Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.104253
Revised: January 17, 2025
Accepted: February 25, 2025
Published online: April 15, 2025
Processing time: 101 Days and 1.5 Hours
Cholangiocarcinoma (CCA), also known as bile duct cancer, is a devastating malignancy primarily affecting the biliary tract.
To assess their performance in clinical diagnosis and monitoring of CCA, plasma methylation and circulating tumor cells were detected.
Plasma samples were collected from Hubei Cancer Hospital (n = 156). Plasma DNA was tested to detect SHOX2, HOXA9, SEPTIN9, and RASSF1A methylation using TaqMan PCR. Circulating tumor cells (CTCs) were detected in the peripheral blood of patients using the United States Food and Drug Administration-approved cell search system before and after clinical therapy. The CCA diagnostic value was estimated using the area under the curve. The independent prognosis risk factors for patients with CCA were estimated using Cox and logistic regression analyses.
The sensitivity and specificity of the four DNA plasma methylations exhibited 64.74% sensitivity and 93.88% specificity for detecting CCA. The receiver operating characteristic curve of the combined value for CCA diagnosis in plasma was 0.828 ± 0.032. RASSF1A plasma methylation was related to the prognosis of patients with CCA. We determined the prognostic hazard ratio for CCA using CTC count, tumor stage, methylation, and carbohydrate antigen 19-9 levels as key factors. Our overall survival nomogram achieved a C-index of 0.705 (0.605-0.805).
SHOX2, HOXA9, SEPTIN9, and RASSF1A plasma methylation demonstrated increased sensitivity for diagnosing CCA. RASSF1A plasma methylation and CTCs were valuable predictors to assess CCA prognosis and recurrence.
Core Tip: This study first analyzed the clinical diagnosis and monitoring value of detecting plasma SHOX2, HOXA9, SEPTIN9, and RASSF1A methylation for cholangiocarcinoma (CCA). We determined that the four DNA plasma methylations exhibited 64.58% sensitivity and 94% specificity for detecting CCA. The hazard ratio of prognosis for the risk of CCA risk was identified using the Circulating tumor cells (CTCs) count, tumor stage, methylation, and carbohydrate antigen 19-9 (CA199) levels as independent prognostic factors. We developed a predictive nomogram for CCA overall survival, age, stage, CTCs, methylation, and CA199, with a C-index of 0.705 (95%CI: 0.605-0.805). This model evaluates risk factors.
- Citation: Yu J, Liu QC, Lu SY, Wang S, Zhang H. Detecting plasma SHOX2, HOXA9, SEPTIN9, and RASSF1A methylation and circulating cancer cells for cholangiocarcinoma clinical diagnosis and monitoring. World J Gastrointest Oncol 2025; 17(4): 104253
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/104253.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.104253
Cholangiocarcinoma (CCA), also known as bile duct cancer, is a devastating malignancy primarily affecting the biliary tract. CCA encompasses the intrahepatic, perihilar, and distal CCA subtypes. The most recent Chinese statistics from 2022 ranked CCA as the second most common primary hepatobiliary malignancy in terms of morbidity and mortality, underscoring its significant clinical burden[1,2]. Unfortunately, the non-specific early symptoms and lack of sensitive diagnostic tools mean that most patients with CCA are diagnosed at an advanced stage, when curative surgical resection is no longer feasible. Furthermore, such patients are prone to frequent metastasis and poor prognosis[3,4]. As a result, the 5-year overall survival (OS) rates are notably low, with patients having a median survival duration of roughly 24 months and facing a staggering 90% mortality rate within five years[3,4].
Identifying CCA early and initiating prompt treatment is essential for enhancing patient prognosis. However, traditional diagnostic methods, such as imaging and tumor marker assessments, have limited sensitivity and specificity, often failing to detect the disease at an early stage[5]. Chronic hepatitis B virus infection is a well-established CCA risk factor with a pivotal etiological role in its development[6]. Despite advances in chemotherapy regimens centered on cisplatin and gemcitabine, CCA remains largely insensitive to chemoradiotherapy and exhibits drug resistance, highlighting the urgent need for more effective diagnostic and monitoring strategies[3].
The role of epigenetic alterations, particularly DNA methylation, in cancer development and progression has garnered significant attention recently[7,8]. Cancer cell genes undergo methylation changes early in carcinogenesis, which persist and dynamically evolve throughout tumor progression. Tumor suppressor genes are frequently silenced by the methylation of CpG islands located upstream of their promoters, disrupting normal cellular processes and promoting malignancy[9,10]. The CpG island hypermethylation of specific genes has been closely linked to cancer initiation and progression, serving as a promising biomarker for early cancer detection and prognosis assessment[11,12].
Circulating tumor DNA (ctDNA) and circulating tumor cells (CTCs) have emerged as non-invasive biomarkers for cancer screening and monitoring[13]. ctDNA is shed by tumor cells into the circulation and contains numerous cancer-specific genetic signatures that reflect the genomic landscape of the primary tumor[14]. CTCs represent micrometastatic dissemination and can be used as real-time indicators of disease status and therapeutic response[15]. The United States Food and Drug Administration (FDA)-approved CellSearch system, which is based on positive immunoselection of epithelial cell adhesion molecule (EpCAM) and negative selection of CD45, enables CTC enumeration and characterization into distinct phenotypes (epithelial, mesenchymal, and hybrid)[16].
Among the myriad of genes undergoing methylation changes in cancer, SHOX2, HOXA9, SEPTIN9, and RASSF1A were specifically chosen for this study due to their established roles in cancer development and progression. SHOX2 and RASSF1A are tumor suppressor genes frequently silenced by methylation in various cancers, and are crucial in the initiation and progression of malignancy[17,18]. The homeobox gene HOXA9 plays a crucial role in modulating mechanisms associated with cancer progression[19]. SEPTIN9 is involved in cytoskeleton organization and cell division and exhibits methylation changes associated with cancer development, particularly in colorectal cancer[20].
Several genes have emerged as potential epigenetic markers in CCA. RASSF1A is a tumor suppressor gene involved in diverse biological processes and is pivotal in tumor development. RASSF1A promoter hypermethylation has been frequently observed in CCA and other tumor types, implicating it as a potential molecular marker[13,18]. Similarly, SEPTIN9, a protein-coding gene critical for cytoskeleton organization and cell division, undergoes methylation changes that affect gene expression and contribute to abnormal cell function, fostering tumor formation and growth in pre-colorectal cancer diseases[20].
Recent studies have further underscored the diagnostic and prognostic value of specific methylation markers in various cancers, including CCA. For example, RASSF1A promoter methylation has exhibited high co-specificity in pan-cancer diagnosis and a certain degree of screening ability[21]. Additionally, the combined detection of SEPTIN9 and SHOX2 methylation was associated with tumor-node-metastasis (TNM) staging, histological grading, and lymphatic infiltration in pre-treatment tumors[22,23]. Furthermore, HOXA9 methylation dynamics predicted the therapeutic efficacy of novel ovarian cancer therapies, bridging the gap in efficacious predictive biomarkers[24].
In particular, recent advancements have indicated the potential of ctDNA methylation markers for improving diagnostic accuracy in CCA. A study by Hu et al[13] demonstrated the potential of RASSF1A promoter methylation as a biomarker for colorectal cancer, emphasizing its potential applicability in other cancers, including CCA. Liang et al[23] recently introduced a new set of DNA methylation biomarkers for identifying malignant pleural effusion, underscoring the versatility of methylation markers across different cancer types. These recent reports highlight the evolving landscape of epigenetic biomarkers in cancer diagnosis and prognosis.
In this study, we aimed to evaluate the clinical utility of combined plasma methylation of SHOX2, HOXA9, SEPTIN9, and RASSF1A, alongside CTC enumeration, in CCA diagnosis and monitoring. We used these epigenetic and cellular biomarkers to develop a non-invasive detection platform that could facilitate early CCA diagnosis, prognosis assessment, and disease monitoring, ultimately guiding personalized therapeutic strategies and improving patient outcomes.
The genomic data used in this study were obtained from the Gene Expression Omnibus database (http://gepia2.cancer-pku.cn/) and The Cancer Genome Atlas (TCGA) database (http://www.ualcan.path.uab.edu/).
The study cohort comprised 156 patients diagnosed with CCA at Hubei Cancer Hospital, China, between June 2020 and December 2022. The patients were included based on pathological evidence in accordance with the World Health Organization criteria and TNM classification. All patients were treatment-naïve at the time of enrollment. The cohort included 11 patients at stage I-II disease and 145 patients at stage III-IV disease. As a control group, 70 healthy individuals were enrolled from Wuhan's TCWM Hospital health check-up clinic. The disease controls were 28 patients with benign bile duct disease (17 with bile duct or gallbladder polyps, 11 with biliary calculus). The Ethics Committee of Hubei Cancer Hospital approved this study (approval number: LLHBCH2023YN-002).
Peripheral blood samples (10 mL per case) were collected from each patient and control by venipuncture into EDTA-coated tubes at the time of diagnosis and prior to any therapeutic intervention. Blood samples for CTC detection were also collected before and after clinical therapy. The samples were immediately processed and stored at 4 °C until further analysis.
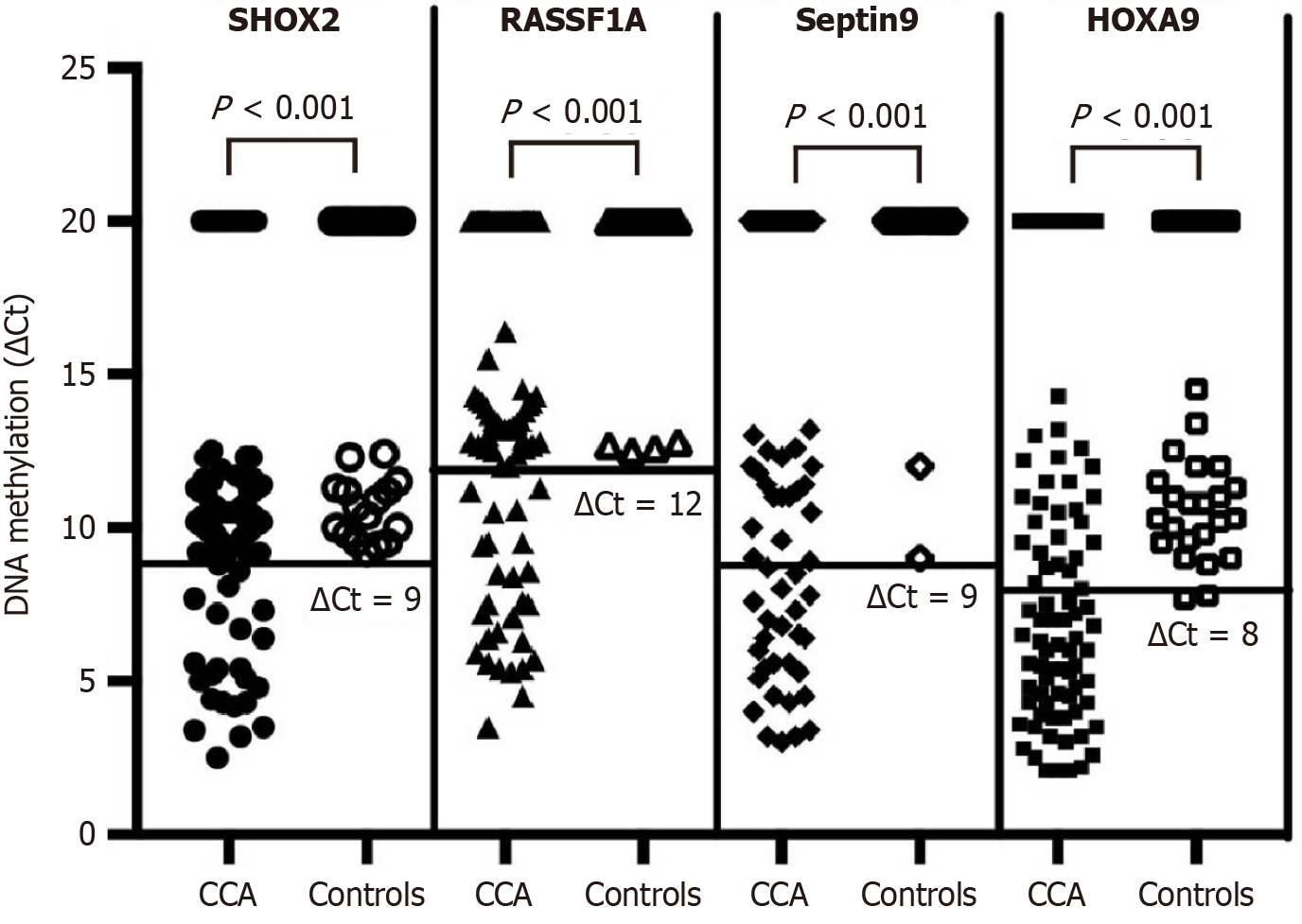
DNA was extracted from the plasma using a commercially available kit according to the manufacturer’s instructions (Tellgen Corporation, Shanghai, China). Before the methylation was detected, the DNA was treated with bisulfite to convert unmethylated cytosines into uracils while preserving the methylated cytosines. The treated DNA was referred to as sDNA (i.e., DNA after bisulfite conversion). This conversion is a crucial step in methylation detection, as it enables the specific detection of methylated DNA sequences through subsequent PCR techniques. Methylation in the genes of interest was detected by subjecting DNA to bisulfite conversion, which converts unmethylated cytosines to uracil while leaving methylated cytosines unchanged. The methylation status of the SHOX2, HOXA9, SEPTIN9, and RASSF1A genes was determined using quantitative real-time PCR with TaqMan probes specific for the CpG islands in the promoters of these genes. The methylation levels were expressed as comparative threshold cycle (ΔCt) values, where ΔCt = Ct (gene of interest) - Ct (internal control, β-actin). β-Actin was selected as the internal reference gene given its relatively stable expression in most tissues and cells, which aids in standardizing and comparing gene expression levels across different samples, and it is a widely used as an internal reference gene. A sample was deemed methylation-positive when satisfying these quantitative thresholds: Ct value for SHOX2 < 32 with DCt ≤ 9; RASSF1A < 35 with DCt ≤ 12; SEPTIN9 < 35 with DCt ≤ 9; HOXA9 < 32 with DCt ≤ 8[22]. The methylation reagents for the four genes were from LungMe Assay (Tellgen Corporation, Shanghai, China).
DNA Extraction and Purification: High-purity DNA was extracted using commercial kits (Tellgen Corporation). The manufacturer's instructions were strictly followed to ensure the quality and purity of the extracted DNA. Methylation detection quality control consists of three parts. First, whole blood is processed to separate the plasma within 2 hours after collection. The separated plasma is stored at -20 °C and tested within 1 month to ensure its quality. Second, an internal control (β-actin) is used as a whole-process quality control to monitor from extraction to bisulfite conversion to PCR detection. Third, PCR positive and negative controls are used to individually assess the PCR system and reactions. These three parts ensure the quality control of methylation detection and provide indications for identifying the causes of detection failure.
CTCs were enriched from peripheral blood samples using the CanPatrol™ CTC enrichment technique (SurExam, Guangzhou, China). Following enrichment, the CTCs were identified by RNA in situ hybridization using a panel of molecular markers. These markers included EpCAM and cytokeratins 8/18/19 as epithelial cell biomarkers, CD45 as a leukocyte biomarker, and vimentin and Twist as mesenchymal cell biomarkers. Before hybridization, probe accessibility was enhanced by permeabilizing blood cells and treating them with a protease. Hybridization was performed using capture probes targeting the selected molecular markers. Fluorescence signals were detected using an automated imaging fluorescence microscope (Zeiss, Oberkochen, Germany). Red and green fluorescent signals represented epithelial and mesenchymal marker expression, respectively, while bright white fluorescent signals identified leukocytes.
Carbohydrate antigen 19-9 (CA199) concentrations in serum were determined by employing a chemiluminescence-based immunoassay kit, supplied by Snibe Diagnostic (Shenzhen, China), adhering to the provided instructions. The normal reference range for CA199 was defined as ≤ 41 U/mL, and values exceeding this threshold were considered abnormal, indicating potential malignancy.
SPSS 25 (IBM) was utilized for statistical analyses. Normality of continuous variables was checked with Shapiro-Wilk. Depending on data distribution, we applied either non-parametric tests (Mann-Whitney U, Kruskal-Wallis H) or parametric ones (ANOVA, independent t-test) for group comparisons. Categorical variables were analyzed using the χ2 test. Spearman's correlation was used to evaluate patient variable relationships. An assessment of the diagnostic precision of single methylation markers, their various combinations, along with CA199, was conducted through the receiver operating characteristic (ROC) curve analysis. We quantified diagnostic efficacy by computing the area under the curve (AUC) of the ROC curve. Independent prognostic factors for CCA survival were identified using univariate and multivariate Cox regression analyses. Statistical significance was set at P < 0.05.
The study cohort comprised 156 patients diagnosed with CCA at Hubei Cancer Hospital between June 2020 and December 2022. Our study cohort comprised 87 male participants (mean age: 58 years, ranging from 40 to 74) and 69 female participants (mean age: 57 years, ranging from 40 to 73). The disease-specific features of this cohort are detailed in Table 1.
| Clinicopathological data | n | SHOX2 positive | SEPT9 positive | HOXA9 positive | RASSF1A positive | Comprehensive methylation Positive | CTC counts/ |
| Tumor location | |||||||
| iCCA | 33 | 24.24% | 24.24% | 42.42% | 27.27% | 57.58% | 10 |
| pCCA | 83 | 21.69% | 27.71% | 56.63% | 27.71% | 66.27% | 12 |
| dCCA | 40 | 27.5% | 32.5% | 50% | 35% | 67.5% | 14 |
| iCCA vs dCCA | 0.795 | 0.604 | 0.638 | 0.614 | 0.467 | 0.342 | |
| Gender | |||||||
| Male | 87 | 22.99% | 34.48% | 47.13% | 32.18% | 68.97% | 13 |
| Female | 69 | 24.64% | 20.29% | 57.97% | 26.09% | 59.42% | 10 |
| Male vs female | 0.851 | 0.702 | 0.199 | 0.481 | 0.24 | 0.760 | |
| Age at diagnosis | |||||||
| ≤ 60 years | 91 | 21.98% | 26.37% | 41.76% | 26.37% | 59.34% | 11 |
| > 60 years | 65 | 26.15% | 30.77% | 66.15% | 33.85% | 72.31% | 14 |
| Median age (years) | 57 | ||||||
| Mean age (years) | 59 | ||||||
| ≤ 60 years vs > 60 years | 0.571 | 0.591 | 0.195 | 0.374 | 0.126 | 0.276 | |
| Tumor stage | |||||||
| I-II | 11 | 9.09% | 9.09% | 18.18% | 9.09% | 36.36% | 3 |
| III-IV | 145 | 24.83% | 29.66% | 54.48% | 31.03% | 66.21% | 13 |
| I-II vs III-IV | 0.237 | 0.144 | 0.020 | 0.176 | 0.046 | 0.001 | |
| Lymphatic invasion | |||||||
| L0 | 105 | 25.71% | 27.62% | 48.57% | 32.38% | 62.86% | 4 |
| L1 | 51 | 19.61% | 29.41% | 58.82% | 23.53% | 68.63% | 16 |
| L0 vs L1 | 0.400 | 0.815 | 0.331 | 0.255 | 0.479 | 0.001 | |
| Follow-up | |||||||
| Follow up available | 43 | ||||||
| Median follow-up months | 14 | ||||||
| Mean follow-up months | 17 | ||||||
| Range (months) | 0-58 | ||||||
| Deceased | 23 | ||||||
| Censored | 20 | ||||||
| Healthy donors | |||||||
| Male | 42 | 3/42 | 2 | 2 | 3 | 3 | 1 |
| Female | 28 | 1/28 | 1 | 1 | 1 | 1 | 0 |
| ≤ 60 years | 51 | 1 | 1 | 0 | 1 | 1 | 0 |
| > 60 years | 19 | 2 | 2 | 2 | 2 | 2 | 1 |
| Benign bile duct patients | |||||||
| Male | 21 | 2/21 | 1 | 1 | 1 | 1 | 0 |
| Female | 7 | 0 | 0 | 0 | 1 | 1 | 0 |
| ≤ 60 years | 23 | 2 | 0 | 1 | 2 | 2 | 0 |
| > 60 years | 5 | 1 | 1 | 1 | 1 | 1 | 0 |
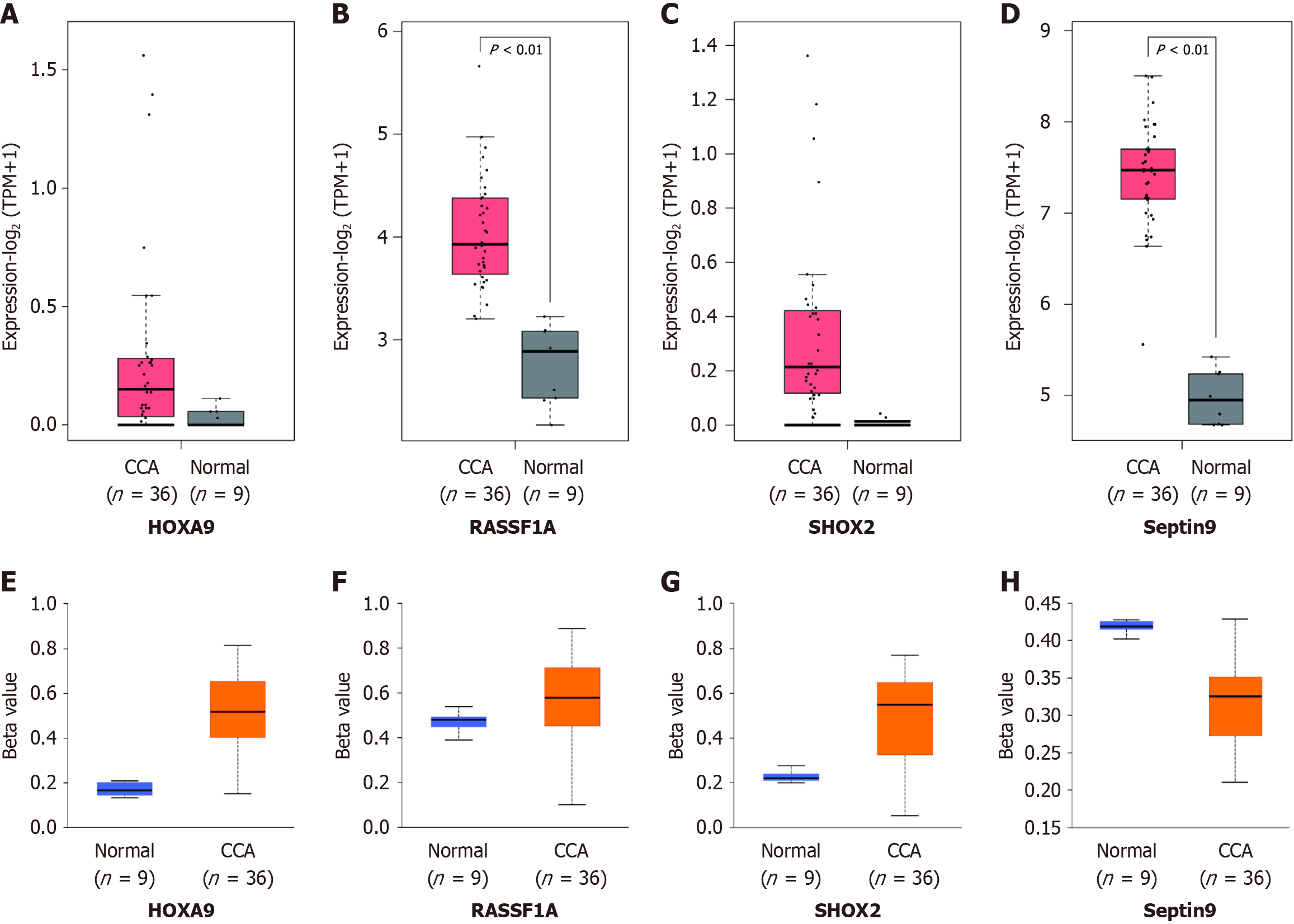
TCGA data analysis revealed that the CCA tissue had significantly higher HOXA9, RASSF1A, SEPTIN9, and SHOX2 expression than normal tissue. Beta values, which span from 0 indicating no methylation to 1 signifying complete methylation, were utilized to represent the DNA methylation status of HOXA9, RASSF1A, SEPTIN9, and SHOX2 gene promoters in both normal tissues (n = 9) and CCA tissues (n = 36). Hypermethylation was signified by beta values between 0.7 and 0.5, whereas hypomethylation was indicated by values from 0.3 to 0.25 (Figure 1).
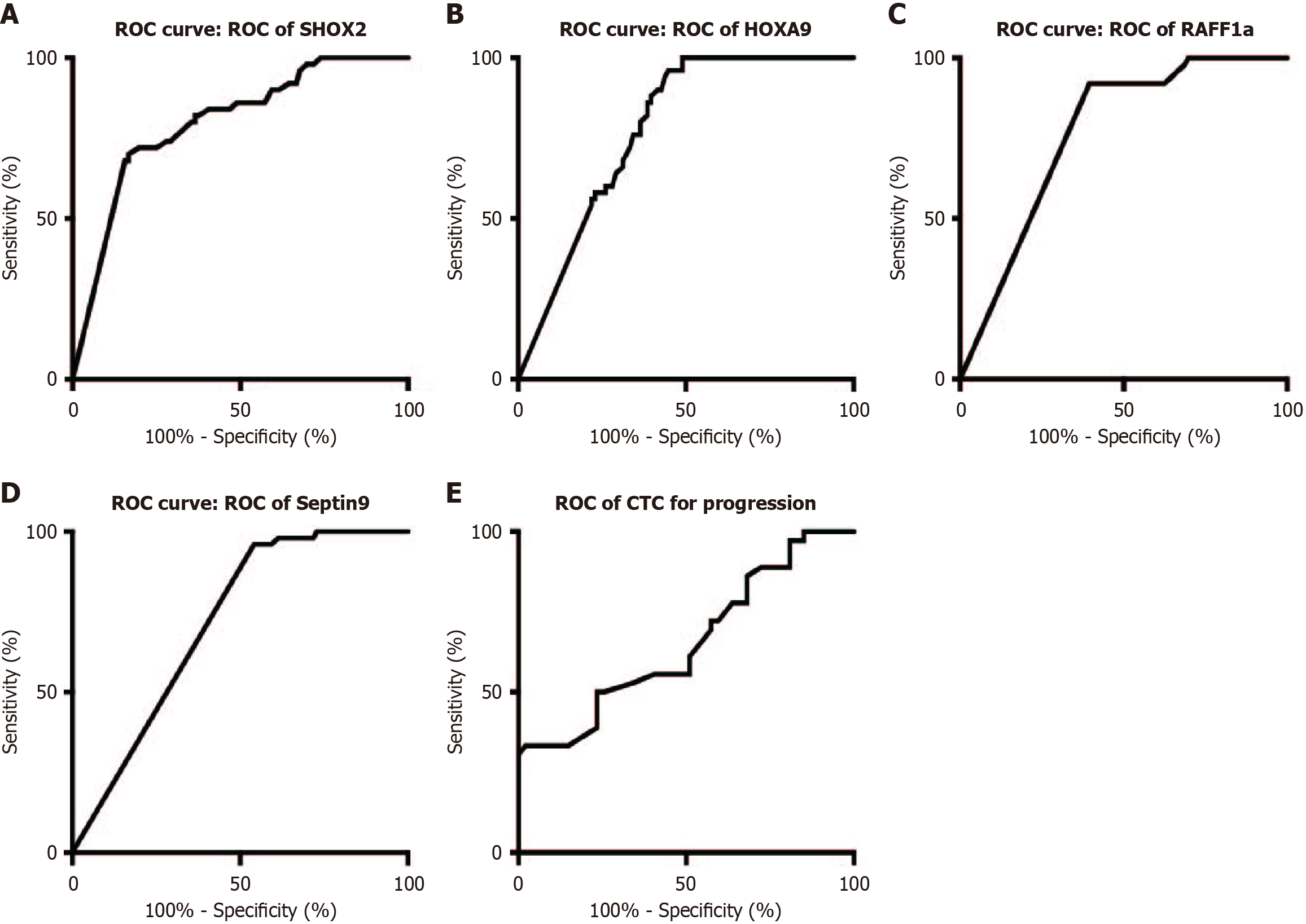
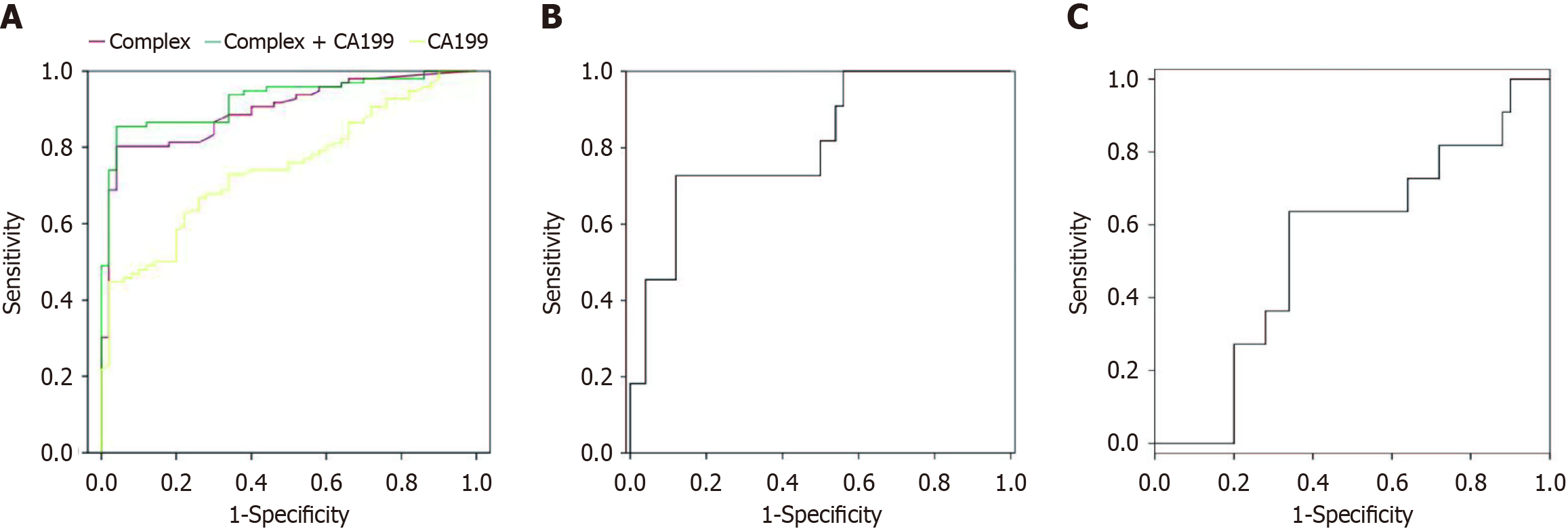
Figure 1 depicts the DNA methylation of the SHOX2, RASSF1A, SEPTIN9, and HOXA9 genes. Utilizing the optimal cutoff for gene-specific methylation, the sensitivity and specificity for CCA detection ranged from 25% to 64.1% and from 93.88% to 95.92%, respectively, as illustrated in Figure 2. The ROC curves for DNA methylation-based CCA diagnosis are shown in Figure 3. For the diagnosis of CCA, the methylation markers SHOX2, HOXA9, SEPTIN9, and RASSF1A exhibited sensitivities of 23.72%, 51.92%, 28.21%, and 29.49%, and specificities of 93.88%, 95.92%, 95.92%, and 93.88%, respectively. The sensitivity and specificity of the four methylations combined for diagnosing CCA were 64.74% and 93.88%, respectively. The AUC for the complex evaluation to diagnose CCA was 0.828 (range: 0.764-0.891) (Figure 4A). In the context of CCA diagnosis, the AUC values for methylation markers SHOX2, HOXA9, SEPTIN9, and RASSF1A were found to be 0.799 (range: 0.724-0.874), 0.777 (range: 0.705-0.850), 0.713 (range: 0.632-0.795), and 0.765 (range: 0.689-0.841), correspondingly. The critical values for SHOX2, HOXA9, SEPTIN9, and RASSF1A methylation for diagnosing CCA were DCtSHOX2 ≤ 9; DCtRASSF1A ≤ 12; DCtSEPTIN9 ≤ 9; and DCtHOXA9 ≤ 8, respectively, which was the same as that in a previous study[23].
CA199 demonstrated a sensitivity of 51.28% and a specificity of 87.76% for detecting CCA, which was significantly lower than the sensitivity of the combined methylation evaluation. The AUC for CA199 was 0.746 ± 0.04, whereas the AUC for the combined methylation evaluation was 0.828 ± 0.032, indicating superior diagnostic performance.
Combining CA199 with the methylation panel improved the sensitivity and specificity to 68.59% and 85.76%, respectively (Table 2).
| Biomarker or combination | Positive test, CCA No. | Negative test, CCA No. | Sensitivity (%) | Early-stage CCA (%) | Specificity (%) | PPV (%) | ||
| SEPTIN9 | 44 | 3 | 112 | 95 | 28.21 | 9.09 | 96.94 | 54.72 |
| HOXA9 | 81 | 3 | 75 | 95 | 51.92 | 18.18 | 96.94 | 69.29 |
| SHOX2 | 37 | 2 | 119 | 96 | 23.72 | 9.09 | 97.96 | 52.36 |
| RASSF1A | 46 | 2 | 110 | 96 | 29.49 | 9.09 | 97.96 | 55.91 |
| CA199 | 80 | 12 | 47 | 86 | 51.28 | 9.09 | 87.76 | 65.35 |
| SEPTIN9 + HOXA9 | 81 | 3 | 75 | 95 | 51.92 | 18.18 | 96.94 | 69.29 |
| SEP + HOXA9 + SHO | 88 | 3 | 68 | 95 | 56.41 | 27.27 | 96.94 | 72.05 |
| SEP + HOXA9 + SHO + RASS | 101 | 6 | 55 | 92 | 64.74 | 36.36 | 93.88 | 75.98 |
| Sep + HOXA9 + SHO + RASS + CA199 | 107 | 12 | 49 | 86 | 68.59 | 45.45 | 87.76 | 75.98 |
Among 156 CCA patients, 11 were diagnosed with stage I-II, while 145 presented with stage III-IV disease, as detailed in Table 1. For the diagnosis of stage I-II and III-IV CCA, the complex evaluation exhibited sensitivities of 36.36% and 75.17%, and specificities of 93.83% and 93.58%, correspondingly. The AUC of the complex evaluation and CA199 to diagnose early CCA was 0.682 ± 0.014 (Figure 4B) and 0.542 ± 0.092 (Figure 4C). Table 2 presents the sensitivity (68.59%) and specificity (85.76%) of the complex evaluation of CA199 combined with methylation.
The ROC curves for CTCs had an AUC of 0.653 (95%CI: 0.533-0.774) for diagnosing tumor progression, with a cut-off value of 10 CTCs/5 mL (Figure 3E). Among the 156 patients, 51 underwent surgery and chemotherapy. Another 105 patients underwent chemotherapy, of which 67 also received immunotherapy as first-line treatment. The remaining 45 patients were treated with targeted therapy. Postoperative CTC counts were monitored in 43 patients, revealing significantly decreased counts after surgical resection. Notably, the CTC counts of 10 out of 22 patients surpassed the threshold 3 months before imaging-confirmed recurrence or metastatic lesions.
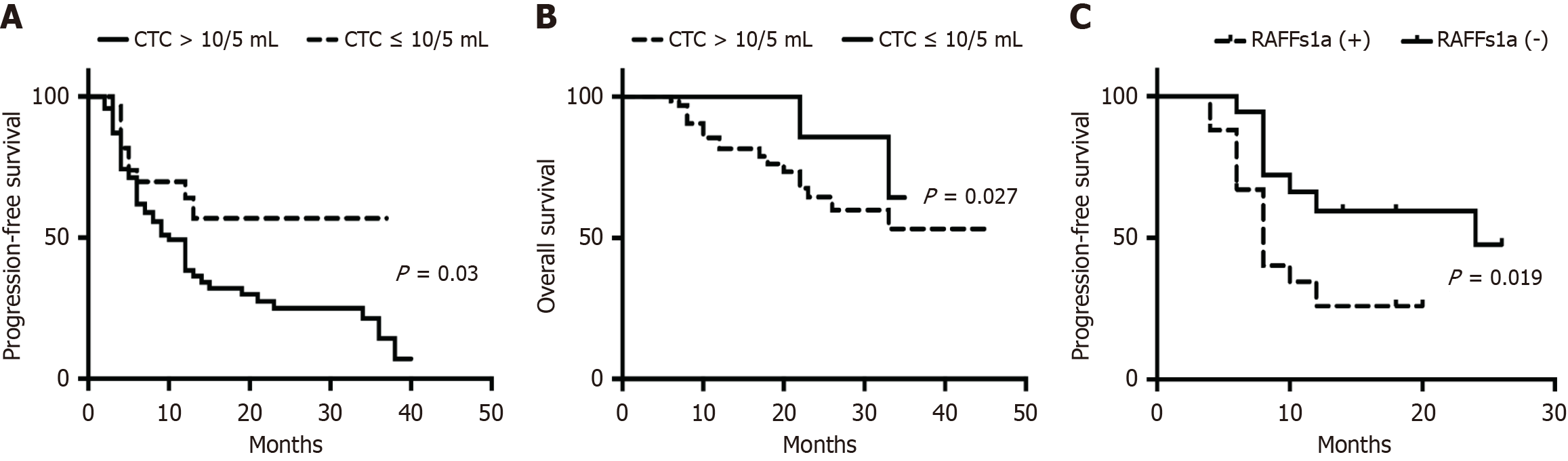
Figure 5 presents the relapse-free survival (RFS) and OS of patients with CCA with varying CTC counts at admission. Patients with > 10 CTCs/5 mL had faster disease progression and a poorer outcome.
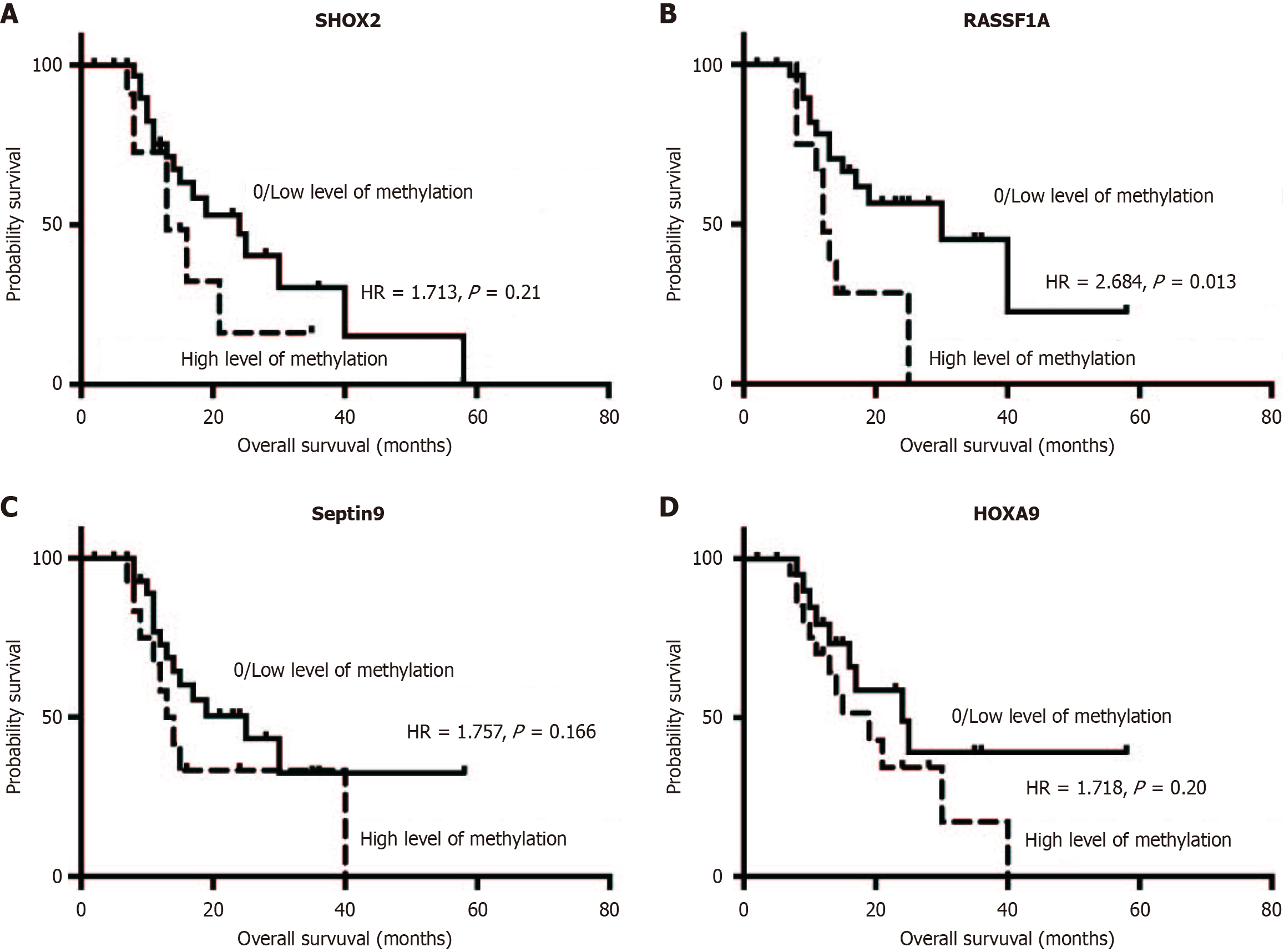
The utility of plasma sDNA methylations and CTCs in monitoring CCA progression was assessed using postoperative evaluation of these biomarkers in 22 patients 1 month following resection. Four patients exhibited a notable decrease in plasma DNA methylation levels. Kaplan-Meier survival analysis demonstrated that patients with high methylation levels (defined as DCtSHOX2 ≤ 9; DCtRASSF1A ≤ 12; DCtSEPTIN9 ≤ 9; DCtHOXA9 ≤ 8) had shorter OS than those with low methylation levels (Figure 6). Notably, patients with advanced CCA exhibiting RASSF1A methylation had a significantly worse RFS (P = 0.002), as illustrated in Figure 5C. Nevertheless, there was no notable link found between CCA metastasis and the methylation status of SEPTIN9, HOXA9, or SHOX2.
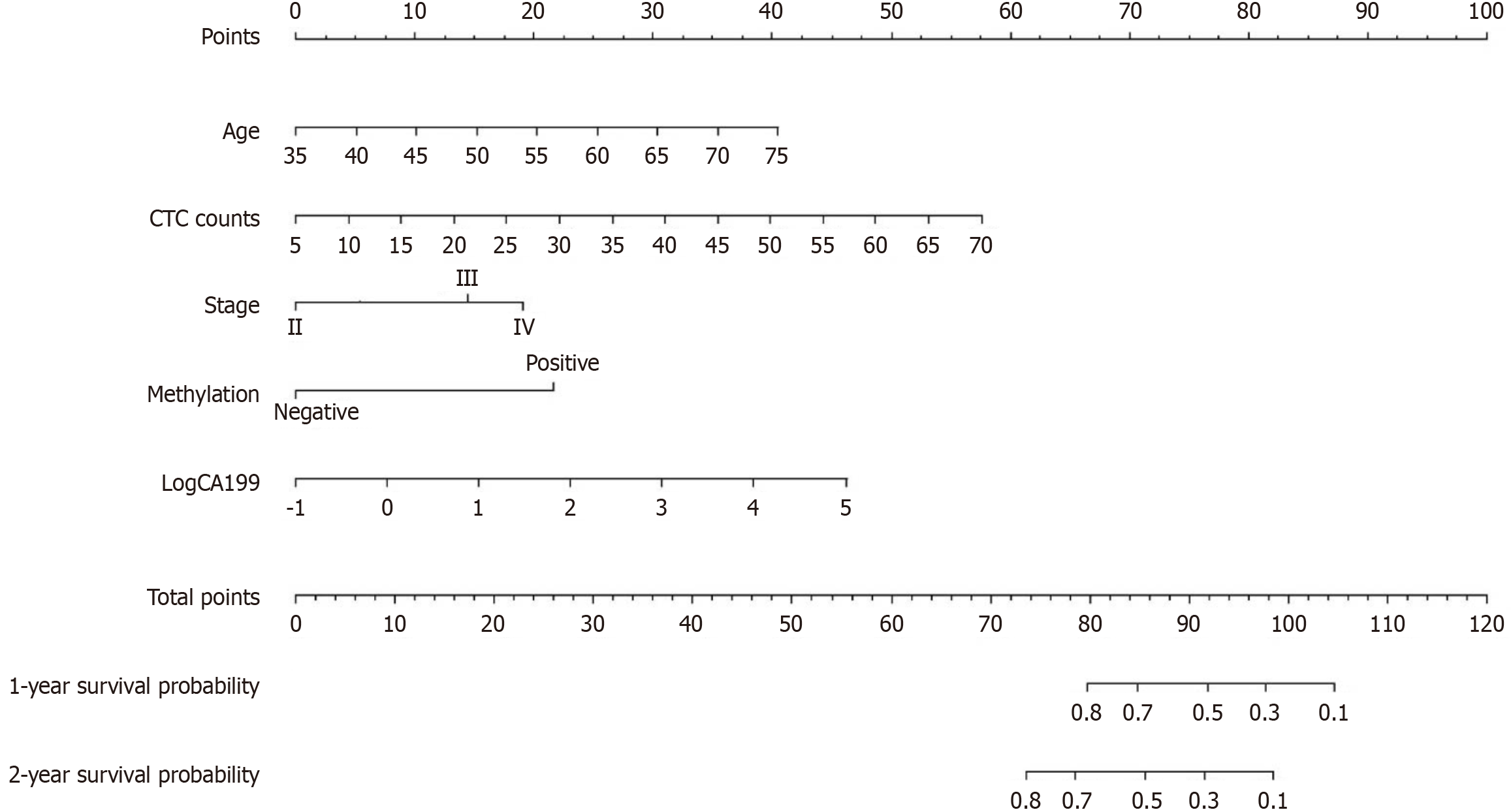
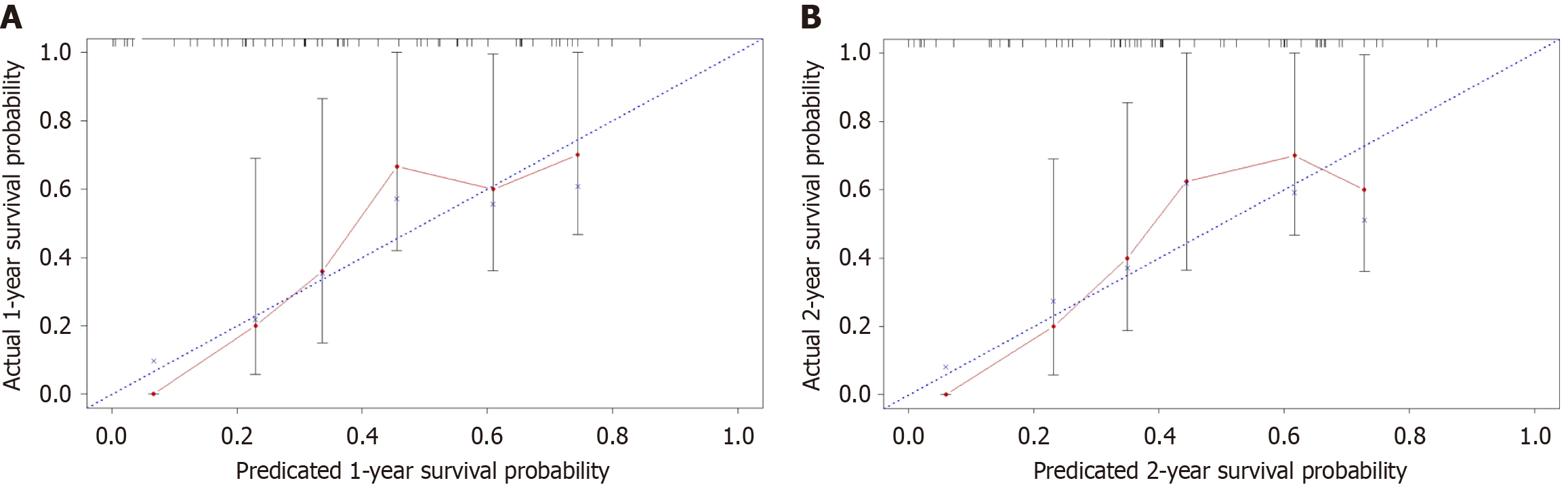
A prognostic nomogram for predicting the OS of patients with CCA was constructed based on multivariate analysis. The nomogram incorporated age, tumor stage, CTC count, methylation status, and CA199 levels. Each predictor in the nomogram was assigned a score, and the sum of these scores indicated the probability of the 1- and 2-year OS (Figure 7). The C-index of this nomogram was 0.705 (95%CI: 0.605-0.805), demonstrating its discriminative ability. Internal cross-validation revealed close approximation between the calibration plots and observed estimates for the 1-year and 2-year OS (Figure 8).
Univariate and multivariate Cox regression analyses identified tumor stage, lymphatic invasion, CTC count > 10/5 mL, and RASSF1A methylation as independent prognostic factors for CCA (Table 3). In detail, the risk estimates, expressed as hazard ratios, for tumor stage, lymphatic invasion, CA199 levels, CTC counts, and RASSF1A methylation status were 3.13 (with a 95%CI: 1.06-9.242, P value 0.039), 2.902 (95%CI: 1.069-7.874, P = 0.037), 2.41 (95%CI: 1.079-5.394, P = 0.028), 3.542 (95%CI: 1.354-9.266, P = 0.027), and 2.684 (95%CI: 1.0-7.557, P = 0.013). These results highlight the importance of combining multiple biomarkers, including plasma DNA methylations and CTCs, for comprehensive CCA management.
| Variables | Number of patients | Univariate | Multivariate | |||
| HR (95%CI) | P value | HR (95%CI) | P value | |||
| Tumor location | (iCCA vs dCCA) | 33 | 1.246 (0.495-3.138) | 0.624 | ||
| Gender | (male vs female) | 43 | 1.536 (0.673-3.861) | 0.284 | ||
| Age at diagnosis | (≤ 60 vs > 60 years) | 43 | 1.527 (0.673-3.467) | 0.273 | ||
| Tumor stage | (I-II vs III-IV) | 43 | 3.13 (1.06-9.242) | 0.039 | 2.871 (1.056-7.653) | 0.03 |
| Lymphatic invasion | (V0 vs V1) | 38 | 2.902 (1.069-7.874) | 0.037 | 3.425 (1.081-8.523) | 0.035 |
| CA199 | ≤ 41 U/mL vs > 41 U/mL) | 43 | 2.41 (1.079-5.384) | 0.028 | ||
| SHOX2 methylation | (SHOX2- vs SHOX2+) | 43 | 1.713 (0.623-4.713) | 0.210 | ||
| SEPT9 methylation | (SEPT9- vs SEPT9+) | 43 | 1.757 (0.698-4.422) | 0.166 | ||
| HOXA9 methylation | (HOXA9- vs HOXA9+) | 43 | 1.718 (0.743-3.975) | 0.200 | ||
| RASSF1A methylation | (RASSF1A- vs RASSF1A+) | 43 | 2.684 (1.0-7.557) | 0.013 | ||
| CTC count > 10/5 mL | (> 10/5 mL vs ≤ 10/5 mL) | 43 | 3.542 (1.354-9.266) | 0.027 | 3.24 (1.465-7.562) | 0.021 |
The significance of ctDNA methylation in clinical oncology has been increasingly recognized due to its potential to detect early-stage cancers with high sensitivity and specificity[21]. The present study demonstrates the value of plasma SHOX2, HOXA9, SEPTIN9, and RASSF1A methylation in CCA diagnosis and monitoring. These markers, together with CTCs, present a promising non-invasive approach to improve early detection and management of CCA.
The high sensitivity (64.74%) and specificity (93.88%) of the combined methylation panel for detecting CCA underscore its potential as a diagnostic tool. Importantly, a detailed comparison with previous studies that used similar markers or techniques for CCA diagnosis revealed that our study achieved comparable or even superior performance. For example, Branchi et al[17] reported 55% sensitivity and 90% specificity using a combination of SHOX2 and SEPT9 methylation for diagnosing biliary tract cancer . With the addition of HOXA9 and RASSF1A methylation, our study reports higher sensitivity (64.74%) while maintaining a high specificity (93.88%). Moreover, the AUC of our complex evaluation (0.828) was higher than that reported by Liang et al[23] (0.801) for a panel of novel methylated DNA markers in pleural effusion. These comparisons highlight the advantages of our multi-marker panel in identifying early disease.
In addition to its diagnostic utility, plasma methylation potentially informs prognosis and treatment monitoring. Our results indicate that RASSF1A methylation is significantly associated with the prognosis of patients with CCA, with hypermethylated patients exhibiting poorer OS. This result aligns with previous studies demonstrating the prognostic significance of RASSF1A methylation in various cancers[13]. Furthermore, our study is unique compared to other studies that explored the prognostic value of individual methylation markers in CCA, given its comprehensive approach, where multiple markers and clinical factors were incorporated into a prognostic nomogram. For example, Peng et al[18] found that RASSF1A hypermethylation was associated with increased risk of hepatocellular carcinoma, but did not construct a prognostic model. Contrastingly, our nomogram, which includes age, tumor stage, CTC count, methylation status, and CA199 levels, achieved a C-index of 0.705, demonstrating its discriminative ability in predicting OS.
The clinical implications of this study extend beyond diagnosis. Due to its brief half-life, ctDNA enables immediate assessment of therapy effectiveness and tumor load, aiding precise tumor classification and early recurrence identification. The decrease in CTC counts post-surgery and their subsequent increase prior to imaging recurrence highlights the potential of CTCs as a complementary tool for monitoring disease progression. In clinical practice, integrating these biomarkers into routine diagnostics or monitoring for patients with CCA could significantly improve patient outcomes. For example, in the early diagnosis of CCA, the combined use of our methylation panel with imaging techniques, such as computed tomography or magnetic resonance imaging, could increase the detection rate of early-stage tumors. Similarly, in the prognosis assessment and monitoring of patients with advanced CCA, the combined use of our prognostic nomogram with biopsy results could provide a more comprehensive evaluation of the patient's condition and guide personalized treatment strategies.
It is also crucial to acknowledge the limitations of our statistical models, particularly regarding the potential for overfitting, which could affect the robustness and real-world applicability of our results. We mitigated this risk by using internal cross-validation and assessed the calibration of our nomogram, and identified a close approximation between the calibration plots and observed estimates for the 1-year and 2-year OS (Figure 8). Additionally, using bootstrap methods could further validate the stability and reliability of our model. Despite these efforts, future studies with larger patient cohorts and longer follow-up durations are necessary to fully evaluate the generalizability of our results. The inclusion of additional biomarkers and clinical factors, such as imaging findings and biopsy results, in our model may also enhance its diagnostic and prognostic accuracy.
The non-invasive nature of liquid biopsy techniques offers several advantages over traditional methods. First, it enhances patient compliance by eliminating the need for invasive procedures, such as biopsy. Second, the high sensitivity and specificity of the methylation panel allow earlier disease detection, which is crucial for improving outcomes in CCA, where late-stage diagnosis is common. Third, the ability to monitor disease progression and treatment response non-invasively could inform timely adjustments to treatment plans, ultimately improving patient survival.
Our study also clarifies the potential applications of these markers in specific patient populations. For example, plasma methylation markers may complement existing detection methods for high-risk groups, such as patients with primary sclerosing cholangitis, who are at an increased risk of developing CCA. Furthermore, the ability to monitor disease progression and treatment response non-invasively could inform personalized treatment strategies, particularly in patients with unresectable disease.
Despite the promising results, the present study was subject to limitations. The relatively small sample size and limited follow-up duration may have affected the robustness of our conclusions. Therefore, our results should be validated through studies with larger patient cohorts and longer follow-up periods. Additionally, a more detailed exploration of the relationship between methylation status and OS, and the potential for overfitting in the nomogram construction, would enhance the clinical applicability of our results.
In conclusion, ctDNA contains abundant cancer cell genetic information, among which methylation variation partly reflects the presence and malignancy of cancer in situ. Here, we constructed a nomogram for predicting the OS of patients with CCA (model: Age + stage + CTC + methylation + CA199), attaining a C-index score of 0.705 with a 95%CI: 0.605-0.805. The model could assess the clinical risk factors to predict the OS of patients with CCA. Clinicians could utilize these data to determine the most effective and tailored treatment approach for CCA patients. The non-invasive procedure and high-quality cancer information render ctDNA methylation and CTCs promising and clinically valuable liquid biopsy projects.
ctDNA contains abundant cancer cell genetic information, among which methylation variation partly reflects the presence and malignancy of cancer in situ. Here, we constructed a nomogram for predicting the OS of patients with CCA (model: Age + stage + CTC + methylation + CA199) with a C-index score of 0.705 with a 95%CI: 0.605-0.805. The model has the capability to evaluate clinical risk factors for forecasting the OS in CCA patients. These data could aid clinicians in selecting an optimal and customized management strategy for treating patients with CCA. The high sensitivity (64.74%) and specificity (93.88%) of the combined methylation panel for detecting CCA underscored its potential as a diagnostic tool. The methylation panel exhibited superior diagnostic performance compared to the tumor marker CA199 (AUC: 0.828 vs 0.746). This result highlighted the advantages of ctDNA methylation-based assays in identifying early disease, as demonstrated by the ability of the panel to detect early-stage CCA with 36.36% sensitivity and 93.83% specificity. Furthermore, RASSF1A plasma methylation and CTCs were valuable predictors for assessing CCA prognosis and recurrence. Despite the promising results presented herein, it is important to recognize that the present study has several limitations, notably a comparatively modest sample size and a restricted period of follow-up. Our results should be validated and refined through studies that involve larger patient cohorts and longer follow-up durations. Additionally, exploring the combination of more epigenetic markers and integrating them with other clinical parameters, such as imaging findings and biopsy results, may enhance the diagnostic and prognostic accuracy for CCA. Furthermore, the development of more sensitive and specific detection methods for ctDNA methylation and CTC enumeration is also crucial for improving the clinical utility of these biomarkers. Investigating the potential mechanisms underlying the methylation alterations of these genes in CCA may also provide deeper insights into its pathogenesis and progression. Ultimately, translating these results into clinical practice, such as integrating plasma methylation and CTC analysis into routine diagnostic algorithms for CCA, could significantly improve patient outcomes through earlier diagnosis, personalized treatment strategies, and more effective disease monitoring.
| 1. | Deng M, Ran P, Chen L, Wang Y, Yu Z, Cai K, Feng J, Qin Z, Yin Y, Tan S, Liu Y, Xu C, Shi G, Ji Y, Zhao JY, Zhou J, Fan J, Hou Y, Ding C. Proteogenomic characterization of cholangiocarcinoma. Hepatology. 2023;77:411-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 2. | Vithayathil M, Khan SA. Current epidemiology of cholangiocarcinoma in Western countries. J Hepatol. 2022;77:1690-1698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 3. | Moris D, Palta M, Kim C, Allen PJ, Morse MA, Lidsky ME. Advances in the treatment of intrahepatic cholangiocarcinoma: An overview of the current and future therapeutic landscape for clinicians. CA Cancer J Clin. 2023;73:198-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 255] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 4. | Parikh ND, Pillai A. Recent Advances in Hepatocellular Carcinoma Treatment. Clin Gastroenterol Hepatol. 2021;19:2020-2024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 5. | Zhang ZJ, Huang YP, Liu ZT, Wang YX, Zhou H, Hou KX, Tang JW, Xiong L, Wen Y, Huang SF. Identification of immune related gene signature for predicting prognosis of cholangiocarcinoma patients. Front Immunol. 2023;14:1028404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Merters J, Lamarca A. Integrating cytotoxic, targeted and immune therapies for cholangiocarcinoma. J Hepatol. 2023;78:652-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777-786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 411] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 8. | Heery R, Schaefer MH. DNA methylation variation along the cancer epigenome and the identification of novel epigenetic driver events. Nucleic Acids Res. 2021;49:12692-12705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Tu J, Chen S, Wu S, Wu T, Fan R, Kuang Z. Tumor DNA Methylation Profiles Enable Diagnosis, Prognosis Prediction, and Screening for Cervical Cancer. Int J Gen Med. 2022;15:5809-5821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 10. | Li Y, Melnikov AA, Levenson V, Guerra E, Simeone P, Alberti S, Deng Y. A seven-gene CpG-island methylation panel predicts breast cancer progression. BMC Cancer. 2015;15:417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Namba S, Sato K, Kojima S, Ueno T, Yamamoto Y, Tanaka Y, Inoue S, Nagae G, Iinuma H, Hazama S, Ishihara S, Aburatani H, Mano H, Kawazu M. Differential regulation of CpG island methylation within divergent and unidirectional promoters in colorectal cancer. Cancer Sci. 2019;110:1096-1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Dementeva N, Kokova D, Mayboroda OA. Current Methods of the Circulating Tumor Cells (CTC) Analysis: A Brief Overview. Curr Pharm Des. 2017;23:4726-4728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Hu F, Chen L, Bi MY, Zheng L, He JX, Huang YZ, Zhang Y, Zhang XL, Guo Q, Luo Y, Tang WR, Sheng MM. Potential of RASSF1A promoter methylation as a biomarker for colorectal cancer: Meta-analysis and TCGA analysis. Pathol Res Pract. 2020;216:153009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Sun M, Liu J, Hu H, Guo P, Shan Z, Yang H, Wang J, Xiao W, Zhou X. A novel panel of stool-based DNA biomarkers for early screening of colorectal neoplasms in a Chinese population. J Cancer Res Clin Oncol. 2019;145:2423-2432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Han YD, Oh TJ, Chung TH, Jang HW, Kim YN, An S, Kim NK. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenetics. 2019;11:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 16. | Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, Levin TR, Lieberman D, Robertson DJ. Colorectal Cancer Screening: Recommendations for Physicians and Patients From the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2017;153:307-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 513] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 17. | Branchi V, Schaefer P, Semaan A, Kania A, Lingohr P, Kalff JC, Schäfer N, Kristiansen G, Dietrich D, Matthaei H. Promoter hypermethylation of SHOX2 and SEPT9 is a potential biomarker for minimally invasive diagnosis in adenocarcinomas of the biliary tract. Clin Epigenetics. 2016;8:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Peng JL, Wu JZ, Li GJ, Wu JL, Xi YM, Li XQ, Wang L. Association of RASSF1A hypermethylation with risk of HBV/HCV-induced hepatocellular carcinoma: A meta-analysis. Pathol Res Pract. 2020;216:153099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Li XF, Zhang HB, Huo Y. High HOXA9 gene expression predicts response to chemotherapy and prognosis of high-grade serous ovarian cancer patients. J Int Med Res. 2022;50:3000605221135864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, Zhang S. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19:450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 21. | de Vos L, Gevensleben H, Schröck A, Franzen A, Kristiansen G, Bootz F, Dietrich D. Comparison of quantification algorithms for circulating cell-free DNA methylation biomarkers in blood plasma from cancer patients. Clin Epigenetics. 2017;9:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Lin Q, Fang X, Chen H, Weng W, Liu B, Kong J. Dual-modality loop-mediated isothermal amplification for pretreatment-free detection of Septin9 methylated DNA in colorectal cancer. Mikrochim Acta. 2021;188:307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Liang C, Liu N, Zhang Q, Deng M, Ma J, Lu J, Yin Y, Wang J, Miao Y, She B, Li Q, Hou G. A detection panel of novel methylated DNA markers for malignant pleural effusion. Front Oncol. 2022;12:967079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 24. | Faaborg L, Andersen RF, Waldstrøm M, Henriksen JR, Adimi P, Jakobsen A, Steffensen KD. Prognostic Impact of Circulating Methylated Homeobox A9 DNA in Patients Undergoing Treatment for Recurrent Ovarian Cancer. Cancers (Basel). 2022;14:1766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |