Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.102913
Revised: January 10, 2025
Accepted: February 21, 2025
Published online: April 15, 2025
Processing time: 117 Days and 9.4 Hours
Colon cancer represents a significant malignant neoplasm within the digestive system, characterized by a high incidence rate and substantial disease burden. The F-box protein 22 (FBXO22) plays a role in forming a specific type of ubiquitin ligase subunit, which is expressed abnormally in various malignant neoplasms and shows a notable relationship with prognosis in patients with cancer. Never
To explore the role of FBXO22 in colon cancer by examining FBXO22 expression patterns and analyzing how the protein affects the prognosis in patients who have undergone surgery.
Samples of cancerous and nearby normal tissues from patients with colon cancer were gathered, along with pertinent clinical data. Expression levels of the FBXO22 gene in both cancerous and paracancerous tissues were assessed through immu
Compared with normal colonic tissues, the FBXO22 gene was highly expressed in colon cancer tissues. Post-operative patients with colon cancer elevated FBXO22 reduced survival and exhibited resistance to various chemotherapeutic agents. FBXO22 expression suppresses the infiltration of anti-tumor immune cells. In vitro, FBXO22 knockdown inhibited the proliferation and migration of colon cancer cells.
The FBXO22 gene is a biomarker of poor prognosis in patients with colon cancer and has potential as a target for immunotherapy and overcoming chemotherapy resistance.
Core Tip: The precise role of the F-box protein 22 (FBXO22) gene in post-operative colon cancer patients remains to be fully elucidated. Utilizing bioinformatics assessments alongside cellular experimentation, researchers observed that the expression of the FBXO22 gene is markedly higher in colon cancer tissues when compared to normal colonic tissues. Patients suffering from colon cancer who present with increased FBXO22 gene levels tend to have decreased survival rates and show resistance to various chemotherapy agents. The elevated levels of the FBXO22 gene are associated with a reduction in the infiltration of immune cells that target tumors. Laboratory experiments demonstrate that reducing the activity of the FBXO22 gene hinders the growth and movement of colon cancer cells. The evidence suggests that the FBXO22 gene may facilitate the advancement of colon cancer, be associated with the onset of resistance to chemotherapy among post-operative colon cancer patients, and could act as an indicator of poor outcomes for these individuals. Consequently, targeting FBXO22 could represent a novel approach for treating colon cancer.
- Citation: Lu XF, Zhang HW, Chang X, Guo YZ. F-box protein 22: A prognostic biomarker for colon cancer associated with immune infiltration and chemotherapy resistance. World J Gastrointest Oncol 2025; 17(4): 102913
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/102913.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.102913
Globally, colorectal cancer ranks as the third most common cancer and is the second most frequent cause of death attributed to cancer, posing a substantial public health challenge[1]. Notwithstanding the implementation of preventive measures and the progress made in medical technology, which have notably enhanced the methods for diagnosing and treating colorectal cancer, the long-term outcomes for affected individuals still fall short of expectations. The timing of diagnosing colorectal cancer has a significant impact on the survival outlook for post-operative patients. Statistics reveal that among post-operative individuals identified with colorectal cancer in its initial stages, the five-year survival probability can soar to as much as 90%. On the other hand, for patients diagnosed in more advanced stages who have lost the chance of surgery, the five-year survival rate drastically drops to just 13.1%. On the other hand, the development of colorectal cancer arises from various factors, including intricate abnormalities in molecular signaling pathways, a variety of genetic mutations, and changes in epigenetic mechanisms. These alterations can lead to unchecked cell growth, a disturbance in the mechanisms regulating programmed cell death, and changes within the tissue microenvironment, which collectively promote the development and advancement of tumors[2,3]. Therefore, studying the detailed molecular mechanisms of colorectal cancer is of great practical importance for clinical practice. These investigations facilitate the identification of biomarkers for early screening, thereby enhancing the rate of early diagnosis. Furthermore, these efforts lead to the identification of predictive biomarkers and unique treatment targets, which play a significant role in formulating more tailored and effective treatment plans for post-operative individuals.
The ubiquitin protease system is an epigenetic modification mechanism widely and intensively studied in biology. This system relies on three core enzymes, E1, E2 and E3, which work together to perform their biological functions[4,5]. Among the F-box protein family, F-box protein 22 (FBXO22) serves as a key member of the E3 ubiquitin ligase complex[6]. The involvement of FBXO22 in various biological mechanisms is notable, as it influences cell cycle dynamics, facilitates the repair of DNA damage, interacts within signaling networks, and regulates programmed cell death. Moreover, FBXO22 is crucial in regulating responses to stress and facilitating the mechanism of cellular autophagy. Numerous investigations have revealed that FBXO22 holds importance across various cancer types, though its distinct functions can differ among them. In the context of lung cancer, FBXO22 is crucial in influencing the metastatic deve
Fetal bovine serum, RPMI-1640 complete medium, and DMEM complete medium, all procured from Thermo Fisher Scientific; anti-FBXO22 antibody, also obtained from Thermo Fisher Scientific; sheep anti-rabbit secondary antibody sourced from Proteintech Group, Inc.; RNA extraction kit; reverse transcription kit; polymerase chain reaction (PCR) kit; siRNA kit, with the siRNA-FBXO22 sequences being S: GGUCCUUGUCUUUGGUUAUTT and AS: AUAACCAAA
Clinical samples and patient information: Tissue samples, including both cancerous and adjacent non-cancerous tissues, were obtained from a cohort of 80 individuals undergoing treatment at the Affiliated Hospital of Hebei University of Engineering, with sample collection taking place from September 2022 to February 2024. These samples were frozen at -80 °C for future use, and the corresponding clinical information of the patients was concurrently collected. All patients were diagnosed with colon cancer based on clinical and histopathological evidence and had not received any treatment prior to surgery. The study received assessment and consent from the Ethics Review Board at the Affiliated Hospital of Hebei University of Engineering.
The Tumor Immunity Evaluation Resource (TIMER2) serves as a web-based platform for assessing immunity within tumors, utilizing data from The Cancer Genome Atlas (TCGA) dataset to compare the levels of FBXO22 mRNA expression in malignant tissues against nearby non-cancerous counterparts across various cancer types. A web-based platform for information services was utilized to assess data from the TCGA database, aiming to determine the differences in FBXO22 mRNA expression between cancerous tissues and their adjacent healthy counterparts across various cancer types. Similar pan-cancer analyses were done by applying the GEN2 web information service platform and data from GEO. Moreover, the web-based platform GEPIA2 was utilized to investigate the expression variations of FBXO22 in colorectal carcinoma samples compared to non-cancerous tissues, leveraging TCGA correlated information from the GTEx database.
To assess the variant type and occurrence of mutations in the FBXO22 gene among colon adenocarcinoma (COAD) patients, the cBioPortal online information service was utilized for analyzing data derived from TCGA patient samples.
The UALCAN web-based information service platform was applied to analyze differences in FBXO22 promoter methylation levels in different subgroups of the colon cancer population, and the DNA methylation interactive visualization database was employed to examine the correlation between FBXO22 expression and the methylation status of its promoter.
Using R software, an examination of 492 samples of colorectal cancer from the TCGA dataset was performed, leading to the discovery of 163 genes exhibiting differential expression related to FBXO22. This set included 43 genes that exhibited increased expression levels and 123 genes that showed reduced expression levels. Following this, the notable genes that exhibited differential expression underwent thorough exploration via analyses such as Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA).
The analysis was conducted using the TIMER 2.0 online information platform to investigate the correlation between the expression levels of FBXO22 and the degree of immune cell infiltration present in the colon cancer tumor microenvironment, involving a complete set of CD8+ T lymphocytes, CD4+ T cells, T-shaped battery controller, B cells, neutrophils, monocyte, macrophage dendritic cells, natural killer (NK) cells polar battery, fibroblasts associated with tumors, immune cell precursors associated with lymphoid lineage 21, which encompass progenitor cells of the myeloid lineage, epithelial cells, eosinophils, progenitor cells for granulocytes and monocytes, progenitor cells originating from the blood cell lineage, T follicular helper battery gamma delta cells T, NK T battery, as well as suppressor cells derived from bone marrow. Furthermore, the web-based platform TISIDB was subsequently utilized to investigate the relationship between the expression levels of FBXO22 and key immune molecules, including those that promote immune responses, and those that inhibit immune responses along with signaling substances.
We utilized the String and GeneMANIA network information service platforms to perform protein-protein interaction analysis for co-expressed genes of FBXO22, and the findings were visualized using Cytoscape software.
Using data from the TCGA dataset and the Genomics of Drug Sensitivity in Cancer training dataset, the R software oncoPredict package was applied to analyze the sensitivity of commonly used drugs for colon cancer patients, The limma package was employed to assess the differential sensitivity of these drugs in relation to high and low expression levels of FBXO22, while the ggplot package was utilized for data visualization.
The specimens obtained were preserved in a solution of 4% paraformaldehyde. Subjected to dehydration using a gradient of ethanol solutions and then encased in paraffin, subsequently, specimens were sectioned to a thickness of 4 micrometers for immunohistochemical analysis. Sections were deparaffinised by applying xylene immersion for 3 times, and then hydrated via ethanol immersion with concentrations ranging from high to low, and rinsed 3 times with distilled water and phosphate buffered saline solution, respectively. The rinsed sections were subjected to incubation with a 3% hydrogen peroxide solution for a duration of 10 minutes at room temperature in order to inactivate endogenous peroxidase activity, and after rinsing as before, the sections were treated for 15 minutes using a microwave oven to heat the citrate solution until the temperature reached 95 °C to repair the antigen. Subsequent to the sealing and washing steps, the sections were subjected to incubation at 4 °C overnight with an antibody targeting FBXO22 (catalogue number, Brand, followed by a one-hour incubation at ambient temperature using a secondary antibody at a dilution of 1:2000. Color development was achieved using DAB, and images were captured under a microscope. The evaluation of FBXO22 protein expression was carried out through the application of H scores.
The overall survival of colon cancer patients was evaluated by analyzing the expression levels of the FBXO22 gene through the Kaplan-Meier Plotter online tool. Survival analysis was conducted through the generation of Kaplan-Meier curves to explore how varying levels of FBXO22 expression correlate with the outcomes of patient survival.
An analysis was conducted using clinical data from 80 patients, focusing on how the levels of FBXO22 expression correspond with various clinical attributes including factors such as age, sex, tumor-node-metastasis classification, body mass index, educational background, history of smoking, and patterns of alcohol use.
The HCT116 and DLD1 cell lines were maintained in 1640 medium enriched with 10% fetal bovine serum, incubated at 37 °C with a 5% carbon dioxide atmosphere. Cells in the logarithmic growth phase were utilized for subsequent transfection and functional assays.
Cells in the logarithmic growth phase were collected and subsequently inoculated into 6-well plates. Once the cells achieved a confluence of 70%-80%, a mixture of siRNA and transfection reagents was introduced into the plates. After twenty-four hours of the transfection process, the cells were gathered, and PCR and western blotting analyses were conducted to ascertain knockdown efficacy. Additionally, proliferation and invasion assays were performed.
The evaluation of the effects of inhibiting the FBXO22 gene on the growth of colon cancer cells was conducted using the CCK-8 assay alongside the Edu cell proliferation detection assay. Following the experimental protocol, the cells were harvested and seeded into 96-well plates, placing 1000 cells in each well. Subsequently, the corresponding reagents were added following the instructions provided in the CCK-8 assay kit, and the absorbance values were measured using an enzyme marker after 2 hours of incubation. These absorbance values were further measured at 0, 24, 36, and 48 hours, respectively. In a similar manner, the cells that underwent treatment were harvested and then plated into 96-well plates, with a cell density of 1000 cells for each well. Upon achieving stable growth of the cells, the proliferation capacity was assessed using the methodology specified in the Edu Proliferation Detection Kit.
The effect of inhibiting the FBXO22 gene on the ability of colon cancer cells to invade was evaluated through the use of both scratch assays and Transwell assays.
Cells that had been transfected were harvested, and total RNA was obtained by applying the TRIzol method following the guidelines provided by the manufacturer. The isolated RNA underwent reverse transcription to synthesize cDNA using the TaKaRa kit, after which the QuantStudio 5 instrument was utilized for the PCR analysis. The quantitative PCR instrument designed for real-time fluorescence analysis.
Cells were harvested and then transfected, total proteins were extracted, electrophoresis, membrane transfer, primary and secondary antibody incubation were followed by chemical colour development, and photographs were taken by instrumental exposure.
The datasets from the two groups were subjected to t-test analysis in this investigation, whereas one-way ANOVA was employed for examining the variances among numerous groups. The relationship between the expression of FBXO22 and the clinical-pathological features of colon cancer was assessed through the χ2 test, while the survival outcomes were analyzed using the Kaplan-Meier method.
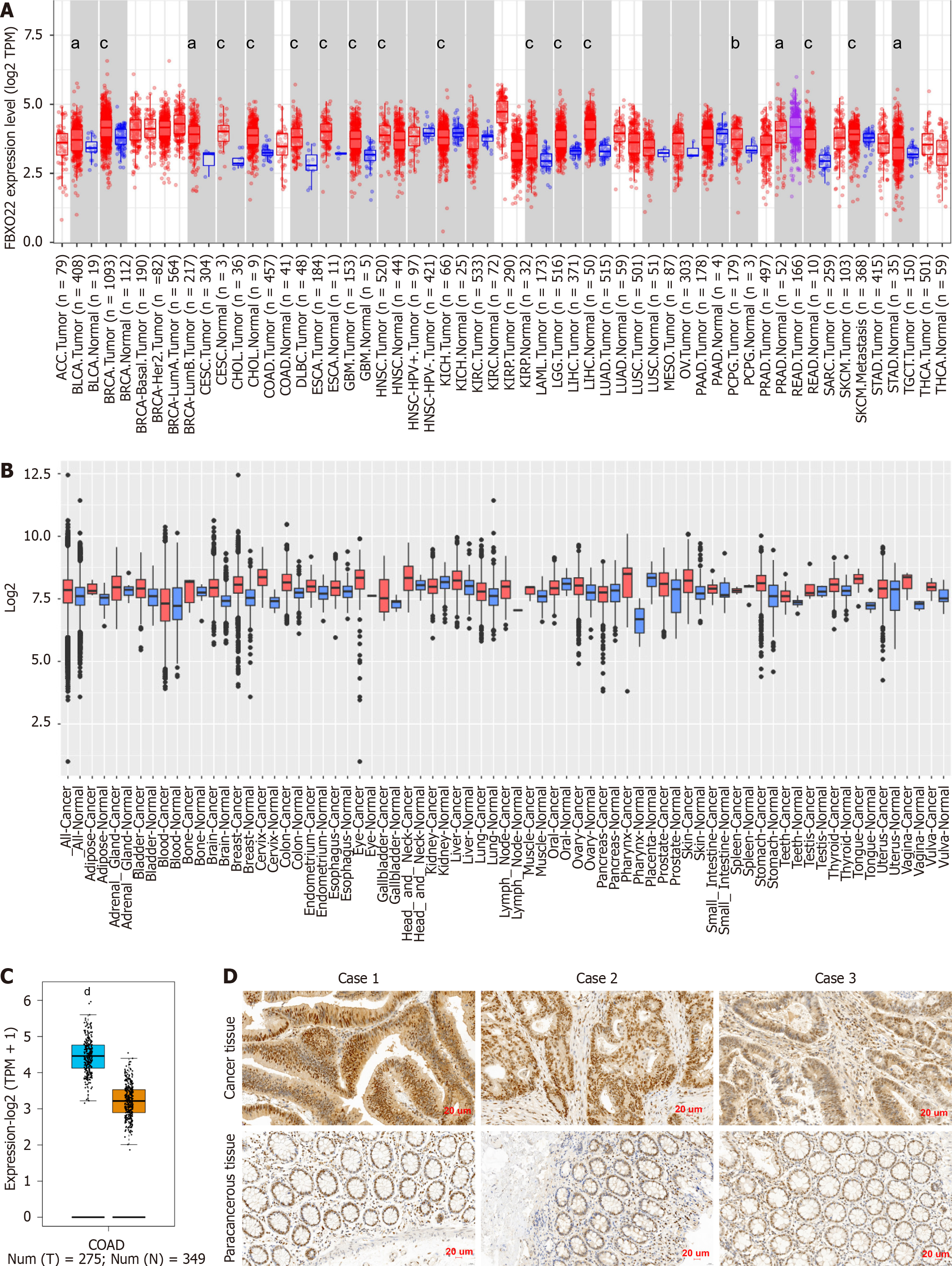
The mRNA expression levels of FBXO22 were examined across various tumors in TCGA by employing the TIMER 2 platform. The objective of this study was to explore possible differences in the expression of FBXO22 mRNA between cancerous tissues and their corresponding normal tissues. The analysis revealed that the FBXO22 gene was present in bladder uroepithelial carcinomas, invasive breast carcinomas, uterine carcinomas, carcinomas of the uterus and cervix, bile duct carcinomas, COAD, and oesophageal carcinomas. In comparison to normal tissues, there was a notable increase in expression levels in several malignancies, such as glioblastoma multiforme, squamous cell carcinoma located in the head and neck region, clear cell carcinoma affecting the kidneys, hepatocellular carcinoma, as well as adenocarcinomas of the lung and stomach, squamous cell carcinoma of the lung, rectal carcinoma, thyroid carcinoma, and endometrial carcinoma originating from the uterine corpus (Figure 1A). Further examination of FBXO22 mRNA expression was conducted using the GENT2 online platform, which indicated upregulated expression in certain cancer types, including colon cancer (Figure 1B). To delve deeper into the expanded dataset, the GEPIA2 web-based information service platform was utilized to analyze FBXO22 mRNA expression in colon cancer vs paraneoplastic tissues in TCGA-matched GTEx data. The results also indicated higher expression of FBXO22 mRNA in colon cancer tissues compared to paraneoplastic tissues (Figure 1C). Subsequently, in order to examine the expression of FBXO22 in colon cancer at the protein level, a comprehensive analysis was conducted utilizing a total of 80 paired tissue samples, comprising both cancerous and adjacent non-cancerous tissues. These samples were subjected to immunohistochemical analysis. The analysis revealed that FBXO22 was expressed at higher levels in colon cancer tissues than in paracancer tissues at the protein level (Figure 1D).
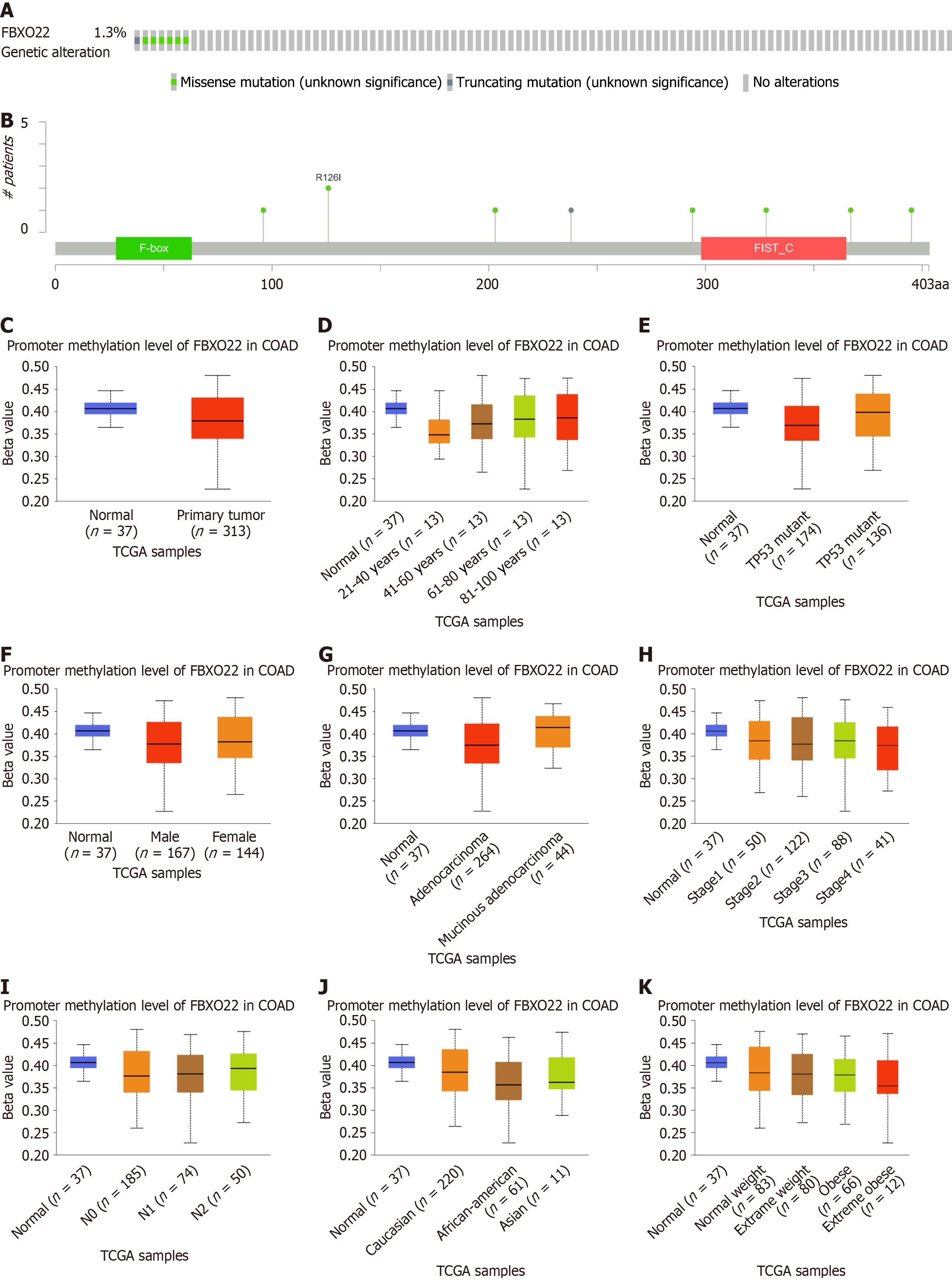
Mutations and changes in DNA methylation are crucial factors in tumorigenesis. In order to explore the mutations associated with FBXO22 in colon cancer, the online platform cBioPortal was used to examine data samples from the TCGA colon cancer archive. Furthermore, the ULACAN tool was employed to assess the FBXO22 promoter methylation level in the TCGA colon cancer data. The findings indicated a reduction in FBXO22 promoter methylation in cancerous primary tissues when compared to adjacent normal tissues. Furthermore, as the tumor stage advanced, there was a notable decline in the FBXO22 promoter methylation level. Moreover, significant differences in FBXO22 promoter methylation levels were observed across different ages, genders, and races (Figure 2).
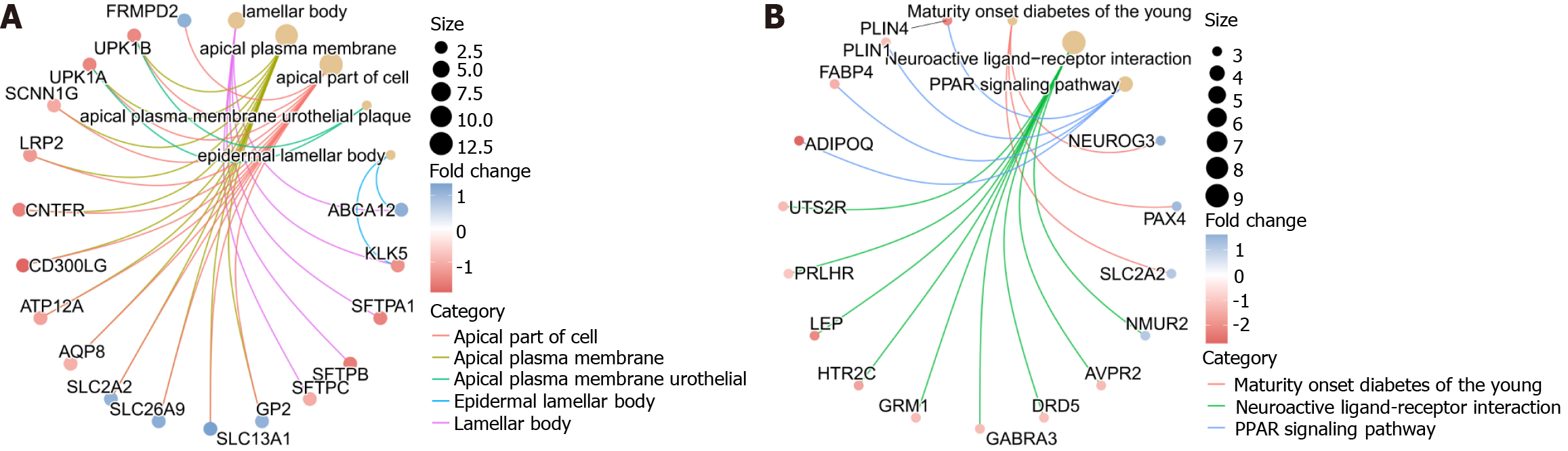
To further explore the function of FBXO22, the expression of other genes between FBXO22 high and low expression groups was differentially analyzed with P < 0.05, |logfold change|> 1. A total of 163 genes displaying differential expression were identified, which included 43 genes that exhibited increased expression and 123 genes that showed decreased expression. GO enrichment analysis of the identified differential genes showed that FBXO22 co-expressed differential genes were mainly enriched with cell membrane structure and function, endosomes and other structures and functions, followed by KEGG and GSEA enrichment analysis of the differential genes, which indicated that FBXO22 was mainly involved in PPAR and other signaling pathways (Figure 3).
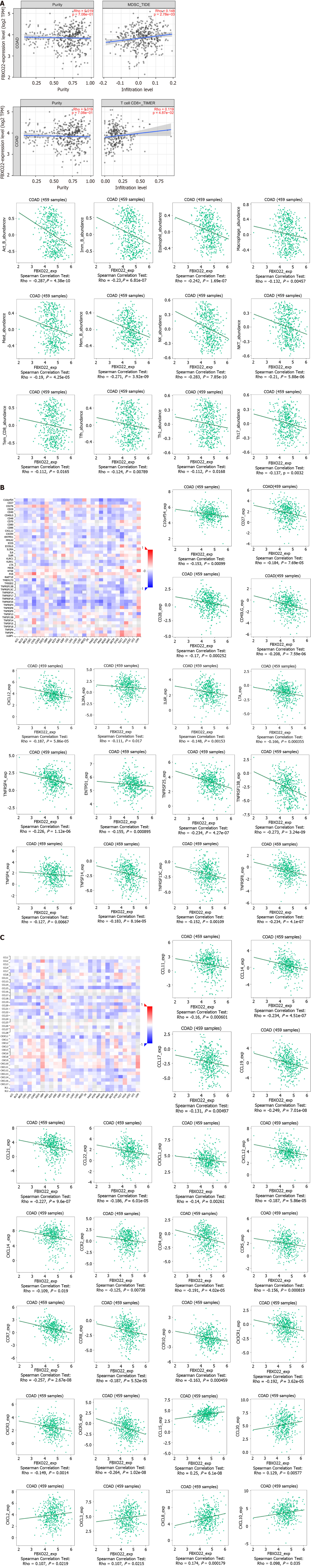
The characteristics of immune infiltration within the tumor microenvironment are intimately associated with tumorigenesis, progression, metastasis, and prognosis. GO enrichment analysis has revealed that FBXO22 may play a role in structures and functions such as cell tops, calcium channel complexes, and multivesicular bodies, which are closely linked to intercellular communication, material transport, and cell signaling. Notably, exosomes derived from multivesicular bodies can exert an influence on the tumor microenvironment. Consequently, the online tools TIMER2.0 and TISIDB were utilized to evaluate the correlation between various immune cell infiltrations, immune molecules, and FBXO22 expression in colon cancer. The results indicated that FBXO22 expression was positively correlated with the infiltration of immunosuppressive cells, such as bone marrow-derived suppressor cells, and negatively correlated with the infiltration of anti-tumor cells, including B cells, T cells, and NK cells. Correlation analysis with immune molecules demonstrated that FBXO22 expression was associated with the expression of most immunosuppressive cells, including C10orf54, CD27, CD28, CD40 LG, C-X-C chemokine ligand 12 (CXCL12), ENTPD1, IL2RA, IL6R, LTA, tumor necrosis factor receptor superfamily, member 4 (TNFRSF4), TNFRSF8, TNFRSF13B, TNFRSF13C, TNFRSF25, and TNFSF14, while most other immune-promoting molecules exhibited positive correlations with their expression. Additionally, correlation analysis of chemokine and FBXO22 expression revealed that the expression of C-C motif chemokine ligand 11 (CCL11), CCL14, CCL17, CCL19, CCL21, CCL22, CX3C chemokine ligand 1, CXCL12, CXCL14, CC chemokine receptor 2 (CCR2), CCR4, CCR5, CCR7, CCR8, CCR10, CX3CR1, CXCR3, and CXCR5 molecules was generally concordant with FBXO22 expression, whereas the expression of CCL15, CCL20, CXCL2, CXCL3, CXCL8, CXCL10, and CXCL11 was negatively correlated (Figure 4).
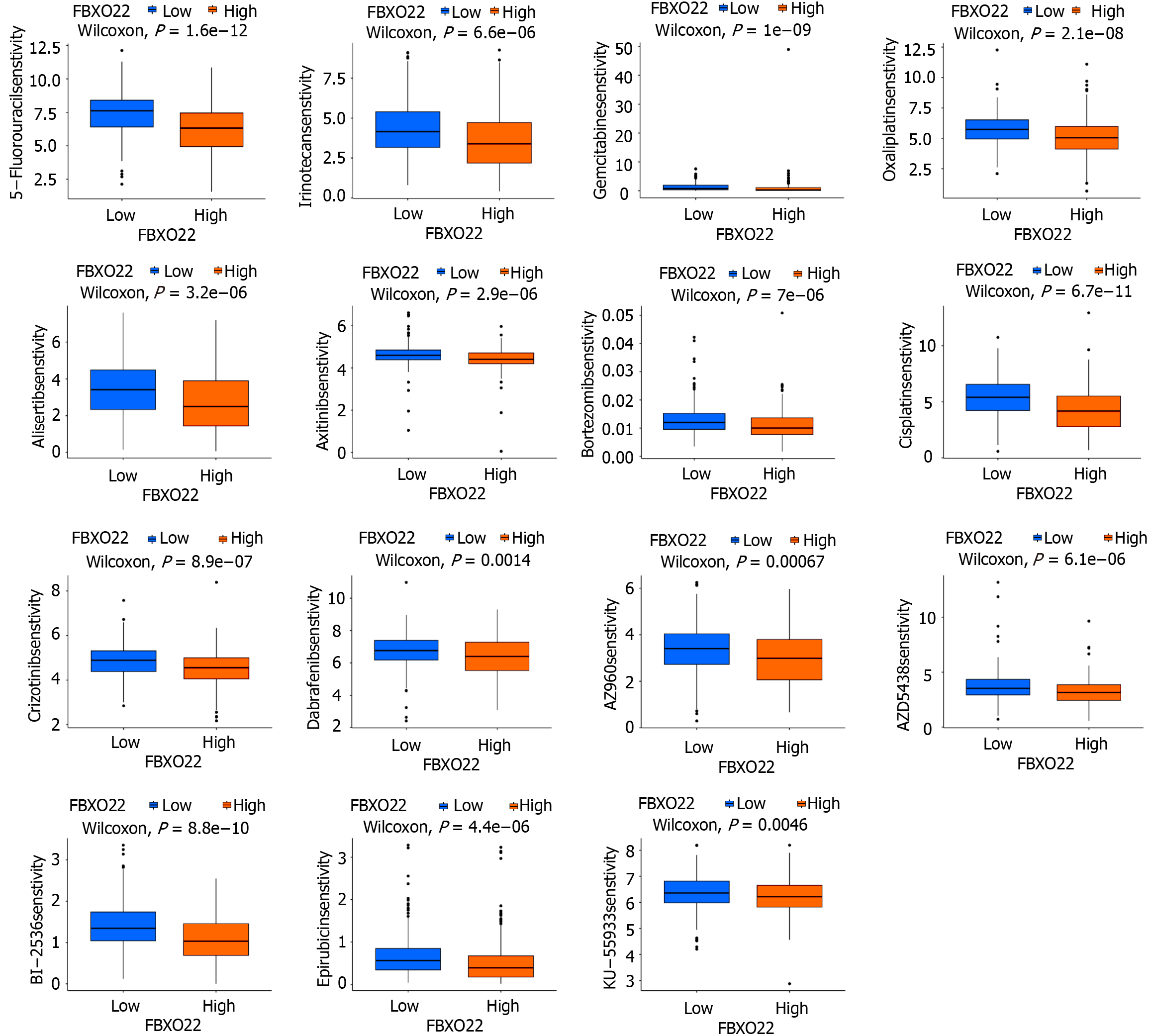
One of the main approaches for treating colon cancer is chemotherapy; therefore, this research sought to determine if there is a difference in drug sensitivity among the two groups of patients with varying levels of FBXO22 expression across 169 different medications. The results revealed that among the 169 drugs analyzed, 67 exhibited differential sensitivities between the two patient groups. Furthermore, 69 drugs, including 5-fluorouracil, oxaliplatin, gemcitabine, and irinotecan, demonstrated reduced sensitivity in patients with high FBXO22 expression compared to those in the group with low FBXO22 expression (Figure 5).
In patients with advanced tumor stages III-IV, the levels of FBXO22 expression were notably elevated when compared to those with earlier stages I-II. This increase was particularly apparent in individuals with lymph node hyperplasia, lower differentiation grades, and tumor sizes measuring 3 cm or larger, contrasting with patients devoid of lymph node spread, exhibiting medium to high differentiation grades, and possessing tumors smaller than 3 cm. These differences were statistically significant (P < 0.05) (Table 1).
| Pathological trait | Number of cases | Expression of FBXO22 | χ2 | P value | |
| High (n = 55) | Low (n = 18) | ||||
| Age | 0.003 | 0.957 | |||
| ≥ 60 | 28 | 21 (38.18) | 7 (38.89) | ||
| < 60 | 45 | 34 (61.82) | 11 (61.11) | ||
| Gender | 0.125 | 0.724 | |||
| Male | 42 | 31 (56.36) | 11 (61.11) | ||
| Female | 31 | 24 (43.64) | 7 (38.89) | ||
| BMI | 0.408 | 0.523 | |||
| ≥ 25 kg/m2 | 29 | 23 (41.82) | 6 (33.33) | ||
| < 25 kg/m2 | 44 | 32 (58.18) | 12 (66.67) | ||
| Educational background | 0.054 | 0.816 | |||
| High school and above | 26 | 20 (36.36) | 6 (33.33) | ||
| Junior high school and below | 47 | 35 (63.64) | 12 (66.67) | ||
| Tumor stages | 6.508 | 0.011 | |||
| I-II | 42 | 27 (49.09) | 15 (83.33) | ||
| III-IV | 31 | 28 (50.91) | 3 (16.67) | ||
| Lymph node metastasis | 10.392 | 0.001 | |||
| Yes | 32 | 30 (54.55) | 2 (22.11) | ||
| No | 41 | 25 (45.45) | 16 (88.89) | ||
| Degree of differentiation | 5.890 | 0.015 | |||
| Low | 30 | 27 (49.09) | 3 (16.67) | ||
| Middle and high | 43 | 28 (50.91) | 15 (83.33) | ||
| Diameter of the tumour | 6.334 | 0.012 | |||
| ≥ 3 cm | 35 | 31 (56.36) | 4 (22.22) | ||
| < 3 cm | 38 | 24 (43.64) | 14 (77.78) | ||
| Smoking history | 1.853 | 0.173 | |||
| Yes | 50 | 40 (72.73) | 10 (55.56) | ||
| No | 23 | 15 (27.27) | 8 (44.44) | ||
| Drinking history | 0.003 | 0.957 | |||
| Yes | 45 | 34 (61.82) | 11 (61.11) | ||
| No | 28 | 21 (38.18) | 7 (38.89) | ||
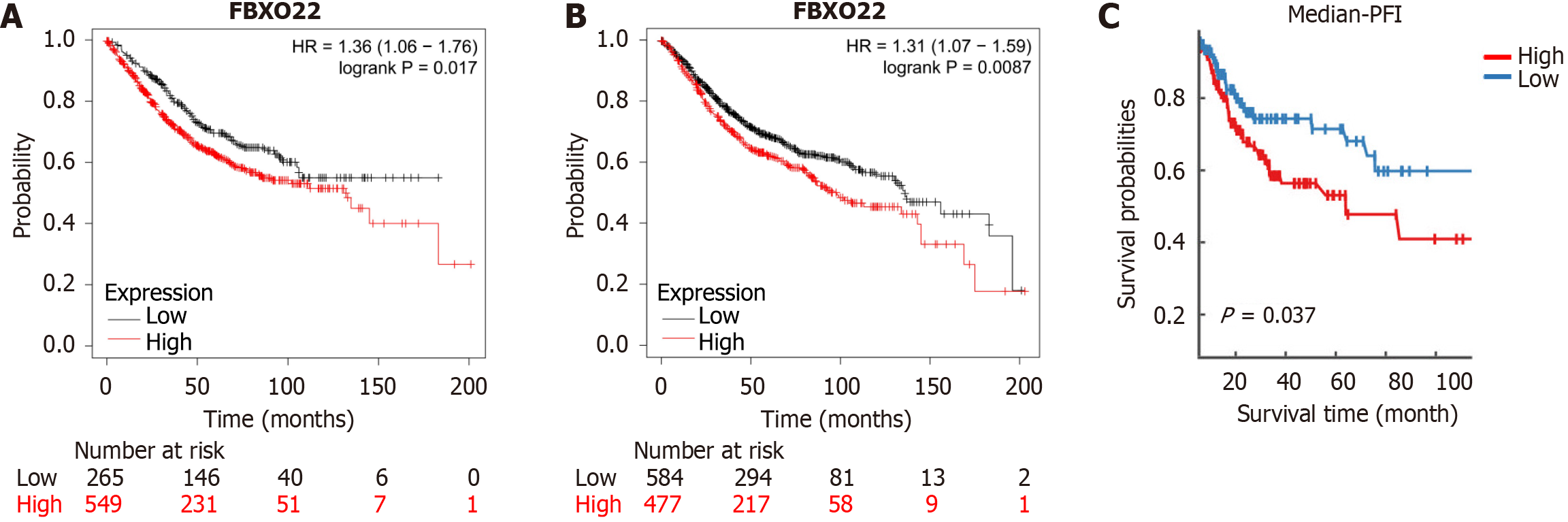
The analysis of survival utilizing the Kaplan-Meier approach revealed that individuals with heightened FBXO22 expression levels encountered significantly lower overall and disease-free survival rates when compared to those with reduced FBXO22 expression levels. The difference observed was statistically significant (P < 0.05) (Figure 6).
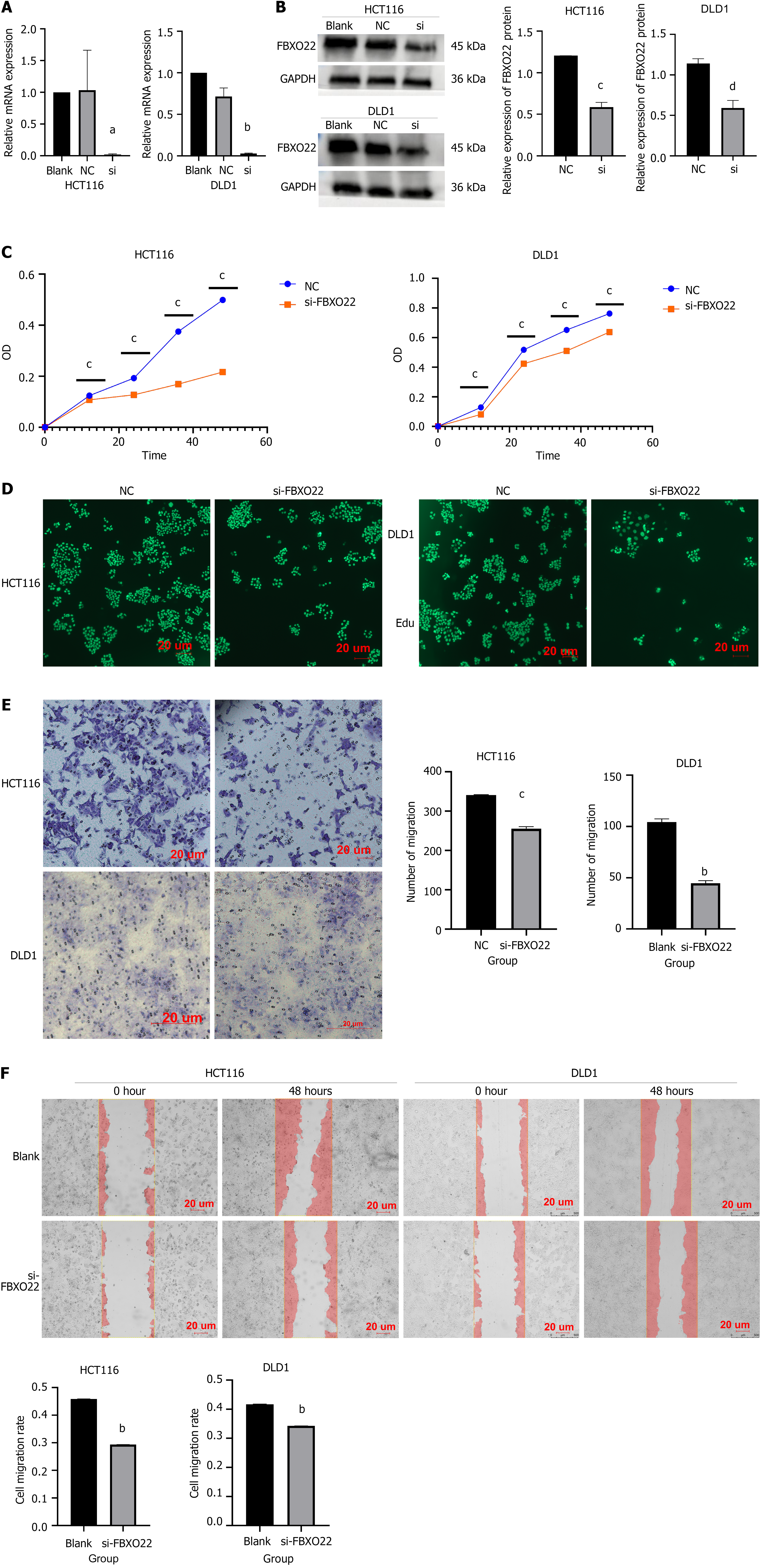
The effects of FBXO22 inhibition on colon cancer cell proliferation and invasion were verified in HCT116 and DLDI cell lines, respectively, and it was found that the proliferative and invasive abilities of the two cell lines were significantly weakened by FBXO22 inhibition, and ImageJ quantitative analysis showed that the difference was statistically significant (P < 0.05) (Figure 7).
With the popularity of colonoscopy and the promotion of the concept of early diagnosis and treatment, a considerable portion of pre-cancerous lesions have been addressed at an early stage, and the incidence of colon cancer has been controlled to a certain extent. However, on the whole, the incidence of colon cancer is still persistently rising[1,17,18]. Meanwhile, due to factors such as the inhibitory and metastatic nature of colon cancer, some colon cancer patients still have a poor prognosis despite active treatment including surgery[2,3,19]. Therefore, it is crucial to examine the fundamental processes that contribute to the emergence and advancement of colon cancer, along with identifying dependable prognostic markers that may improve the treatment outcomes for individuals diagnosed with this illness.
Recent research has indicated that the FBXO22 protein is linked to the progression of several types of malignant tumors[10,11,13-16,20,21]. Nonetheless, the significance of FBXO22 levels in colon cancer and its potential role as a marker for prognosis in postoperative patients remain to be clarified. This investigation aimed to explore the function of FBXO22 within the context of colon cancer. It was observed that in colon cancer tissues, the levels of FBXO22 expression were markedly higher when compared to those in normal tissues. To further analyse the possible reasons for the abnormal expression of FBXO22 in colon cancer, the mutation and methylation of FBXO22 in colon cancer were analysed using an online web-based service platform. The findings indicated that the variation in FBXO22 expression associated with colon cancer could potentially be linked to its missense mutation and decreased methylation levels, suggesting a role for FBXO22 in the development and advancement of colon cancer. In order to assess the potential of FBXO22 as a postoperative prognostic biomarker for colon cancer, this investigation examined the correlation between FBXO22 expression levels and patient survival outcomes, specifically overall survival and disease-specific survival. This analysis was conducted utilizing various datasets and employed the Kaplan-Meier method in conjunction with the DNA methylation interactive visualization database online platform. The findings indicated that postoperative colon cancer patients exhibiting elevated levels of FBXO22 expression experienced reduced survival rates in comparison to those with lower expression levels. These findings imply that the expression of FBXO22 could serve as an effective predictor of operative patient outcomes, potentially aiding in the creation of targeted therapies for colon cancer.
To delve deeper into the potential mechanisms by which FBXO22 may contribute to colon cancer development, analyses such as GO, KEGG, and GSEA enrichment were conducted on the differentially expressed genes from the TCGA-COAD cohort. This study compared patients with elevated levels of FBXO22 mRNA against those with reduced expression. The findings indicated that FBXO22 plays a role in several pathways associated with cancer, notably including the PPAR, mitogen-activated protein kinases, and phosphatidylinositol 3-kinase/protein kinase B signaling pathways. Recent studies have shown that the phosphatidylinositol 3-kinase/protein kinase B signaling pathway is associated with oxaliplatin resistance in colon cancer[22]. The PPAR signaling pathway is essential for numerous biological functions such as the breakdown of fatty acids[23,24], bile acid production, glycerophospholipid metabolism[25-27], lipid processing, adipocyte differentiation, adaptive thermogenesis, cell viability[28-30], ubiquitination, and glucose production, all of which are governed by the expression of target genes[31-35]. The activation of PPAR-delta, a crucial element of the PPAR signaling pathway, has been shown to reduce colon carcinogenesis[36]. This suggests that FBXO22 may play a negative regulatory role in the PPAR signaling pathway, thereby promoting the progression of colon cancer.
The examination of the relationship between FBXO22 levels and clinical-pathological features revealed a greater incidence of FBXO22 positivity in individuals diagnosed with tumor stages III-IV, lymph node hyperplasia, a low degree of differentiation, and tumor diameter ≥ 3 cm compared to patients with tumor stages I-II, absence of lymph node hyperplasia, tumors with moderate and high levels of differentiation, and tumor diameter < 3 cm. It was suggested that FBXO22 was highly expressed in colon cancer, the rate of positive expression was greater in individuals with tumor stages III-IV, lymph node hyperplasia, low difference, and tumor diameter ≥ 3 cm. The expression level of FBXO22 was able to predict the prognosis of the postoperative patients to a certain degree and provide guidance for clinical treatment. The reason is that FBXO22 can participate in multiple processes of tumor genesis and development. Therefore, patients with high FBXO22 expression generally have high tumor stages, lymph node metastasis, and low differentiation degrees, which can directly affect the prognosis of postoperative patients and increase the risk of postoperative recurrence and metastasis. FBXO22 can be used as a new target for treatment in order to improve the antitumor regimen and improve the postoperative patient’s prognosis.
The composition of the tumor microenvironment includes various elements such as neoplastic cells, vascular structures, immune system components, stromal cells from the mesenchyme, inflammatory mediators, and the matrix outside the cells. These elements significantly influence the advancement of tumors and the effectiveness of clinical treatments[37,38]. Immune cells infiltrating the tumor microenvironment greatly shape the advancement of tumors and the effectiveness of immunotherapies. The correlation between the expression of FBXO22 and the abundance of tumor cell infiltration in the microenvironment of colon cancer showed that myeloid-derived suppressor cells, quiescent dendritic cells, neutrophils, monocytes, and other tumor-promoting immune cells were positively correlated with the abundance of tumor cell infiltration in colon cancer and FBXO22 expression. Whereas the abundance of anti-tumor immune cells, such as B cells, T cells, and NK cells, was negatively correlated with FBXO22 expression. Myeloid-derived suppressor cells have an immunosuppressive role in tumors[39], which not only promote tumor immune escape by inhibiting T cell, B cell, and NK cell functions, but also directly promote tumor cell proliferation and metastasis through the secretion of cytokines, chemokines, and a variety of enzymes. Neutrophils in tumors can provide pro-angiogenic factors to promote tumor angiogenesis on the one hand, and on the other hand, they can encapsulate circulating tumor cells to promote the formation of metastatic foci of their exudates. B cells[40], T cells[41,42], and NK cells are the main cells in anti-tumor immunity. Therefore, the results of this study suggest that the expression of FBXO22 may increase the infiltration of pro-tumor immune cells and decrease anti-tumor immune cells, impairing the anti-tumor immunity of colon cancer, indicating a deterioration of the prognosis.
FBXO22 predicts sensitivity to chemotherapeutic agents. Chemotherapy is one of the main modalities of colon cancer treatment[3]. Analysing the sensitivity of chemotherapeutic drugs between high and low FBXO22 expression groups using the oncoPredict package showed that most chemotherapeutic drugs had worse sensitivity in the high FBXO22 expression group, such as 5-fluorouracil and capecitabine. In this study, the median FBXO22 expression level served as the benchmark for categorizing patients into high and low FBXO22 expression groups. Patients with high FBXO22 expression, defined as those with H-scores exceeding 144.13, exhibited reduced sensitivity to clinically administered chemotherapeutic agents, suggesting a potential for increased resistance. This finding paves the way for the preliminary prediction of chemotherapy resistance in patients through immunohistochemistry. However, to further clarify the influence of FBXO22 expression on drug sensitivity, it is essential to conduct experimental studies to determine whether FBXO22 expression indeed confers resistance to specific drugs in colon cancer cells. This will constitute our next research endeavor.
To conclude, FBXO22 holds promise as both a potential prognostic marker and a target for treatment in patients diagnosed with advanced stages of colon cancer and who have lost the chance of surgery. Nonetheless, this research does have certain limitations. To begin with, while bioinformatics analyses provide valuable insights, they do not substitute for experimental verification; this research primarily assessed the role of FBXO22 in colon cancer through straightforward in vitro testing. Additional investigations are required to understand the role of FBXO22 in the progression of colon cancer. Additionally, while this investigation identified a notable link between FBXO22 and tumor immunity in colon cancer, the specific mechanisms by which FBXO22 influences immune cell infiltration remain uncertain, necessitating further experimental inquiries. In the future, as we further elucidate the specific mechanisms by which FBXO22 influences immune cells through experimental studies, and clarify the precise pathways through which FBXO22 induces chemoresistance, it will be promising to explore the potential of combining FBXO22 gene targeting with immunotherapy. Additionally, enhancing the efficacy of chemotherapy may be achieved by integrating this targeted therapy with standard chemotherapeutic regimens.
The findings of this study indicate that there is a significant rise in the levels of FBXO22 mRNA and protein in colon cancer cases, which is linked to a poorer prognosis for postoperative patients. Additionally, our findings suggest that FBXO22 may serve as a potential indicator for colon cancer outcomes and is linked to factors such as immune cell infiltration and resistance to chemotherapy.
I would like to extend my sincere appreciation to my classmates for their invaluable support and guidance during the challenges I faced in my experiments. Secondly, I am also deeply thankful to the laboratory instructors for their meticulous guidance on the technical details of my experiments.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64436] [Article Influence: 16109.0] [Reference Citation Analysis (176)] |
| 2. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (2)] |
| 3. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3008] [Article Influence: 501.3] [Reference Citation Analysis (3)] |
| 4. | Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6346] [Cited by in RCA: 6680] [Article Influence: 247.4] [Reference Citation Analysis (0)] |
| 5. | Yang Q, Zhao J, Chen D, Wang Y. E3 ubiquitin ligases: styles, structures and functions. Mol Biomed. 2021;2:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 194] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 6. | Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573-2580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 574] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 7. | Lignitto L, LeBoeuf SE, Homer H, Jiang S, Askenazi M, Karakousi TR, Pass HI, Bhutkar AJ, Tsirigos A, Ueberheide B, Sayin VI, Papagiannakopoulos T, Pagano M. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell. 2019;178:316-329.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 423] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 8. | Zhang L, Chen J, Ning D, Liu Q, Wang C, Zhang Z, Chu L, Yu C, Liang HF, Zhang B, Chen X. FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J Exp Clin Cancer Res. 2019;38:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 9. | Ma J, Wu Y, Cheng S, Yang W, Zhong L, Li Q, Fang L. FBXO22 Accelerates Pancreatic Cancer Growth by Deactivation of the Hippo Pathway via Destabilizing LATS2. Dig Dis Sci. 2023;68:1913-1922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Guo F, Liu J, Han X, Zhang X, Lin T, Wang Y, Bai J, Han J. FBXO22 Suppresses Metastasis in Human Renal Cell Carcinoma via Inhibiting MMP-9-Mediated Migration and Invasion and VEGF-Mediated Angiogenesis. Int J Biol Sci. 2019;15:647-656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Zhang Y, Li W, Guo S, Wu Z, Zhang L, Liu Y, Li X, Guo X, Cao J, Yang C, Wang Z. FBXO22 Mediates the NGF/TRKA Signaling Pathway in Bone Metastases in Prostate Cancer. Am J Pathol. 2023;193:1248-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Lin M, Zhang J, Bouamar H, Wang Z, Sun LZ, Zhu X. Fbxo22 promotes cervical cancer progression via targeting p57(Kip2) for ubiquitination and degradation. Cell Death Dis. 2022;13:805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Li M, Zhao X, Yong H, Shang B, Lou W, Wang Y, Bai J. FBXO22 Promotes Growth and Metastasis and Inhibits Autophagy in Epithelial Ovarian Cancers via the MAPK/ERK Pathway. Front Pharmacol. 2021;12:778698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Li S, He J, Liao X, He Y, Chen R, Chen J, Hu S, Sun J. Fbxo22 inhibits metastasis in triple-negative breast cancer through ubiquitin modification of KDM5A and regulation of H3K4me3 demethylation. Cell Biol Toxicol. 2023;39:1641-1655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 15. | Liu Y, Pan B, Qu W, Cao Y, Li J, Zhao H. Systematic analysis of the expression and prognosis relevance of FBXO family reveals the significance of FBXO1 in human breast cancer. Cancer Cell Int. 2021;21:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Ge MK, Zhang N, Xia L, Zhang C, Dong SS, Li ZM, Ji Y, Zheng MH, Sun J, Chen GQ, Shen SM. FBXO22 degrades nuclear PTEN to promote tumorigenesis. Nat Commun. 2020;11:1720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 17. | Paragomi P, Zhang Z, Abe SK, Islam MR, Rahman MS, Saito E, Shu XO, Dabo B, Pham YT, Chen Y, Gao YT, Koh WP, Sawada N, Malekzadeh R, Sakata R, Hozawa A, Kim J, Kanemura S, Nagata C, You SL, Ito H, Park SK, Yuan JM, Pan WH, Wen W, Wang R, Cai H, Tsugane S, Pourshams A, Sugawara Y, Wada K, Chen CJ, Oze I, Shin A, Ahsan H, Boffetta P, Chia KS, Matsuo K, Qiao YL, Rothman N, Zheng W, Inoue M, Kang D, Luu HN. Body Mass Index and Risk of Colorectal Cancer Incidence and Mortality in Asia. JAMA Netw Open. 2024;7:e2429494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 18. | Sadeghi S, Barzi A. USPSTF Recommendations and Colorectal Cancer in Younger Adults-Current Challenges and Future Opportunities. JAMA Netw Open. 2024;7:e2436305. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Nakamura Y, Watanabe J, Akazawa N, Hirata K, Kataoka K, Yokota M, Kato K, Kotaka M, Kagawa Y, Yeh KH, Mishima S, Yukami H, Ando K, Miyo M, Misumi T, Yamazaki K, Ebi H, Okita K, Hamabe A, Sokuoka H, Kobayashi S, Laliotis G, Aushev VN, Sharma S, Jurdi A, Liu MC, Aleshin A, Rabinowitz M, Bando H, Taniguchi H, Takemasa I, Kato T, Kotani D, Mori M, Yoshino T, Oki E. ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat Med. 2024;30:3272-3283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 20. | Zheng Y, Chen H, Zhao Y, Zhang X, Liu J, Pan Y, Bai J, Zhang H. Knockdown of FBXO22 inhibits melanoma cell migration, invasion and angiogenesis via the HIF-1α/VEGF pathway. Invest New Drugs. 2020;38:20-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Zhu XN, Wei YS, Yang Q, Liu HR, Zhi Z, Zhu D, Xia L, Hong DL, Yu Y, Chen GQ. FBXO22 promotes leukemogenesis by targeting BACH1 in MLL-rearranged acute myeloid leukemia. J Hematol Oncol. 2023;16:9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 22. | Zhou X, Han J, Zuo A, Ba Y, Liu S, Xu H, Zhang Y, Weng S, Zhou Z, Liu L, Luo P, Cheng Q, Zhang C, Chen Y, Shan D, Liu B, Yang S, Han X, Deng J, Liu Z. THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer. Mol Cancer. 2024;23:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Reference Citation Analysis (0)] |
| 23. | Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating UDP-glucuronosyltransferase 2B4 enzyme. J Biol Chem. 2003;278:32852-32860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Basilotta R, Lanza M, Casili G, Chisari G, Munao S, Colarossi L, Cucinotta L, Campolo M, Esposito E, Paterniti I. Potential Therapeutic Effects of PPAR Ligands in Glioblastoma. Cells. 2022;11:621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Chen H, Tan H, Wan J, Zeng Y, Wang J, Wang H, Lu X. PPAR-γ signaling in nonalcoholic fatty liver disease: Pathogenesis and therapeutic targets. Pharmacol Ther. 2023;245:108391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 128] [Reference Citation Analysis (0)] |
| 26. | Derosa G, Sahebkar A, Maffioli P. The role of various peroxisome proliferator-activated receptors and their ligands in clinical practice. J Cell Physiol. 2018;233:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 360] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 27. | Fenner MH, Elstner E. Peroxisome proliferator-activated receptor-gamma ligands for the treatment of breast cancer. Expert Opin Investig Drugs. 2005;14:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Finck BN. The PPAR regulatory system in cardiac physiology and disease. Cardiovasc Res. 2007;73:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 159] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 29. | Ishtiaq SM, Arshad MI, Khan JA. PPARγ signaling in hepatocarcinogenesis: Mechanistic insights for cellular reprogramming and therapeutic implications. Pharmacol Ther. 2022;240:108298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 30. | Okine BN, Gaspar JC, Finn DP. PPARs and pain. Br J Pharmacol. 2019;176:1421-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 31. | Qiu YY, Zhang J, Zeng FY, Zhu YZ. Roles of the peroxisome proliferator-activated receptors (PPARs) in the pathogenesis of nonalcoholic fatty liver disease (NAFLD). Pharmacol Res. 2023;192:106786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 102] [Reference Citation Analysis (0)] |
| 32. | Sertznig P, Seifert M, Tilgen W, Reichrath J. Peroxisome proliferator-activated receptors (PPARs) and the human skin: importance of PPARs in skin physiology and dermatologic diseases. Am J Clin Dermatol. 2008;9:15-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 33. | Trottier J, Perreault M, Rudkowska I, Levy C, Dallaire-Theroux A, Verreault M, Caron P, Staels B, Vohl MC, Straka RJ, Barbier O. Profiling serum bile acid glucuronides in humans: gender divergences, genetic determinants, and response to fenofibrate. Clin Pharmacol Ther. 2013;94:533-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Wang Y, Lei F, Lin Y, Han Y, Yang L, Tan H. Peroxisome proliferator-activated receptors as therapeutic target for cancer. J Cell Mol Med. 2024;28:e17931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 35. | Wang S, Dougherty EJ, Danner RL. PPARγ signaling and emerging opportunities for improved therapeutics. Pharmacol Res. 2016;111:76-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 173] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 36. | Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-delta attenuates colon carcinogenesis. Nat Med. 2004;10:481-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 169] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 37. | DePeaux K, Delgoffe GM. Metabolic barriers to cancer immunotherapy. Nat Rev Immunol. 2021;21:785-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 343] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 38. | Hiam-Galvez KJ, Allen BM, Spitzer MH. Systemic immunity in cancer. Nat Rev Cancer. 2021;21:345-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 325] [Cited by in RCA: 847] [Article Influence: 211.8] [Reference Citation Analysis (0)] |
| 39. | Lasser SA, Ozbay Kurt FG, Arkhypov I, Utikal J, Umansky V. Myeloid-derived suppressor cells in cancer and cancer therapy. Nat Rev Clin Oncol. 2024;21:147-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 140] [Reference Citation Analysis (0)] |
| 40. | Laumont CM, Nelson BH. B cells in the tumor microenvironment: Multi-faceted organizers, regulators, and effectors of anti-tumor immunity. Cancer Cell. 2023;41:466-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 94] [Reference Citation Analysis (0)] |
| 41. | Hu Y, Hu Q, Li Y, Lu L, Xiang Z, Yin Z, Kabelitz D, Wu Y. γδ T cells: origin and fate, subsets, diseases and immunotherapy. Signal Transduct Target Ther. 2023;8:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 127] [Reference Citation Analysis (0)] |
| 42. | Oh DY, Fong L. Cytotoxic CD4(+) T cells in cancer: Expanding the immune effector toolbox. Immunity. 2021;54:2701-2711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 261] [Article Influence: 65.3] [Reference Citation Analysis (0)] |