P < 0.001, 95%CI: -9.691 to -6.976) (Figure 4B). Histological analyses revealed increased immune cell infiltration and reduced angiogenesis in the inhibitor groups (Figure 4C). Immunoblotting of CD34 showed reduction in the miR-425-5p inhibitor group (P < 0.05, 95%CI: -5.024 to -2.309), the miR-135b-3p inhibitor group (P < 0.01, 95%CI: -6.358 to -3.642), and the combined inhibition group (P < 0.001, 95%CI: -9.024 to -6.309), further confirming the anti-angiogenic effects (Figure 4D).
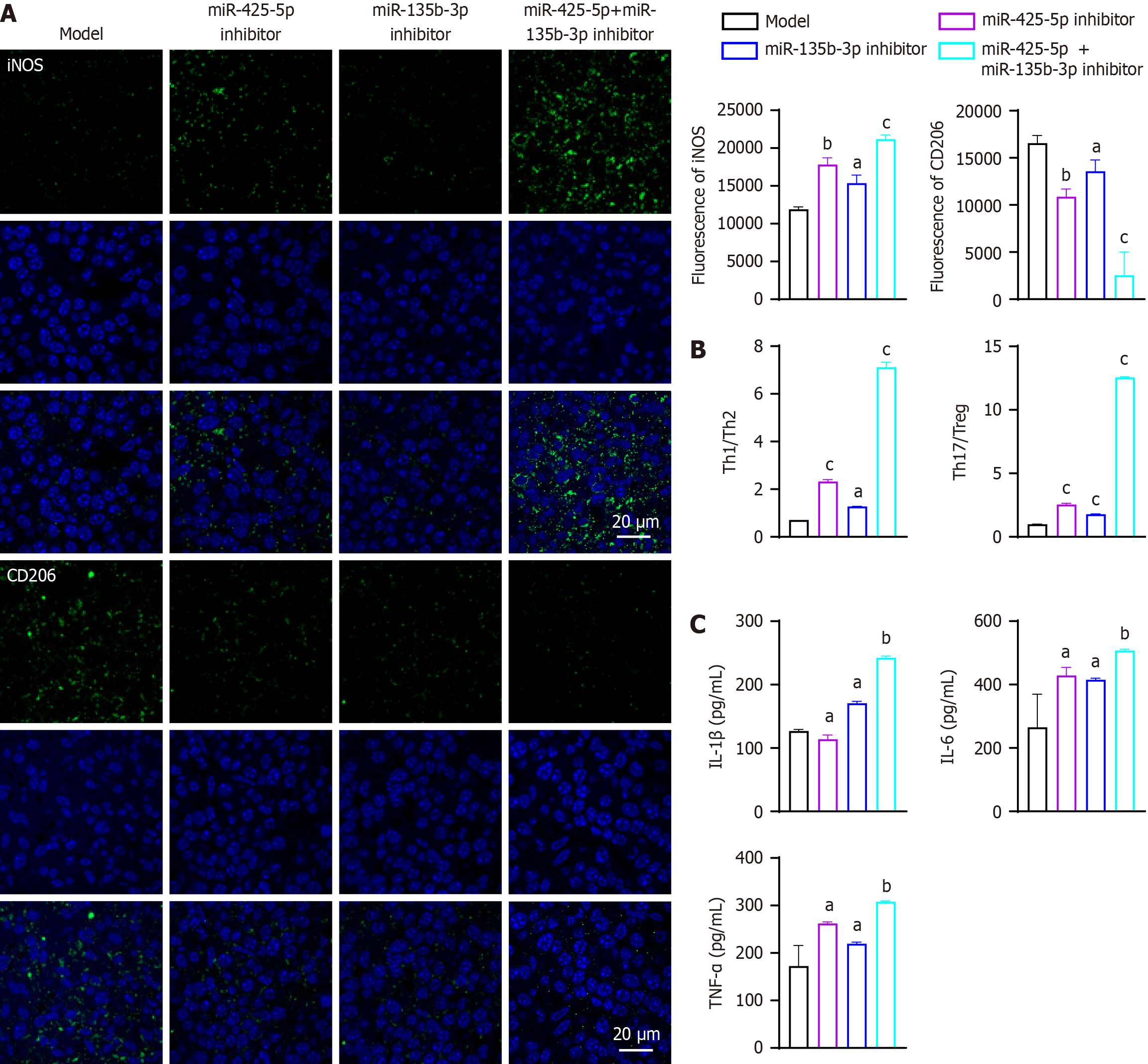
Further analyses showed a pronounced shift in the immune microenvironment. Macrophages exhibited a proinflammatory polarization, with increased M1 marker (iNOS) expression and decreased M2 marker (CD206) expression shown by immunofluorescence staining (Figure 5A). Specifically, compared to the model group, iNOS expression was signi
Flow cytometry of tumor-infiltrating T cells indicated increased Th1 (CD4+IFNγ+) and Th17 (CD4+IL-17A+) populations, along with reduced Th2 (CD4+IL-4+) and Treg (CD4+CD25+Foxp3+) populations (Figure 5B). Compared to the model group, Th1/Th2 ratios were significantly increased in the miR-425-5p inhibitor group (P < 0.001, 95%CI: 1.157-2.075), miR-135b-3p inhibitor group (P < 0.05, 95%CI: 0.1080-1.026), and combined inhibition group (P < 0.001, 95%CI: 5.949-6.867). Th17/Treg ratios showed increases in the miR-425-5p inhibitor group (P < 0.001, 95%CI: 1.325-1.787), miR-135b-3p inhibitor group (P < 0.001, 95%CI: 0.5619-1.025), and combined inhibition group (P < 0.001, 95%CI: 11.33-11.79).
Correspondingly, ELISA results demonstrated elevated levels of proinflammatory cytokines IL-1β, IL-6 and TNF-α in the inhibitor groups, with the highest levels in the combined inhibitor group (Figure 5C). Compared to the model group, IL-1β levels were slightly decreased in the miR-425-5p inhibitor group (P < 0.05, 95%CI: -19.79 to -6.363), and significantly increased in the miR-135b-3p inhibitor group (P < 0.05, 95%CI: 36.93-50.36), and combined inhibition group (P < 0.01, 95%CI: 108.7-122.2). IL-6 levels showed significant increases in the miR-425-5p inhibitor group (P < 0.05, 95%CI: 83.94-242.5), miR-135b-3p inhibitor group (P < 0.05, 95%CI: 70.66-229.2), and combined inhibition group (P < 0.01, 95%CI: 162.9-321.4). TNF-α levels were significantly elevated in all inhibitor groups (miR-425-5p inhibitor: P < 0.05, 95%CI: 53.76-125.3; miR-135b-3p inhibitor: P < 0.05, 95%CI: 11.00-82.51; combined inhibition: P < 0.01, 95%CI: 99.32-170.8).
To validate these findings, the same in vivo experiments were performed using SW620 cells. The results were consistent with those observed in HCT116 cells, further supporting the roles of exosomal miR-425-5p and miR-135b-3p in promoting tumor growth, angiogenesis, and immune evasion. These validation results are presented in Supplementary Figures 4 and 5. These findings underscore that the inhibition of miR-425-5p and miR-135b-3p can effectively reprogram the tumor microenvironment to a less aggressive phenotype.
Our study identifies and characterizes the roles of exosomal miR-425-5p and miR-135b-3p in promoting CRC progression through immune modulation and vascular dynamics. Our findings reveal that both miRNAs are markedly enriched in CRC tumor-derived exosomes and synergistically orchestrate a tumor-permissive microenvironment. Inhibition of these miRNAs suppresses tumor growth, reduces metastasis, and shifts the tumor microenvironment to a less aggressive state. These results highlight their potential as therapeutic targets and biomarkers in CRC.
Exosomal miR-425-5p was found to be of crucial significance in immune modulation, promoting M2 macrophage po
In parallel, miR-135b-3p appears to facilitate tumor metastasis by enhancing vascular permeability and angiogenesis. Its inhibition leads to the upregulation of tight junction proteins such as ZO-1, occludin, and claudin-5, thereby restoring endothelial barrier integrity. These results are consistent with previous studies in breast cancer, where miR-135b-3p has been implicated in promoting angiogenesis[21]. Our findings offer novel insights into its function in CRC by linking it to vascular permeability and endothelial barrier dysfunction. The observed effects may involve VEGF pathway modulation, given its established role in vascular permeability and miRNAs are known to post-transcriptionally regulate its expre
The in vivo experiments underscore the synergistic effects of targeting both miRNAs. Combined inhibition of miR-425-5p and miR-135b-3p resulted in the most pronounced reduction in tumor growth and metastasis, along with significant changes in immune and vascular dynamics. These findings suggest that combinatorial targeting of these miRNAs could provide a more effective therapeutic strategy compared to single-agent approaches.
Our study also highlights the potential clinical utility of these miRNAs as biomarkers. Their elevated levels in CRC-derived exosomes, compared to normal tissues, suggest their use in non-invasive diagnostic or prognostic applications. The result of miR-425-5p aligns with previous studies in CRC tumors or serums[25-28,39], and we recommend miR-135b-3p as a new biomarker for CRC. Furthermore, their roles in immune suppression and vascular permeability make them attractive targets for combination therapies, such as immune checkpoint inhibitors or anti-angiogenic agents.
Despite these promising findings, there are limitations to our study. One limitation of our study is the relatively limited number of patient samples, which may affect the generalizability of the findings. Although our cohort includes well-characterized CRC specimens and the results were corroborated in multiple CRC cell lines and in vivo models, we recognize that expanding the sample size - especially including patients from various clinical stages and with different molecular subtypes - would further strengthen the conclusions. Future studies are planned to validate these results in larger and more diverse cohorts.
The use of cell-line-derived exosomes may not fully recapitulate patient tumor heterogeneity. Additionally, while functional enrichment analyses provided insights into potential pathways, direct validation of these mechanisms remains to be explored. While our study provides compelling evidence that exosomal miR-425-5p and miR-135b-3p play critical roles in modulating immune responses and vascular permeability in CRC, we recognize that further experimental validation of the underlying molecular mechanisms is necessary. Specifically, additional experiments such as gene knockout/overexpression studies, dual-luciferase reporter assays, and immunoprecipitation experiments would be valuable to confirm the involvement of the NOD-like receptor and VEGF signaling pathways, as well as to identify direct downstream targets of these miRNAs. Due to constraints in time and resources, these experiments were not included in the present study. However, we consider this an important avenue for future research, which will help to more fully elucidate the molecular mechanisms by which these exosomal miRNAs contribute to CRC progression. Expanding this work to include patient-derived exosomes and pathway-specific experiments will further strengthen the translational relevance of these findings.
This study establishes exosomal miR-425-5p and miR-135b-3p as key mediators of CRC progression, shaping immune evasion and vascular dysfunction. Their dual targeting offers a promising strategy to disrupt tumor-microenvironment crosstalk, paving the way for innovative therapeutic interventions in CRC.
| 1. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3015] [Article Influence: 502.5] [Reference Citation Analysis (3)] |
| 2. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (2)] |
| 3. | Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharmacol Sci. 2023;44:222-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 282] [Reference Citation Analysis (0)] |
| 4. | Ionescu VA, Gheorghe G, Bacalbasa N, Chiotoroiu AL, Diaconu C. Colorectal Cancer: From Risk Factors to Oncogenesis. Medicina (Kaunas). 2023;59:1646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 5. | Huang Y, Kanada M, Ye J, Deng Y, He Q, Lei Z, Chen Y, Li Y, Qin P, Zhang J, Wei J. Exosome-mediated remodeling of the tumor microenvironment: From local to distant intercellular communication. Cancer Lett. 2022;543:215796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 6. | Zhang Y, Tang S, Gao Y, Lu Z, Yang Y, Chen J, Li T. Application of exosomal miRNA mediated macrophage polarization in colorectal cancer: Current progress and challenges. Oncol Res. 2023;32:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 7. | Savardashtaki A, Shabaninejad Z, Movahedpour A, Sahebnasagh R, Mirzaei H, Hamblin MR. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics. 2019;11:1627-1645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 8. | Tang Y, Zong S, Zeng H, Ruan X, Yao L, Han S, Hou F. MicroRNAs and angiogenesis: a new era for the management of colorectal cancer. Cancer Cell Int. 2021;21:221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 9. | Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, He X, Zhong X, Li G, Chen Z, Li D. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 537] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 10. | Yang C, Dou R, Wei C, Liu K, Shi D, Zhang C, Liu Q, Wang S, Xiong B. Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol Ther. 2021;29:2088-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 153] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 11. | Pei W, Wei K, Wu Y, Qiu Q, Zhu H, Mao L, Shi X, Zhang S, Shi Y, Tao S, Mao H, Pang S, Wang J, Liu M, Wang W, Yang Q, Chen C. Colorectal cancer tumor cell-derived exosomal miR-203a-3p promotes CRC metastasis by targeting PTEN-induced macrophage polarization. Gene. 2023;885:147692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 12. | Liang Y, Li J, Yuan Y, Ju H, Liao H, Li M, Liu Y, Yao Y, Yang L, Li T, Lei X. Exosomal miR-106a-5p from highly metastatic colorectal cancer cells drives liver metastasis by inducing macrophage M2 polarization in the tumor microenvironment. J Exp Clin Cancer Res. 2024;43:281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 13. | Feng Y, Jin C, Wang T, Chen Z, Ji D, Zhang Y, Zhang C, Zhang D, Peng W, Sun Y. The Uridylyl Transferase TUT7-Mediated Accumulation of Exosomal miR-1246 Reprograms TAMs to Support CRC Progression. Adv Sci (Weinh). 2024;11:e2304222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 14. | Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, Shen C, Ma Y, Jiang S, Ma D, Tong T, Zhang X, Gao Z, Zhu X, Fang JY, Chen H, Hong J. Enterotoxigenic Bacteroidesfragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged miR-149-3p. Gastroenterology. 2021;161:1552-1566.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 189] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 15. | Ning T, Li J, He Y, Zhang H, Wang X, Deng T, Liu R, Li H, Bai M, Fan Q, Zhu K, Ying G, Ba Y. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol Ther. 2021;29:2723-2736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 16. | Hosseini M, Baghaei K, Hajivalili M, Zali MR, Ebtekar M, Amani D. The anti-tumor effects of CT-26 derived exosomes enriched by MicroRNA-34a on murine model of colorectal cancer. Life Sci. 2022;290:120234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Rode MP, Silva AH, Cisilotto J, Rosolen D, Creczynski-Pasa TB. miR-425-5p as an exosomal biomarker for metastatic prostate cancer. Cell Signal. 2021;87:110113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Wu Z, Guo J, Zhang Y, Liu J, Ma H, Tang Y. MiR-425-5p accelerated the proliferation, migration, and invasion of ovarian cancer cells via targeting AFF4. J Ovarian Res. 2021;14:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Meng Z, Zhang R, Wu X, Piao Z, Zhang M, Jin T. LncRNA HAGLROS promotes breast cancer evolution through miR-135b-3p/COL10A1 axis and exosome-mediated macrophage M2 polarization. Cell Death Dis. 2024;15:633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Wang Z, Shao C, Lu G, Xie M, Wang J, Duan H, Li X, Yu W, Duan W, Yan X. Melatonin may suppress lung adenocarcinoma progression via regulation of the circular noncoding RNA hsa_circ_0017109/miR-135b-3p/TOX3 axis. J Pineal Res. 2022;73:e12813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 21. | Kim YJ, Zhao Y, Myung JK, Yi JM, Kim MJ, Lee SJ. Suppression of breast cancer progression by FBXL16 via oxygen-independent regulation of HIF1α stability. Cell Rep. 2021;37:109996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Zhang JY, Su XP, Li YN, Guo YH. MicroRNA-425-5p promotes the development of prostate cancer via targeting forkhead box J3. Eur Rev Med Pharmacol Sci. 2019;23:547-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 23. | Wu S, Liu S, Cao Y, Chao G, Wang P, Pan H. Downregulation of ZC3H13 by miR-362-3p/miR-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging (Albany NY). 2022;14:2304-2319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 24. | Jian S, Kong D, Tian J. Expression of miR-425-5p in Pancreatic Carcinoma and Its Correlation with Tumor Immune Microenvironment. J Invest Surg. 2023;36:2216756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 25. | Urh K, Žlajpah M, Zidar N, Boštjančič E. Identification and Validation of New Cancer Stem Cell-Related Genes and Their Regulatory microRNAs in Colorectal Cancerogenesis. Biomedicines. 2021;9:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi LW, Wu L, Cheng W, Zhu J, Zhang L, Zhang H, Chen Y, Zhu W, Wang T, Liu P. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget. 2017;8:17081-17091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 27. | Tan Y, Lin JJ, Yang X, Gou DM, Fu L, Li FR, Yu XF. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019;60:67-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Urh K, Zidar N, Tomažič A, Boštjančič E. Intratumor heterogeneity of cancer stem cellrelated genes and their potential regulatory microRNAs in metastasizing colorectal carcinoma. Oncol Rep. 2022;48:193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 29. | Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 249] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 30. | Wang L, Yang L, Zhuang T, Shi X. Tumor-Derived Exosomal miR-29b Reduces Angiogenesis in Pancreatic Cancer by Silencing ROBO1 and SRGAP2. J Immunol Res. 2022;2022:4769385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 31. | Bazzoni G, Dejana E. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol Rev. 2004;84:869-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 864] [Cited by in RCA: 987] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 32. | Lu Y, Wu X, Wang J. Correlation of miR-425-5p and IL-23 with pancreatic cancer. Oncol Lett. 2019;17:4595-4599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | McGeachy MJ, Cua DJ. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin Immunol. 2007;19:372-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 111] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 34. | Wilke CM, Kryczek I, Wei S, Zhao E, Wu K, Wang G, Zou W. Th17 cells in cancer: help or hindrance? Carcinogenesis. 2011;32:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 35. | Tsankov BK, Luchak A, Carr C, Philpott DJ. The effects of NOD-like receptors on adaptive immune responses. Biomed J. 2024;47:100637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Zhou Y, Yu S, Zhang W. NOD-like Receptor Signaling Pathway in Gastrointestinal Inflammatory Diseases and Cancers. Int J Mol Sci. 2023;24:14511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 37. | Patel SA, Nilsson MB, Le X, Cascone T, Jain RK, Heymach JV. Molecular Mechanisms and Future Implications of VEGF/VEGFR in Cancer Therapy. Clin Cancer Res. 2023;29:30-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 210] [Article Influence: 105.0] [Reference Citation Analysis (0)] |
| 38. | Zhang L, Zhang Y, Li X, Gao H, Chen X, Li P. CircRNA-miRNA-VEGFA: an important pathway to regulate cancer pathogenesis. Front Pharmacol. 2023;14:1049742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Lenart M, Siemińska I, Szatanek R, Mordel A, Szczepanik A, Rubinkiewicz M, Siedlar M, Baj-Krzyworzeka M. Identification of miRNAs Present in Cell- and Plasma-Derived Extracellular Vesicles-Possible Biomarkers of Colorectal Cancer. Cancers (Basel). 2024;16:2464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |