Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103679
Revised: January 16, 2025
Accepted: February 11, 2025
Published online: April 15, 2025
Processing time: 119 Days and 2.6 Hours
Early screening methods for gastric cancer (GC) are lacking; therefore, the disease often progresses to an advanced stage when patients first start to exhibit typical symptoms. Endoscopy and pathological biopsy remain the primary diagnostic approaches, but they are invasive and not yet widely applicable for early popu
To identify effective plasma miRNA biomarkers and investigate the clinical value of combining multiple miRNAs for early detection of GC.
Plasma samples from multiple centres were collected. Differentially expressed genes among healthy controls, early-stage GC patients, and advanced-stage GC patients were identified through small RNA sequencing (sRNA-seq) and validated via real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR). A Wilcoxon signed-rank test was used to investigate the differences in miRNAs. Sequencing datasets of GC serum samples were retrieved from the Gene Expression Omnibus (GEO), ArrayExpress, and The Cancer Genome Atlas databases, and a multilayer perceptron-artificial neural network (MLP-ANN) model was constructed for the key risk miRNAs. The pROC package was used to assess the discriminatory efficacy of the model.
Plasma samples of 107 normal, 71 early GC and 97 advanced GC patients were obtained from three centres, and serum samples of 8443 normal and 1583 GC patients were obtained from the GEO database. The sRNA-seq and RT-qPCR experiments revealed that miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p were significantly increased in early GC patients compared with healthy controls and in advanced GC patients compared with early GC patients (P < 0.05). An MLP-ANN model was constructed for the six key miRNAs. The area under the curve (AUC) within the training cohort was 0.983 [95% confidence interval (CI): 0.980–0.986]. In the two validation cohorts, the AUCs were 0.995 (95%CI: 0.987 to nearly 1.000) and 0.979 (95%CI: 0.972–0.986), respectively.
Potential miRNA biomarkers, including miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p, were identified. A GC classifier based on these miRNAs was developed, benefiting early detection and population screening.
Core Tip: This was an in-house small RNA sequencing analysis of five healthy, five early gastric cancer (GC) and five advanced GC plasma samples, and the top 15 differentially expressed genes were verified in 275 plasma samples via real-time quantitative reverse transcription polymerase chain reaction. Six key miRNAs, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p, were ultimately identified. A multilayer perceptron-artificial neural network classifier incorporating these six miRNAs was innovatively constructed based on 10 026 serum samples via machine learning techniques and is anticipated to become a novel biomarker for GC.
- Citation: Ma FC, Zhang GL, Chi BT, Tang YL, Peng W, Liu AQ, Chen G, Gao JB, Wei DM, Ge LY. Blood-based machine learning classifiers for early diagnosis of gastric cancer via multiple miRNAs. World J Gastrointest Oncol 2025; 17(4): 103679
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103679.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103679
Gastric cancer (GC), which originates from the gastric mucosa, is one of the most aggressive neoplasms of the digestive system and ranks fifth worldwide in both incidence and mortality[1]. The hazard factors for GC include infection with Helicobacter pylori, older age, a diet high in salt, unhealthy eating habits, gene mutations and psychological factors; all of which can lead to a high incidence rate. The absence of early screening techniques often contributes to GC progressing to an advanced stage when patients exhibit the typical symptoms, which include indigestion, anorexia, early satiety, abdominal pain and weight loss[2]. Currently, endoscopy and pathological biopsy remain the primary diagnostic approaches because of their ability to offer high diagnostic accuracy. However, these methods are invasive and not wi
miRNAs are a category of noncoding RNAs with single-stranded structures involved in regulating gene expression in different types of tumours and play roles as oncogenes or tumour suppressors via posttranscriptional regulatory mechanisms[4]. Studies have shown that miRNAs represent a category of highly conserved RNA molecules that remain stable in plasma. The dysregulation of these genes is closely associated with tumorigenesis and tumour progression[5]. Therefore, plasma-derived miRNAs can accurately reflect cellular changes during tumour progression, making them promising candidate biomarkers for early diagnosis. For example, a diagnostic kit based on seven miRNAs has already been integrated into guidelines as a haematological biomolecular marker to assist in the early diagnosis of hepatocellular carcinoma (86.1% sensitivity, 76.8% specificity)[6,7]. Research on GC has revealed that aberrant expression of either single or multiple miRNAs is related to clinicopathological features and patient prognosis. For example, miR-10 is overexpressed in GC and is positively correlated with tumour dimensions, the extent of invasion and lymph node metastasis[8]. Upregulation of miR-142-5p, miR-20b, miR-214, miR-150 and miR-375 or downregulation of miR-125-5p, miR-451, let-7g and miR-433 are linked to unfavourable outcomes in patients with GC[9]. Additionally, the integration of seven miRNAs (miR-126, miR-10b, miR-30a-5p, miR-21, miR-223, let-7a and miR-338) can predict either overall survival or recurrence-free survival in GC patients[10]. Despite extensive research on miRNAs as biomarkers for GC, no established miRNA diagnostic kit for early GC detection is currently available in clinical practice. Therefore, investigating the molecular characteristics and efficient miRNA markers is essential for GC, potentially improving the early diagnosis and screening opportunities for GC.
In the past 10 years, artificial intelligence (AI) has been gradually developed and practiced in various clinical tasks, including detection, diagnosis, classification and prognosis of early tumours. AI is an algorithm programme that identifies the relationships between input and output variables through training. These programmes include machine learning and deep learning techniques[11]. To simulate the structure and function of the biological nervous system, an artificial neural network (ANN) is a mathematical approach used for deep learning. Common ANN algorithms include multilayer perceptron (MLP), recurrent neural networks, and convolutional neural networks[12]. Compared with traditional mathematical models, ANNs demonstrate superior nonlinear fitting ability for complex relationships between independent variables and outcomes[13], hence improving their applicability in medical practice. In this study, the ANNs approach was utilized to investigate the predictive significance of combining multiple miRNAs for GC.
The objective of the present study was to identify differentially expressed miRNAs through the collection of plasma samples from healthy controls, early GC patients and advanced GC patients and to analyse the results via small RNA sequencing (sRNA-seq) and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR). A classifier utilizing an MLP-ANN approach was constructed for the critical differentially expressed miRNAs and validated with external datasets. This model investigated the clinical significance of a serum multi-miRNA classifier for early GC.
Participants opportunistically screened for untreated early GC were selected from three centres between January 2021 and December 2023: Guangxi Medical University Cancer Hospital, Guilin People’s Hospital and Youjiang Medical University Affiliated Hospital. The inclusion criteria were: (1) No other conflicting diseases or tumour history in the health examination population; (2) Patients received no treatment before sample collection; and (3) Early and advanced GCs were definitively diagnosed on the basis of histological standards. The research received approval from the Ethics Committees of Guilin People’s Hospital (No. 2020-102KY), Guangxi Medical University Cancer Hospital (No. KY2020148) and Youjiang Medical University Affiliated Hospital (No. YYFY-LL-2024-005). All participants provided signed informed consent. The plasma samples were promptly frozen in liquid nitrogen and maintained at -80°C.
The miRNA microarray or RNA-seq datasets of GC were searched and collected from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/), ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress) and Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) databases. The inclusion criteria were: (1) GC; (2) Blood or plasma samples; and (3) Sample sizes in both the case and control groups no fewer than five. Samples from participants that had undergone any form of treatment were excluded.
The main reagents and consumables used for the experiments were: Hieff® qPCR SYBR green master mix (low Rox Pius) and MolPure® serum/plasma miRNA kit from Yeasen (https://www.yeasen.com/, Shanghai, China); miRNA first strand complementary DNA (cDNA) synthesis kit (Tail Reaction) purchased from Monad (http://www.monadbiotech.
Experimental specimens and preparation: Total RNA was extracted from the plasma samples of five healthy individuals, five early GC patients and five advanced GC patients via TriQuick Reagent. The assessment of RNA purity and concentration was conducted via agarose gel electrophoresis in conjunction with an ultramicro nucleic acid detector. The miRNA first strand cDNA synthesis kit was used for reverse transcription to synthesise the first strand, followed by PCR amplification and size selection. sRNA fragments were screened via polyacrylamide gel electrophoresis. The sRNA library was constructed by excising the gel and recovering the small RNA fragments. The PCR products were purified via the AMPure XP system, after which the library quality was evaluated. The TruSeq PE cluster kit v4-cBot-HS developed by Illumina was utilized to cluster the indexed samples on the cBot Cluster Generation system. Subsequent to cluster generation, the sequences of the sRNA library were obtained via the Illumina sequencing platform.
Data processing: Perl software was used to eliminate reads that included adapters, poly-N sequences and sequences of suboptimal quality from the unprocessed fastp data, thereby generating clean reads. The Bowtie tool was used to align the processed reads with several databases, including GtRNAdb (https://gtrnadb.ucsc.edu/), Silva (https://www.arb-si
The transcription status of the candidate differential miRNAs was verified through RT-qPCR. After the plasma samples were lysed and centrifuged, total RNA was isolated via the RNA adsorption column method. The miRNA RT enzyme mix was used for reverse transcription to synthesise the cDNA template, which was subsequently stored at -20°C. The PCR mixture was prepared following the instructions of the Hieff® qPCR SYBR green PCR kit, and miRNA quantitative analysis was conducted via the RT-PCR system. The U6 gene, which is stably expressed in tissue, was used as the internal control for calculating the differences in miRNA expression. The CT value represents the number of cycles when the fluorescence signal reaches the pre-established threshold, transitioning from background to exponential growth during PCR amplification. The relative quantification results obtained through qPCR were presented via a 2–ΔΔCT value, which was applied to examine the transcriptionally expressed status of 15 distinctively expressed miRNAs in the plasma of healthy individuals, early GC patients and advanced GC patients. The calculation formulas are as follows: ΔCT = CTtarget gene - CTinternal control gene, - ΔΔCT = - (ΔCTexperimental group - ΔCTcontrol group).
The highly expressed differentially expressed genes identified through sRNA-seq and RT-qPCR were considered key genes. The expression status of key miRNAs was trained with R version 4.3.2 via the RSNNS, MASS and NeuralNetTools packages, and the MLP-ANN GC classifier model was constructed and validated with external datasets. The pROC package was utilized to conduct a receiver operating characteristic (ROC) analysis on the MLP-ANN model within both the training and validation sets and assess its performance by computing the area under the curve (AUC), sensitivity and specificity. An AUC value ranging from 0.5 to 0.7 denoted low discriminatory capacity, whereas a value between 0.7 and 0.8 reflected moderate discriminatory capacity. AUC between 0.8 and 0.9 signified high discriminatory capacity, and AUC exceeding 0.9 denoted very high discriminatory capacity.
Statistical analyses were conducted via statistical product and service solutions software version 22.0 and R software version 4.3.2. Differential analysis of clinicopathological features, including age, sex, American Joint Committee on Cancer (AJCC) stage and tumor node metastasis (TNM) classification stage, was performed via the χ2 test. The differences in the expression levels of miRNAs among healthy controls, early-stage GC patients and advanced-stage GC patients were assessed via the Wilcoxon signed-rank test. The pROC package was used to construct ROC curves to assess the ability of the MLP-ANN classifier to distinguish the efficacy of the miRNAs in GC. Two-tailed P < 0.05 was considered significant.
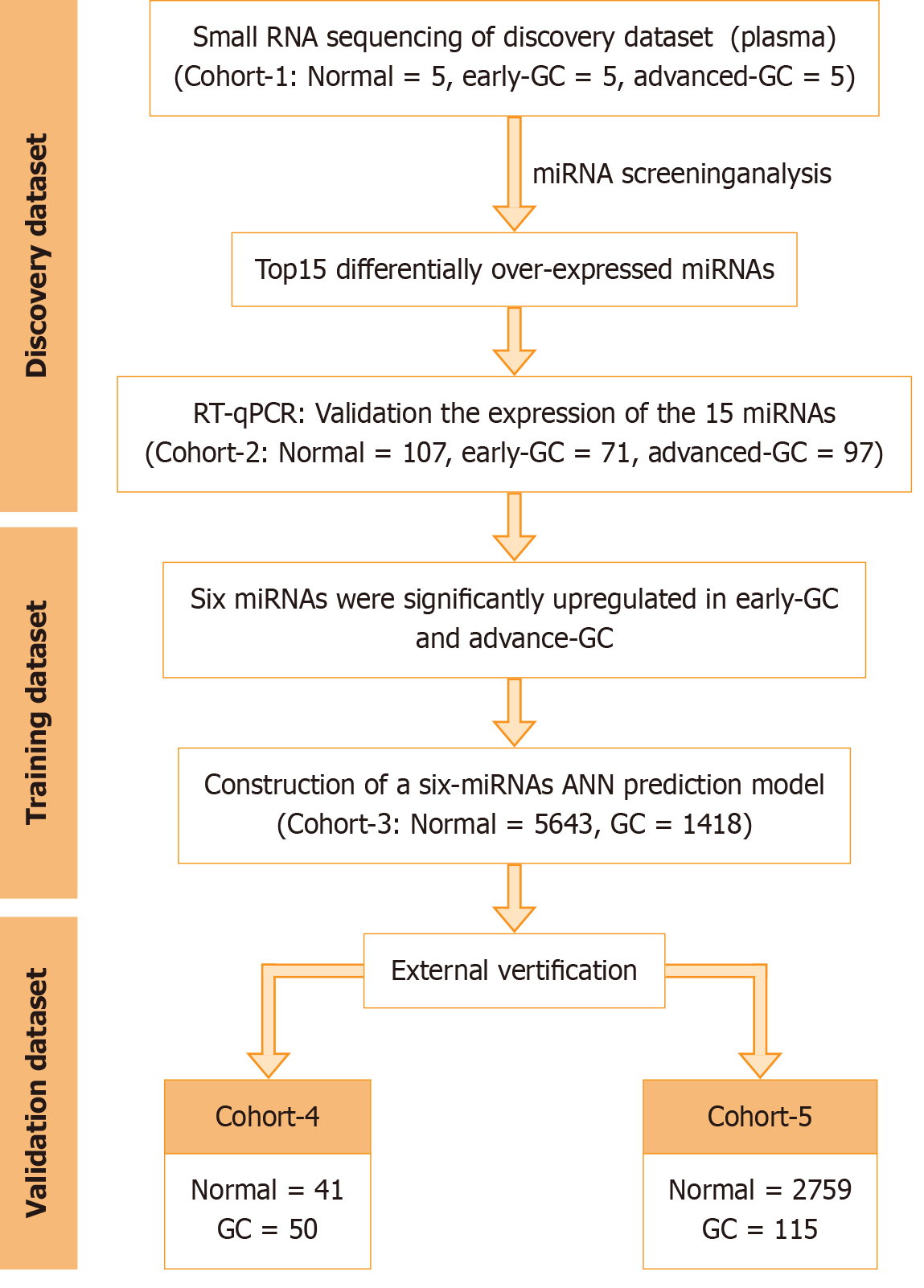
This study included discovery, training and validation phases (Figure 1).
In total, 107 healthy individuals, 71 early GC patients and 97 advanced GC patients participated; all of whom met the established inclusion standards (Table 1). The control group comprised 37 males and 70 females, whereas the early GC group comprised 38 males and 33 females. In contrast, the advanced GC group comprised 67 males and 30 females. Compared with that in the control group, the sex distribution in the early GC and advanced GC groups was significantly different (P < 0.05). In the early GC group, the number of individuals aged ≥ 60 years was substantially greater than that in the control group. Nevertheless, no substantial difference in age was noted between the advanced GC and control groups. It was pointed out that the differences in age and gender composition would not impact the expression levels of miRNAs in plasma (Supplementary Figures 1–5). Based on the AJCC staging standard, the healthy control group had no tumour status, the patients diagnosed with early GC were defined as those with T1N0M0, and the patients diagnosed with advanced GC were defined as those with T2-4, N0-3 and M0-1 stages.
| Characteristics | Control (n = 107) | Early GC (n = 71) | Advanced GC (n = 97) | 1P value | 2P value |
| Gender | |||||
| Male | 37 (34.6) | 38 (53.5) | 67 (69.1) | 0.012 | < 0.001 |
| Female | 70 (65.4) | 33 (46.5) | 30 (30.9) | ||
| Age (year) | |||||
| < 60 | 101 (94.4) | 52 (73.2) | 86 (88.7) | < 0.001 | 0.139 |
| ≥ 60 | 6 (5.6) | 19 (26.8) | 11 (11.3) | ||
| AJCC stage | |||||
| I | 0 | 71 (100) | 37 (38.1) | ||
| II | 0 | 0 | 5 (5.2) | ||
| III | 0 | 0 | 17 (17.5) | ||
| IV | 0 | 0 | 38 (39.2) | ||
| T | |||||
| 1 | 0 | 71 (100) | 0 | ||
| 2 | 0 | 0 | 42 (43.3) | ||
| 3 | 0 | 0 | 6 (6.2) | ||
| 4 | 0 | 0 | 49 (50.5) | ||
| N | |||||
| 0 | 0 | 71 (100) | 42 (43.3) | ||
| 1 | 0 | 0 | 12 (12.4) | ||
| 2 | 0 | 0 | 22 (22.7) | ||
| 3 | 0 | 0 | 21 (21.6) | ||
| M | |||||
| 0 | 0 | 71 (100) | 66 (68.0) | ||
| 1 | 0 | 0 | 31 (32.0) |
Three serum datasets were screened from public databases (Table 2), including GSE211692 (1418 GC samples and 5643 control samples), GSE112264 (50 GC samples and 41 control samples) and GSE106817 (115 GC samples and 2759 control samples).
| Dataset | Platform | Year | Country | GC (n) | Control (n) |
| Cohort-3: GSE211692 | GPL21263 | 2022 | Japan | 1418 | 5643 |
| Cohort-4: GSE112264 | GPL21263 | 2019 | Japan | 50 | 41 |
| Cohort-5: GSE106817 | GPL21263 | 2018 | Japan | 115 | 2759 |
The standards for screening were established as a fold change > 2 accompanied by adjusted P < 0.05. A total of 234 differential miRNAs were detected in the early GC group compared with the healthy control group, including 131 upregulated and 103 downregulated genes. In addition, 259 differentially expressed miRNAs were identified in the advanced GC compared with the healthy control group, including 124 upregulated and 135 downregulated genes. The upregulated miRNAs in both the early and advanced GC groups intersected, and subsequently, the 15 miRNAs with the most sig
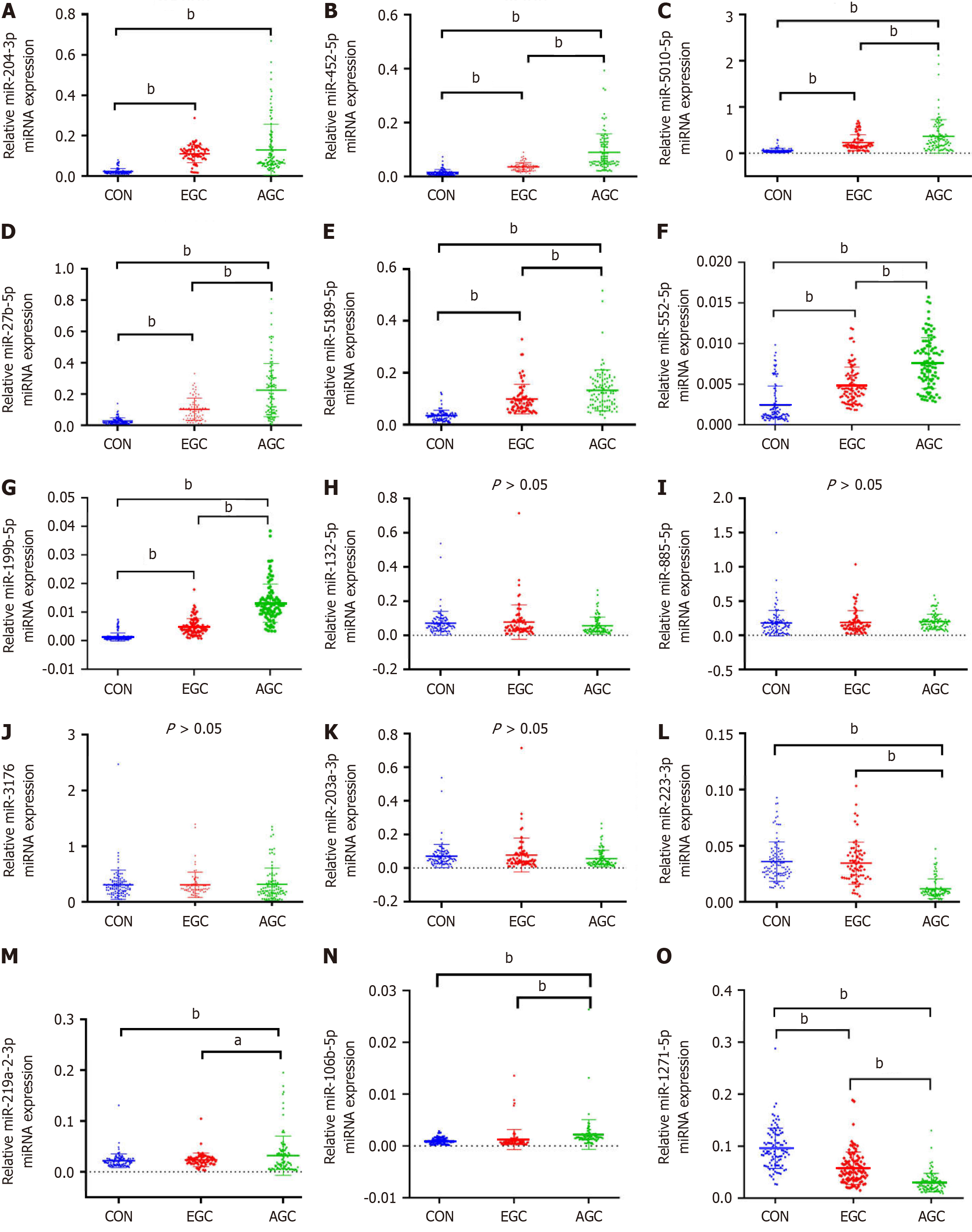
The details of all primer sequences utilized in these RT-qPCR experiments are presented in Supplementary Table 1. In contrast to those in the control group, miR-204-3p, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p were notably elevated in both the early and advanced GC groups (P < 0.01, Figure 2A–G). The levels of miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p were substantially increased in advanced GC patients compared with early GC patients (P < 0.01). Therefore, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p may serve as risk-associated miRNAs in GC evolution and could serve as plasma biomar
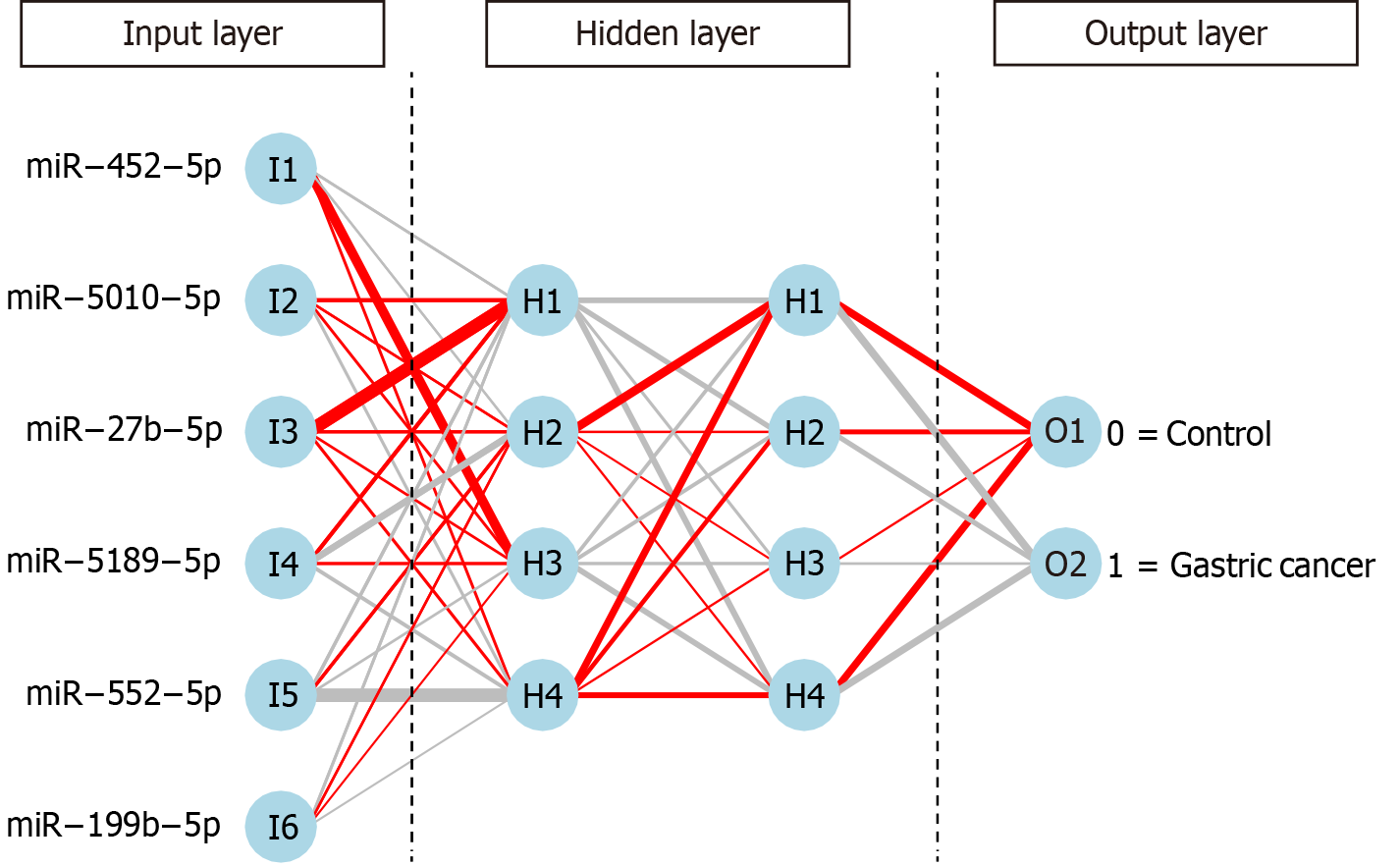
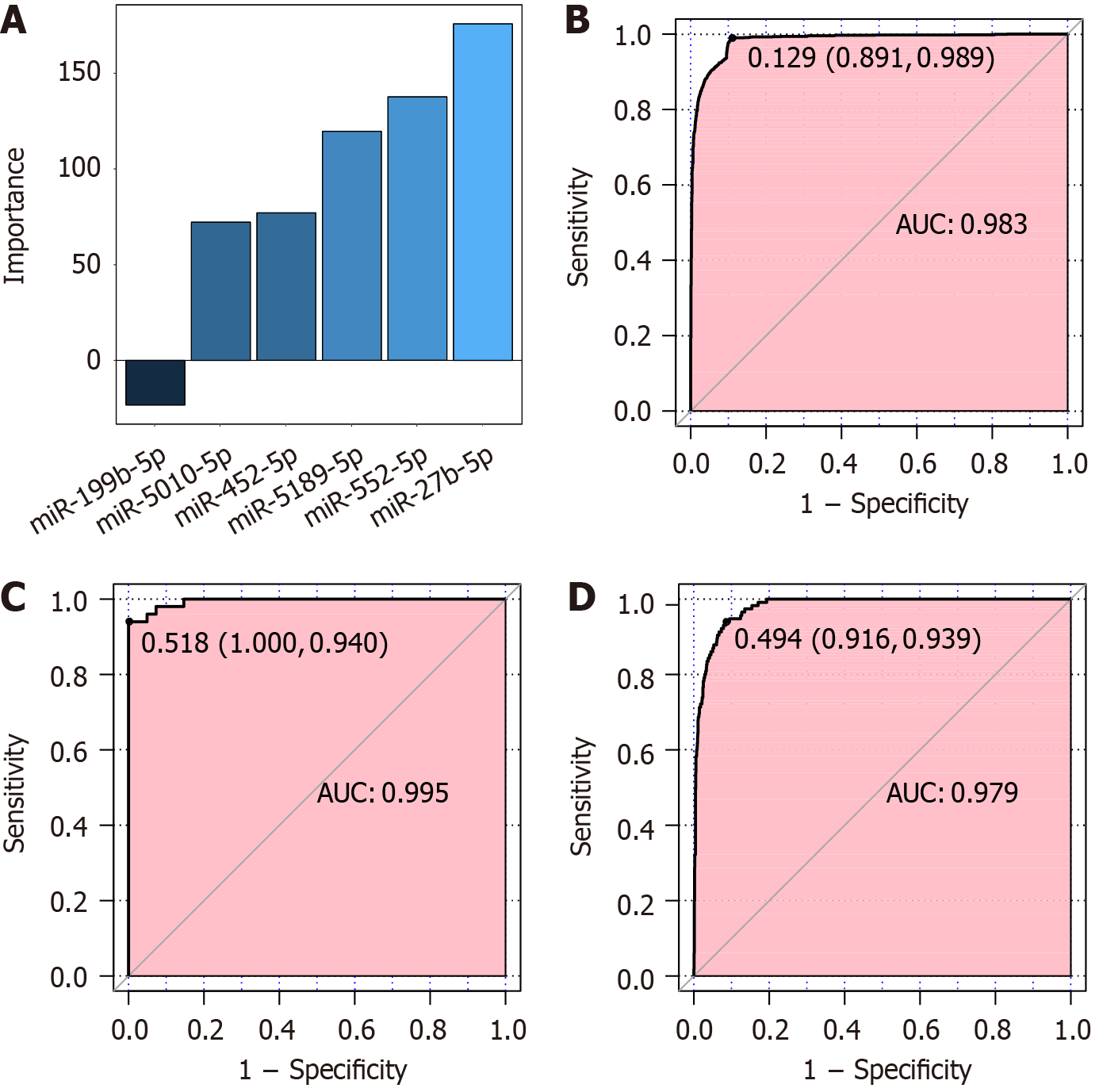
Six critical miRNAs, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p, served as the input layer, and an MLP-ANN model was constructed (Figures 3 and 4). For the MLP-ANN model, the input layer consisted of six miRNAs as factors, the middle two layers were hidden layers, and the output layer indicated the outcomes (control or GC). The independent variable-miR-27b-5p-had the greatest influence on the model (Figure 4A). In training set cohort-3, sensitivity for predicting GC was 0.989 [95% confidence interval (CI): 0.976–0.994], specificity was 0.891 (95%CI: 0.775–0.900), and AUC was 0.983 (95%CI: 0.980–0.986) (Figure 4B). In validation set cohort-4, sensitivity for predicting GC was 0.940 (95%CI: 0.860 to nearly 1.000), with a specificity approaching 1.000 (95%CI: 0.878 to nearly 1.000), along with an AUC of 0.995 (95%CI: 0.987 to nearly 1.000) (Figure 4C). In validation set cohort-5, sensitivity for predicting GC was 0.939 (95%CI: 0.878–0.974), specificity was 0.916 (95%CI: 0.861–0.946), and AUC was 0.979 (95%CI: 0.972–0.986) (Figure 4D).
Early GC often develops insidiously and lacks typical clinical symptoms, resulting in many patients receiving a diagnosis at an advanced stage. Studies have indicated that > 90% of patients with early GC survive for ≥ 5 years[14], whereas < 30% of patients with advanced GC achieve the same survival rate[15,16]. Therefore, conducting large-scale screening for early GC is important because it can identify early GC lesions in a timely manner, enhance the diagnostic accuracy of early GC, lower the tumour stage at diagnosis, and reduce the economic burden on patients and improve their survival rate and quality of life. Currently, the screening methods for GC include endoscopy, Helicobacter pylori detection, serological tumour marker detection and genetic detection (e.g., circulating malignant cells or circulating tumour cell DNA)[17]. Endoscopy is the most direct and accurate screening method; however, patient acceptance is low because it is invasive and expensive[9]. Recently, because miRNAs have stable structures, extensive studies have investigated their potential as new plasma or serum biological markers for tumours[18]. In the current study, plasma samples were collected from healthy controls, early-stage GC patients and advanced-stage GC patients across multiple centres. Key miRNAs were identified through sRNA-seq and RT-qPCR experiments, and an MLP-ANN predictive model was constructed to explore its clinical value in distinguishing GC.
The present study performed sRNA-seq on plasma samples from five healthy persons, five early GC patients and five advanced GC patients and carried out RT-qPCR validation on plasma samples from 107 healthy individuals, 71 early GC patients and 97 advanced GC patients. Six miRNAs, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p, which may be involved in the molecular process underlying the advancement and evolution of GC, were identified. It was previously reported that the level of miR-27b-5p was elevated in GC tissues and MKN74 cells, indicating that miR-27b-5p plays a pivotal role in GC progression[19,20]. Exosomal miR-552-5p may cause damage to natural killer cells through its interaction with the programmed cell death-1/programmed cell death ligand 1 signalling pathway, which may contribute to immune resistance in GC[21]. miR-199b-5p exerts carcinogenic effects by targeting the HHIP gene and may represent a viable therapeutic target[22]. Because these six miRNAs showed markedly elevated levels in both early and advanced GC, among which miR-27b-5p, miR-552-5p and miR-199b-5p have been proposed to have oncogenic effects, we identified these six miRNAs as key risk miRNAs and potential plasma biological markers for the early diagnosis of GC.
Studies have shown that, compared with a single miRNA, combining multiple miRNAs has greater predictability and stability for predicting event outcomes[23,24]. Machine learning has the capacity to investigate and address diverse types of data because of its flexibility and scalability in comparison with traditional statistical methods[25,26], assisting in identifying previously unknown relationships between risky variables and diseases. Hence, machine learning has been widely studied for disease diagnosis, risk stratification and survival prediction. Therefore, in the present study, an MLP-ANN model was constructed to investigate the clinical significance of combining six critical miRNAs for GC differentiation. The AUC for the model within the training group was 0.983 (sensitivity: 0.989; specificity: 0.891). Additionally, the AUCs within the validation groups were 0.995 (sensitivity: 0.940, specificity: nearly 1.000) and 0.979 (sensitivity: 0.939, specificity: 0.916), respectively, suggesting that this six-miRNA MLP-ANN classifier possesses discriminatory ability and may become a tool for the early population screening of patients with GC. Currently, biomarkers based on the combination of multiple miRNAs have been revealed for GC. For example, the combined detection of a five-miRNA (miR-18a, miR-93, miR-146b, miR-181b and miR-335) biomarker achieved an AUC of 0.90 in GC patients. In addition, by reducing the number of miRNAs, a three-miRNA (miR-18a, miR-181b and miR-335) biomarker panel had an AUC of 0.86 in distinguishing patients with GC[24]. A logistic regression model utilizing six miRNAs (miR-10b-5p, miR-132-3p, miR-185-5p, miR-195-5p, miR-20a-3p and miR-296-5p) in serum achieved AUCs of 0.764 within the training dataset and 0.702 within the verification dataset[27]. Nevertheless, the application of combined miRNA biomarker detection in GC remains underdeveloped, and its discriminatory ability is still limited. The present research innovatively constructed a six-miRNA classifier utilizing a machine learning approach, proposing it as an innovative biomarker that has significant potential for the detection and population screening of GC.
In summary, the present research conducted sRNA-seq on 15 plasma samples, and the differential genes acquired were verified in 275 plasma samples via RT-qPCR. Six key miRNAs including miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p were ultimately identified. An MLP-ANN model incorporating these six miRNAs was innovatively constructed based on 10 026 serum samples via a machine learning method. The AUC for the training cohort was 0.983, whereas the AUCs were 0.995 and 0.979 for the two validation cohorts. The model has been shown to be an innovative and efficient biomarker for GC. However, several limitations exist. For example, the exploration of the six-miRNA model as an early diagnostic marker was restricted because of insufficient advanced GC datasets. Additionally, the accuracy of this six-miRNA classifier needs to be validated in further diverse datasets and public health centres to assess its feasibility for practical application.
In the present research, six potential plasma miRNA biomarkers for GC, miR-452-5p, miR-5010-5p, miR-27b-5p, miR-5189-5p, miR-552-5p and miR-199b-5p, were identified through sRNA-seq and RT-qPCR on a large number of serum samples. In addition, a GC classifier composed of these six miRNAs was constructed via a machine learning method, providing new insights for the early diagnosis and population screening of GC.
The authors thank the Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 7889] [Article Influence: 7889.0] [Reference Citation Analysis (2)] |
| 2. | Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 2827] [Article Influence: 565.4] [Reference Citation Analysis (5)] |
| 3. | de Mello RA, Amaral GA, Neves NM, Lippo EG, Parini F, Xu S, Tolia M, Charalampakis N, Tadokoro H, Castelo-Branco P, Zhu J. Current and potential biomarkers in gastric cancer: a critical review of the literature. Future Oncol. 2021;17:3383-3396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, Xie H, Peng X, Yin W, Tao Y, Wang X. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. 2020;16:2628-2647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 383] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 5. | Pan YJ, Wan J, Wang CB. MiR-326: Promising Biomarker for Cancer. Cancer Manag Res. 2019;11:10411-10418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 6. | Zhou J, Sun H, Wang Z, Cong W, Zeng M, Zhou W, Bie P, Liu L, Wen T, Kuang M, Han G, Yan Z, Wang M, Liu R, Lu L, Ren Z, Zeng Z, Liang P, Liang C, Chen M, Yan F, Wang W, Hou J, Ji Y, Yun J, Bai X, Cai D, Chen W, Chen Y, Cheng W, Cheng S, Dai C, Guo W, Guo Y, Hua B, Huang X, Jia W, Li Q, Li T, Li X, Li Y, Li Y, Liang J, Ling C, Liu T, Liu X, Lu S, Lv G, Mao Y, Meng Z, Peng T, Ren W, Shi H, Shi G, Shi M, Song T, Tao K, Wang J, Wang K, Wang L, Wang W, Wang X, Wang Z, Xiang B, Xing B, Xu J, Yang J, Yang J, Yang Y, Yang Y, Ye S, Yin Z, Zeng Y, Zhang B, Zhang B, Zhang L, Zhang S, Zhang T, Zhang Y, Zhao M, Zhao Y, Zheng H, Zhou L, Zhu J, Zhu K, Liu R, Shi Y, Xiao Y, Zhang L, Yang C, Wu Z, Dai Z, Chen M, Cai J, Wang W, Cai X, Li Q, Shen F, Qin S, Teng G, Dong J, Fan J. Guidelines for the Diagnosis and Treatment of Primary Liver Cancer (2022 Edition). Liver Cancer. 2023;12:405-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 188] [Reference Citation Analysis (0)] |
| 7. | Xie DY, Zhu K, Ren ZG, Zhou J, Fan J, Gao Q. A review of 2022 Chinese clinical guidelines on the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr. 2023;12:216-228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 89] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 8. | Wang YY, Ye ZY, Zhao ZS, Li L, Wang YX, Tao HQ, Wang HJ, He XJ. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum Pathol. 2013;44:1278-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Jelski W, Mroczko B. Molecular and Circulating Biomarkers of Gastric Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 10. | Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 268] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 11. | Chen ZH, Lin L, Wu CF, Li CF, Xu RH, Sun Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun (Lond). 2021;41:1100-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 12. | Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 392] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 13. | Tu JV. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol. 1996;49:1225-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1086] [Cited by in RCA: 657] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 14. | Norwood DA, Montalvan-Sanchez E, Dominguez RL, Morgan DR. Gastric Cancer: Emerging Trends in Prevention, Diagnosis, and Treatment. Gastroenterol Clin North Am. 2022;51:501-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 15. | Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, Ogunbiyi OJ, Azevedo E Silva G, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP; CONCORD Working Group. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2711] [Cited by in RCA: 3404] [Article Influence: 486.3] [Reference Citation Analysis (1)] |
| 16. | Early Diagnosis and Treatment Group; Chinese Society of Oncology; Chinese Medical Association. [Chinese expert consensus on early diagnosis and treatment of gastric cancer (2023 edition)]. Zhonghua Xiaohuawaike Zazhi. 2024;23:23-26. [DOI] [Full Text] |
| 17. | Matsuoka T, Yashiro M. Novel biomarkers for early detection of gastric cancer. World J Gastroenterol. 2023;29:2515-2533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 35] [Reference Citation Analysis (1)] |
| 18. | Ho PTB, Clark IM, Le LTT. MicroRNA-Based Diagnosis and Therapy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 337] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 19. | Kim YJ, Jeong S, Jung WY, Choi JW, Hwang KC, Kim SW, Lee YC. miRNAs as potential biomarkers for the progression of gastric cancer inhibit CREBZF and regulate migration of gastric adenocarcinoma cells. Int J Med Sci. 2020;17:693-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Kim YJ, Hwang KC, Kim SW, Lee YC. Potential miRNA-target interactions for the screening of gastric carcinoma development in gastric adenoma/dysplasia. Int J Med Sci. 2018;15:610-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Tang CW, Yang JH, Qin JW, Wu HJ, Cui HP, Ge LY, Liu AQ. Regulation of the PD-1/PD-L1 Axis and NK Cell Dysfunction by Exosomal miR-552-5p in Gastric Cancer. Dig Dis Sci. 2024;69:3276-3289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 22. | Chen S, Wu H, Zhu L, Jiang M, Wei S, Luo J, Liu A. MiR-199b-5p Promotes Gastric Cancer Progression by Regulating HHIP Expression. Front Oncol. 2021;11:728393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Swanson K, Wu E, Zhang A, Alizadeh AA, Zou J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell. 2023;186:1772-1791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 234] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 24. | Izumi D, Zhu Z, Chen Y, Toden S, Huo X, Kanda M, Ishimoto T, Gu D, Tan M, Kodera Y, Baba H, Li W, Chen J, Wang X, Goel A. Assessment of the Diagnostic Efficiency of a Liquid Biopsy Assay for Early Detection of Gastric Cancer. JAMA Netw Open. 2021;4:e2121129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 25. | Chi H, Chen H, Wang R, Zhang J, Jiang L, Zhang S, Jiang C, Huang J, Quan X, Liu Y, Zhang Q, Yang G. Proposing new early detection indicators for pancreatic cancer: Combining machine learning and neural networks for serum miRNA-based diagnostic model. Front Oncol. 2023;13:1244578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 26. | Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262-e273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 653] [Article Influence: 130.6] [Reference Citation Analysis (0)] |
| 27. | Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W, Chen Y, Fan Y, Qi L, Yin Y, Zhu W, Shu Y, Liu P. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |