Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.102084
Revised: January 27, 2025
Accepted: February 14, 2025
Published online: April 15, 2025
Processing time: 166 Days and 13.8 Hours
MEX3A is a member of the human homologous gene MEX-3 family. It has been shown to promote cell proliferation and migration in various cancers, indicating its potential clinical significance. However, the role of MEX3A in hepatocellular carcinoma (HCC) remains largely unexplored, with limited reports available in the literature.
To investigate expression and clinical significance of MEX3A in HCC and explore its potential role in tumor progression.
We analyzed MEX3A mRNA expression in HCC and adjacent tissues using data from The Cancer Genome Atlas (TCGA). The correlation between MEX3A expression and overall survival (OS) was evaluated. Immunohistochemistry was performed on HCC surgical specimens to validate MEX3A expression and its association with clinical parameters, including hepatitis B virus (HBV) positivity, tumor differentiation and tumor size. Additionally, MEX3A knockdown HCC cell lines were constructed to explore the biological functions of MEX3A. Cell proliferation was assessed using cell counting kit-8 and clone formation assays, while cell cycle progression was analyzed by flow cytometry. The effects of MEX3A on the Wnt/β-catenin signaling pathway were examined by western blotting and immunofluorescence. Cell migration was evaluated using scratch and Transwell assays. Finally, the role of the transcription factor RORA in mediating MEX3A effects was explored by silencing RORA and analyzing its impact on cell proliferation and protein expression.
TCGA data analysis revealed that MEX3A mRNA expression was significantly higher in HCC tissues compared to adjacent tissues. Higher MEX3A expression was associated with poorer OS. These findings were validated in HCC surgical specimens. Immunohistochemistry confirmed elevated MEX3A expression in HCC tissues and showed positive correlations with Ki-67 and vimentin levels. MEX3A expression was closely related to HBV positivity, tumor differentiation and tumor size. Mechanistic studies demonstrated that MEX3A knockdown inhibited cell proliferation and cell cycle progression, as shown by reduced expression of β-catenin, c-Myc and cyclin D1. Additionally, MEX3A knockdown inhibited the nuclear entry of β-catenin, thereby suppressing the activation of downstream oncogenic pathways. MEX3A depletion significantly reduced the migratory ability of HCC cells, likely through downregulation of the epithelial-mesenchymal transition pathway. Transcription factor analysis identified RORA as a potential mediator of MEX3A effects. Silencing RORA antagonized the effects of MEX3A on cell proliferation and the expression of β-catenin, c-Myc and cyclin D1.
MEX3A promotes cell proliferation in HCC by regulating the RORA/β-catenin pathway. Our findings suggest that MEX3A could serve as a prognostic marker and therapeutic target for HCC.
Core Tip: This study found that MEX3A was highly expressed in hepatocellular carcinoma (HCC) and associated with poor prognosis. Immunohistochemistry showed that MEX3A expression was positively correlated with Ki-67 and vimentin, and closely related to clinical parameters. Knockdown of MEX3A inhibited cell proliferation and cell cycle progression; attenuated expression of β-catenin, c-Myc and cyclin D1; inhibited nuclear entry of β-catenin; and downregulated epithelial-mesenchymal transition to inhibit cell migration. Transcription factor analysis found that RORA might mediate the effects of MEX3A. In conclusion, MEX3A may promote proliferation of HCC cells by regulating the RORA/β-catenin pathway.
- Citation: Ji PX, Zhang P, Zhou HL, Yu H, Fu Y. MEX3A promotes cell proliferation by regulating the RORA/β-catenin pathway in hepatocellular carcinoma. World J Gastrointest Oncol 2025; 17(4): 102084
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/102084.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.102084
Primary liver cancer is one of the most common malignant tumors and it ranks sixth in incidence and third in mortality among all malignancies[1]. Hepatocellular carcinoma (HCC) accounts for 75%-85% of all liver cancer cases[2]. Due to its insidious onset, insignificant early symptoms, easy metastasis and recurrence, the treatment efficacy of HCC is unsatisfactory. According to data from the National Cancer Institute, the incidence of liver cancer remained stable in males but continued to rise slowly in females during 2013 to 2019. The 5-year relative survival rate for liver cancer patients diagnosed during this period was approximately 22%[3]. Cancer treatment has made significant progress in recent years, with advances in surgery, radiotherapy, chemotherapy, targeted therapy and immunotherapy. These treatments have improved patient outcomes and survival rates, but challenges remain, including the need for more personalized and effective therapies to address the complexity and heterogeneity of cancer[4]. Therefore, it is important to find effective prognostic markers and therapeutic targets to realize the early diagnosis and treatment of HCC.
MEX3A is a member of the human homologous gene MEX3 family (MEX3A, MEX3B, MEX3C and MEX3D), located on human chromosome 1q22. The MEX3A protein was first discovered in Caenorhabditis elegans (C. elegans) by Buchet-Poyau et al[5] and this protein acts as a translation regulator to inhibit the translation of plasminogen activator inhibitor-1 in the early embryos of C. elegans. With the in-depth study of this gene, it has been discovered that MEX3A is also closely related to the occurrence of many diseases, such as cancer. Expression of MEX3A in cancer tissues is significantly higher than that in adjacent tissues in bladder and ovarian cancers[6,7]. Jiang et al[8] found that knockdown of MEX3A gene expression inhibited the proliferation, migration and clone formation ability of gastric cancer cells.
These studies indicated that MEX3A functions as an oncogene, promoting cancer development. Hu et al[9] found 13 genes, including MEX3A, whose high expression was related to poor prognosis in HCC by using The Cancer Genome Atlas (TCGA) database-derived cohort transcriptome profiles. Ding et al[10] found that the MEX3A had a higher mutation frequency in HCC patients, and its expression was often elevated in high-risk patients. However, the function and mechanism of MEX3A in HCC has not been well studied.
β-Catenin is a key component of the Wnt signaling pathway and plays crucial roles in various diseases other than cancer. In osteoporosis, studies have shown that icariin exerts anti osteoporotic effects in ovariectomized rats through the Wnt/β-catenin pathway, highlighting its potential therapeutic application[11]. In pulmonary fibrosis, Wnt/β-catenin signaling is implicated in the pathogenesis, with research indicating that a WNT mimetic can reduce fibrosis and improve lung function in bleomycin-induced models[12]. β-Catenin plays a crucial role in HCC. It can promote the proliferation, migration and invasion of HCC cells by activating the Wnt/β-catenin signaling pathway. For example, mutations in the CTNNB1 gene encoding β-catenin can increase glutamine production, which affects tumor epigenetics and other related pathways, thereby driving HCC progression[13]. Additionally, the expression of β-catenin in HCC tissues is negatively correlated with the degree of pathological differentiation, and its high expression indicates poor prognosis. Research has shown that the average survival time of patients with positive β-catenin expression is significantly shorter than that of those with negative expression[14,15]. In this study, we confirmed high expression of MEX3A in HCC tissues and preliminarily explored the function and molecular mechanism of MEX3A in the proliferation and migration of HCC. We found that the RORA/β-catenin signaling pathway and epithelial-mesenchymal transition (EMT) pathway were implicated in MEX3A-mediated cell growth and migration.
In our study, 26 patients with histologically confirmed HCC tissues were obtained from Taizhou People's Hospital. Tumor and adjacent nontumor tissues were collected. All specimens were obtained at the time of surgery, then analyzed immunohistochemically. Our study was approved by the Ethics Committee of Taizhou People's Hospital (KY2019040). We confirmed that all patients provided informed consent.
RNA sequencing data and the corresponding clinical data of HCC patients were downloaded from the TCGA database (http://cancergenome.nih.gov/), which contained information of 369 HCC tissues and 52 adjacent nontumor liver tissues. MEX3A expression data from the tumor and adjacent nontumor tissues were analyzed using the ggstatsplot package of R software (version 3.4.1).
Kaplan-Meier analysis was performed to evaluate the association between MEX3A expression levels and overall survival (OS) in HCC patients. Patients were stratified into two groups based on high and low MEX3A expression levels. The criteria for stratification were determined by the median expression level of MEX3A in HCC tissues. Specifically, the high-expression group included patients with MEX3A expression levels above the median (n = 170), while the low-expression group included those with MEX3A expression levels below the median (n = 171). The analysis was performed using R software with the survival package. The hazard ratios and 95% confidence intervals were calculated to quantify the risk of death associated with high MEX3A expression levels.
HCC tissues were divided into MEX3A-high and MEX3A-low expression groups (top and bottom 10% of MEX3A expression levels) and differentially expressed genes were analyzed with the R package edge R[16]. Using the hallmark gene set in the MSigDB database[17] as the reference gene set, GSEA analysis was performed by the R package cluster Profiler[18] and plotted using the R package enrich plot[19].
Samples were divided into high/low expression groups (10%) according to MEX3A expression, and differential gene expression analysis was performed by the R package edge R[16], and the sets of differentially expressed genes that were up- and downregulated (in the same way as the previous one) were obtained. Transcription factor analysis was performed by the R package RcisTarget[20], and tabulation was performed by using the R package DT (https://CRAN.R-project.org/package=DT).
Spearman correlations between the expression of RORA, c-Myc, SRY-Box Transcription Factor 9 (SOX9) and MEX3A of HCC tissues were analyzed and visualized by the R package ggpubr (https://CRAN.R-project.org/package=ggpubr).
The PLKO.1 (#10879) plasmid purchased from add gene was linearized by Age I (Thermo Scientific, MA, United States) and EcoRI (Thermo). The two synthesized shRNAs (shMEX3A-1: CCGGAGGCAAGGCTGCAAGATTAAGCTCGAGCTTAATCTTGCAGCCTTGCCTTTTTTG; shMEX3A-2: CCGGCACGCAAGCCATCCGAATATTCTCGAGAATATTCGGATGGCTTGCGTGTTTTTG) were annealed and inserted into the linearized vector by T4 DNA ligase (Thermo Scientific). After successful sequencing, they were used for subsequent transfection experiments. The siRNA of RORA (5’-GCUUGUAUGCAGAAGUACA-3’) was synthesized by Tsingke Biotechnology (Beijing, China).
Human hepatoma cell lines HepG2 and MHCC-97H were donated from Minghua Wang's group at Soochow University. Both cell lines were cultured in Dulbecco's modified Eagle' smedium (Hyclone, Utah, United States) containing 10% fetal bovine serum (BI, Israel) and 1% penicillin-streptomycin (Thermo Scientific) in a humidified incubator at 37°C and 5% CO2, and passages were performed every 2 days.
HCC cells (2 × 105) were inoculated into six-well plates, transfected 12-24 hours after inoculation; 2 μg plasmid and Lipofectamine 3000 Transfection Reagent (Invitrogen, CA, United States) were premixed into six-well plates, and the medium was changed after 6 hours. After 24 hours of transfection, the cells were subjected to selection with 2 μg/mL puromycin for 72 hours, the concentration was reduced to 1 μg/mL for long-term culture. After four to five generations of screening, the stable strain was constructed and used for subsequent experiments.
The constructed negative control and knockdown cell lines were digested with trypsin and diluted into a cell suspension. A cell counting plate was used to count the cell number, and 103 cells were inoculated in each well of a 96-well plate. Four duplicate wells were set in each experimental group and the control group, and seeded into the 96-well to allow the cells to adhere. Cell counting kit-8 (CCK-8) (NCM biotech, Jiangsu, China) was put back into the cell incubator for 2 hours incubation, and absorbance at 450 nm wavelength was detected by a microplate reader (Thermo Scientific). Subsequently, the 96-well plates were taken out every 24 hours for repeat detection. The cell growth curve was drawn with the cell culture time as the horizontal axis and OD 450 as the vertical axis.
The constructed stable strain was digested to a cell suspension, and diluted to a final concentration of 500 cells/mL. The cell suspension was inoculated into a 12-well plate at 1 mL per well. Three replicates were set up for the experimental and control groups. The plates were shaken gently to disperse the cells evenly and placed in a 37 °C, 5% CO2 incubator for static culture. Medium was replaced every 3-4 days, and the plates were cultured continuously for 2 weeks. The plates were washed, fixed and stained with 0.1% crystalline violet solution. The formed clones were observed and counted with Image J. GraphPad was performed for statistical analysis.
Cells were treated with serum-free medium for 24 hours to achieve synchronization, and serum-containing medium was then added for 48 hours. Cells were digested, washed and fixed overnight at -20 °C, then stained with propidium iodide at 37 °C for 30 minutes, and RNase was added to remove RNA at the same time. Finally, flow cytometry was performed for cell cycle detection.
The cells were seeded into a six-well plate to reach 90% confluence. Artificial gaps were created by scraping with a 200 mL pipette tip. The wound area was photographed with a phase contrast microscope at 0 and 24 hours post-scratch. The images were analyzed by Image J software 1.35 and mobility was calculated.
The cells of the experimental and control groups were digested and resuspended in serum-free medium. The cell concentration was adjusted to 5 × 105 cells /mL. The Transwell chamber was placed in a 24-well plate, and 600 μL medium containing20% serum was added to the lower chamber. Uniformly mixed cell suspension (200 μL) was taken and added dropwise into the upper chamber, and cultured for 24 hours. The chamber was taken out, fixed and stained. Five randomly selected fields of view were observed under the microscope and photographs were taken.
The reverse transcription system was prepared according to the instructions of the Novizinkit (Vazyme, Jiangsu, China). The reaction product cDNA was placed on ice for subsequent operations or stored at -80 °C. Real-time fluorescent quantitative PCR (RT-qPCR) was used to detect the mRNA levels of genes in cells. ACTB was used as an internal reference, and the quantitative primers used were all designed by NCBI (https://www.ncbi.nlm.nih.gov/). The primer sequences are shown in Supplementary Table 1.
The cells were harvested and the protein was extracted. The protein concentration was corrected by the BCA protein quantification kit (Beyotime, Shanghai, China). Equal amounts of protein (30 mg) were loaded on each lane and separated by SDS/PAGE, followed by transfer to PVDF membranes. The membranes (Millipore, MA, United States) were blocked in 5% skimmed milk and incubated with primary antibody overnight at 4 °C. The antibodies againstMEX3A, c-Myc, β-catenin, cyclin D1, E-cadherin, N-cadherin, p21, RORA, β-actin and GAPDH were all obtained from Abcam (London, United Kingdom). After washing with TBST, the membranes were incubated with secondary antibody for 90 minutes at 37 °C. Finally, bands were visualized with an ECL and imaged by Tanon imaging system.
Tissue sections (4 mm) were heated at 60 °C for 30 minutes and treated with 3% H2O2 at 25 °C for 10 minutes. Slides were soaked in distilled water for 10 minutes, blocked with10% BSA at 25 °C for 30 minutes, and incubated with the primary antibody MEX3A (1: 400 dilution) overnight. Tissue sections were rinsed twice for 5 minutes with phosphate-buffered saline (PBS). These sections were incubated for 1 hour at 25 °C and diaminobenzidine was added. The sections were stained with hematoxylin for 30 sto stain the nuclei. According to the intensity of staining and the percentage of positively stained cells, expression of MEX3A protein was divided into low and high. After determining expression of MEX3A, immunohistochemical staining was repeated for Ki-67 (with a 1:500 dilution) and vimentin (with a 1:200 dilution) in the low and high MEX3A expression groups. The antibodies against Ki-67 and vimentin were all obtained from Abcam.
The cells were washed with PBS. The tissue was fixed with 4% (v/v) paraformaldehyde for 15 minutes, and 0.2% Triton X-100 was used to permeabilize the tissue for 20 minutes. The cells were blocked with 10% goat serum (v/v) for 1 hour. Subsequently, tissues were incubated with primary antibodies at 37 °C for 2 hand exposed to secondary antibody (1: 50; Bio world Technology) at 20 °C for 2 hours. Nuclear counterstaining was then performed using 4′,6-diamidino-2-phenylindole (Beyotime) for 5 minutes. The stained samples were imaged under a fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany).
At least three independent experiments were performed for each experiment, and each experiment was set up with at least three replicates. The mean ± SD was used as the measurement data. Image J software was used for cell counting, GraphPad prism 8 software was used for statistical analysis, and t-test was used to compare the means between different groups. P < 0.05 was considered to be statistically significant.
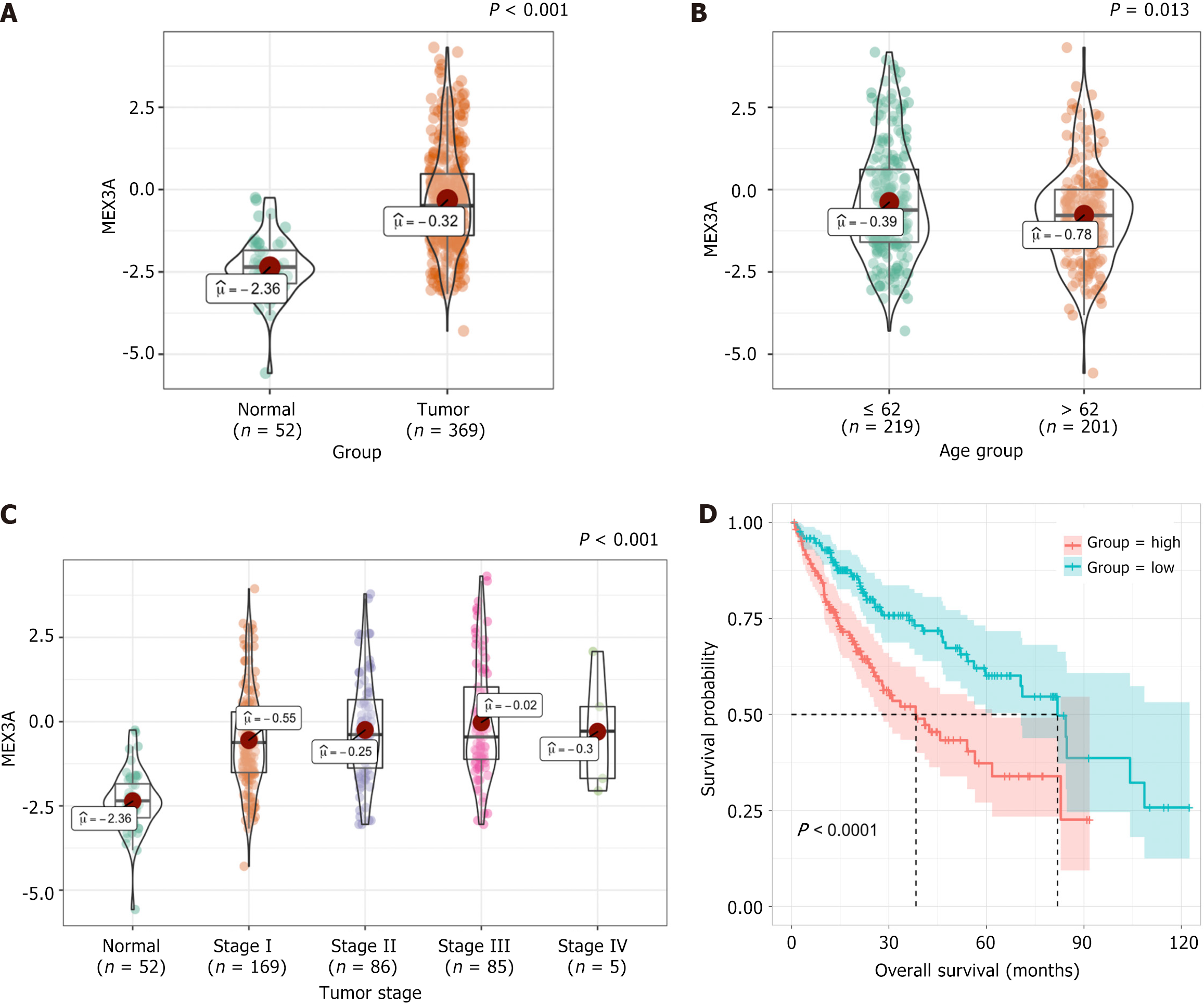
The TCGA (https://cancergenome.nih.gov/) database was used to query MEX3A mRNA expression. Expression of MEX3A in HCC tissues was significantly higher than that in normal tissues (Figure 1A). Expression of MEX3A was significantly associated with age (Figure 1B) and clinical stage, which was increased in Stages I-IV (Figure 1C). Kaplan-Meier analysis revealed that patients with high expression of MEX3A exhibited a shorter OS than those with low expression. These results suggest that elevated MEX3A expression is a potential prognostic marker for poor outcomes in HCC (Figure 1D).
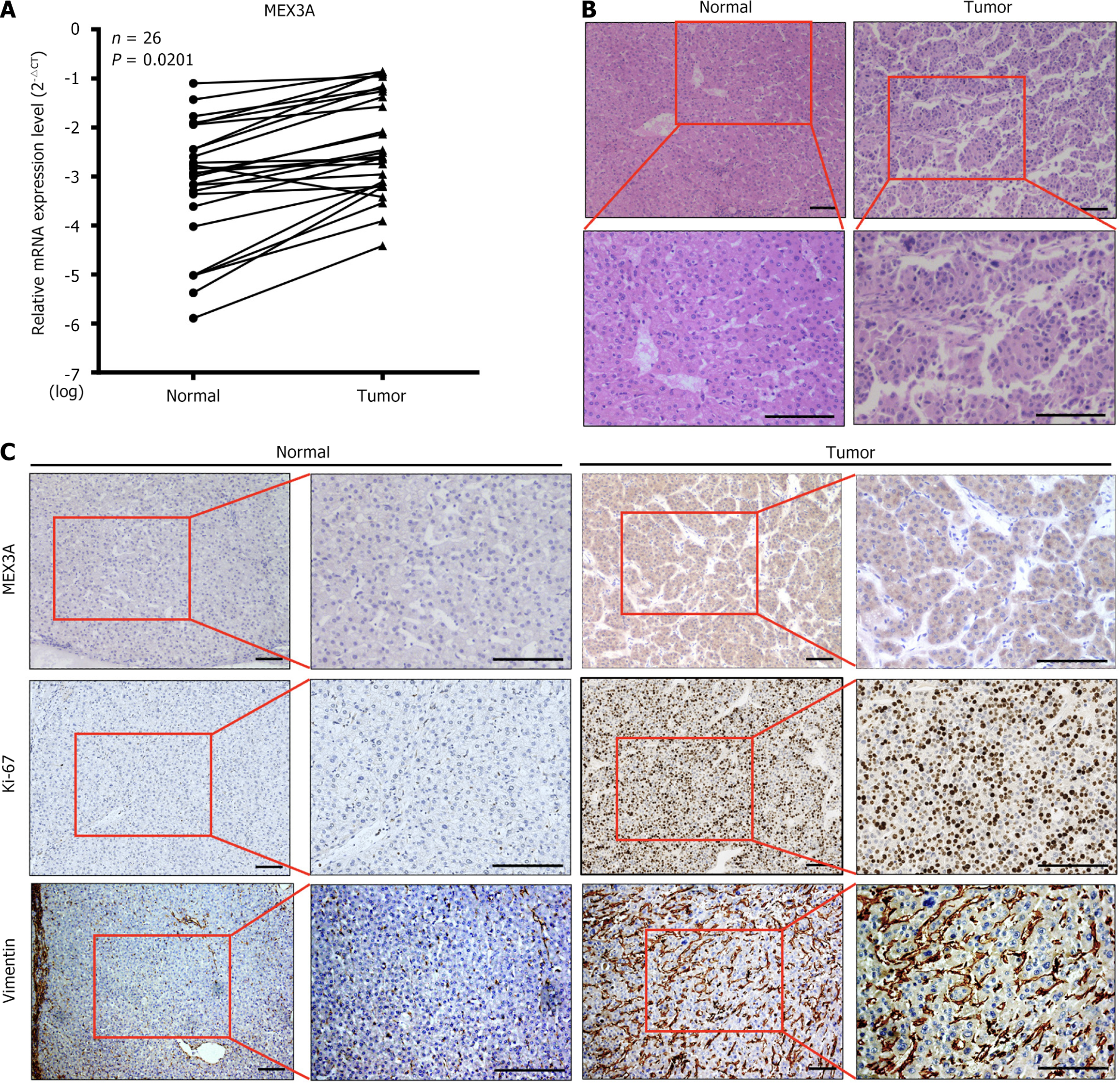
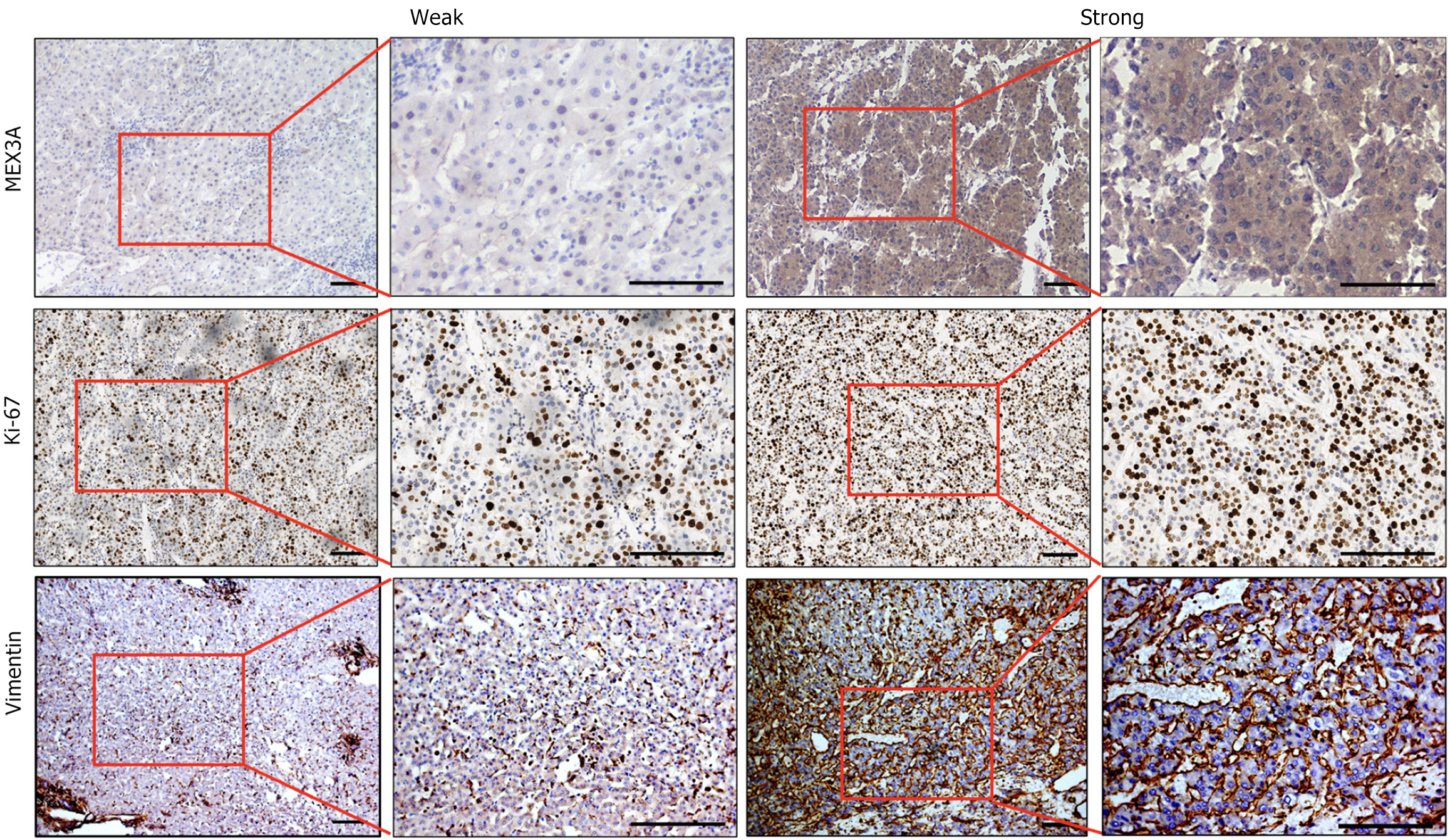
RT-qPCR was performed on 26 surgical specimens of HCC. Expression of MEX3A mRNA in cancer tissues was higher than that in adjacent noncancerous tissues (Figure 2A), which was consistent with the results from the TCGA database. Hematoxylin and eosin staining demonstrated that HCC tissues consisted of tumor cells that resembled hepatocytes. Tumor cells showed marked variation in cellular and nuclear size, shape and staining (Figure 2B). Immunohistochemical staining was used to measure expression of MEX3A protein in tissues. Among the 26 pairs of HCC and adjacent nontumor tissues, MEX3A was positively expressed in all HCC tissues, while negatively expressed in adjacent normal tissues. The cell proliferation marker Ki-67 is expressed in the nucleus, whereas mesenchymal marker vimentin is predominantly expressed in the intercellular substance. Their levels in cancer tissues were higher than those in adjacent tissues (Figure 2C). The number of MEX3A-positive cells in all cancer tissues was > 30%, and in 14 cases, the number was > 50%. The strong positive rate of MEX3A protein expression in the HCC tissues was 65.4% (17/26), and the weak positive rate was 34.6% (9/26) (Table 1). We further selected representative samples with high and low expression of MEX3A for Ki-67 and vimentin staining. Expression of Ki-67 and vimentin was consistent with that of MEX3A (Figure 3). This result suggests that the increase of MEX3A promotes proliferation and metastasis of HCC, and MEX3A could be used as a biomarker for diagnosis of HCC.
| Parameters | MEX3A expression | ||||
| n = 26 | Low (9) | High (17) | P value | ||
| Gender | Male | 24 | 8 | 16 | 1 |
| Female | 2 | 1 | 1 | ||
| Age (year) | < 65 | 14 | 5 | 9 | 1 |
| 65 | 12 | 4 | 8 | ||
| TNM stage | T1, T2 | 7 | 1 | 6 | 0.3574 |
| T3, T4 | 19 | 8 | 11 | ||
| HBV | + | 16 | 3 | 13 | 0.0461a |
| - | 10 | 6 | 4 | ||
| AFP (ng/mL) | < 400 | 19 | 6 | 13 | 0.6613 |
| 400 | 7 | 3 | 4 | ||
| Tumor diameter | < 5 | 10 | 7 | 3 | 0.0085a |
| 5 | 16 | 2 | 14 | ||
| Differentiation | High | 9 | 6 | 3 | 0.0277a |
| Low | 17 | 3 | 14 | ||
| Hepatocirrhosis | + | 16 | 5 | 11 | 0.6924 |
| - | 10 | 4 | 6 | ||
| Vascular invasion | + | 21 | 7 | 14 | 1 |
| - | 5 | 2 | 3 | ||
The relationship between MEX3A expression and clinical features of 26 cases of HCC was statistically analyzed. High expression of MEX3A was correlated with the tumor diameter (P = 0.0085), differentiation (P = 0.0277) and hepatitis B virus (HBV) positivity (P = 0.0461), but there was no significant association between MEX3A expression and gender, age, tumor-node-metastasis stage, serum a-fetoprotein level, liver cirrhosis and vascular invasion (Table 1). Studies have found that SOX9 and c-Myc contribute to the stemness characteristics in HCC[21,22]. Spearman correlations analysis found a significant positive correlation between expression of MEX3A and HCC differentiation factors SOX9 and c-Myc (Supplementary Figure 1), suggesting that MEX3A correlates with HCC differentiation. In conclusion, MEX3A may be a potential prognostic biomarker for HCC.
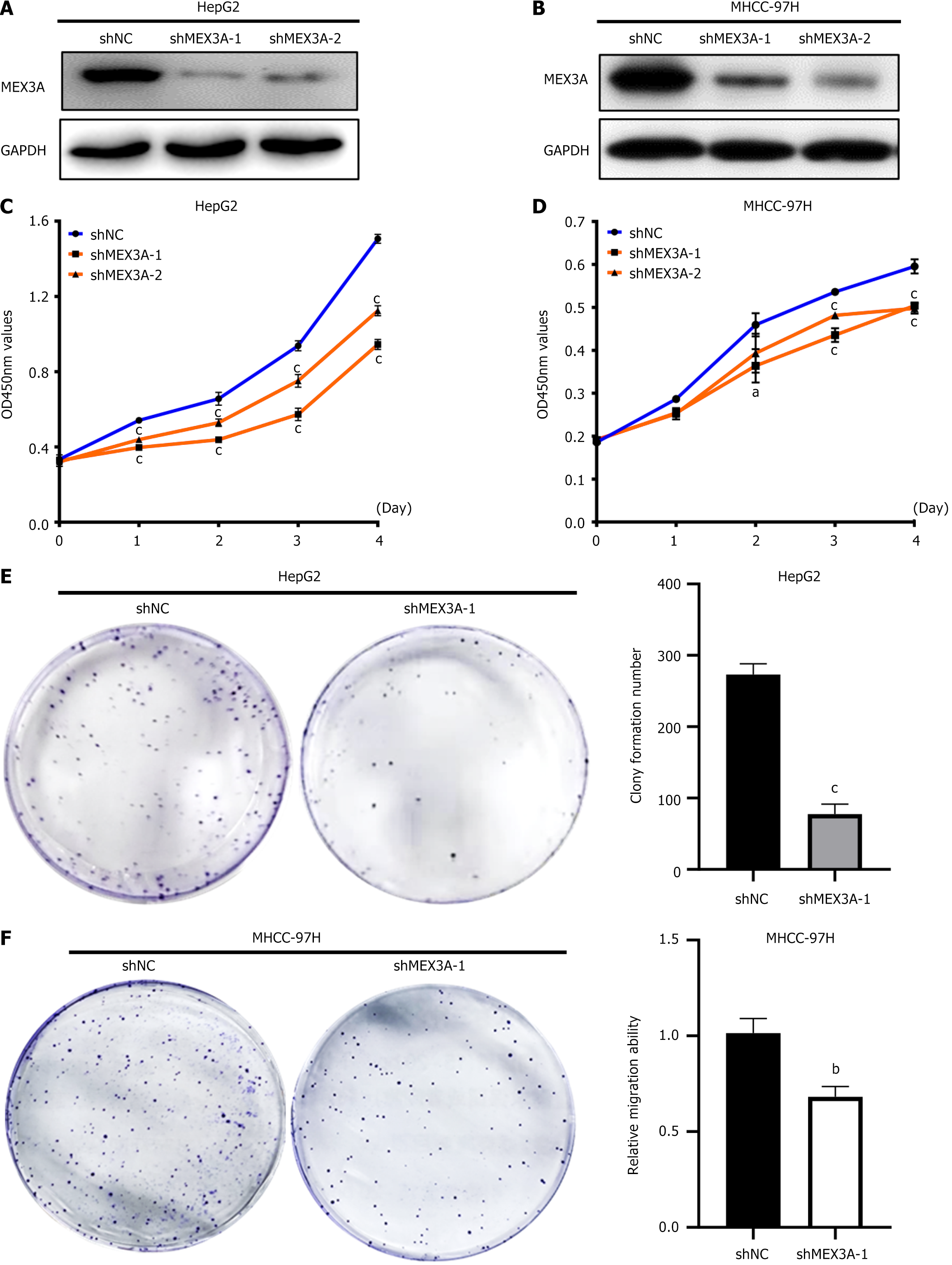
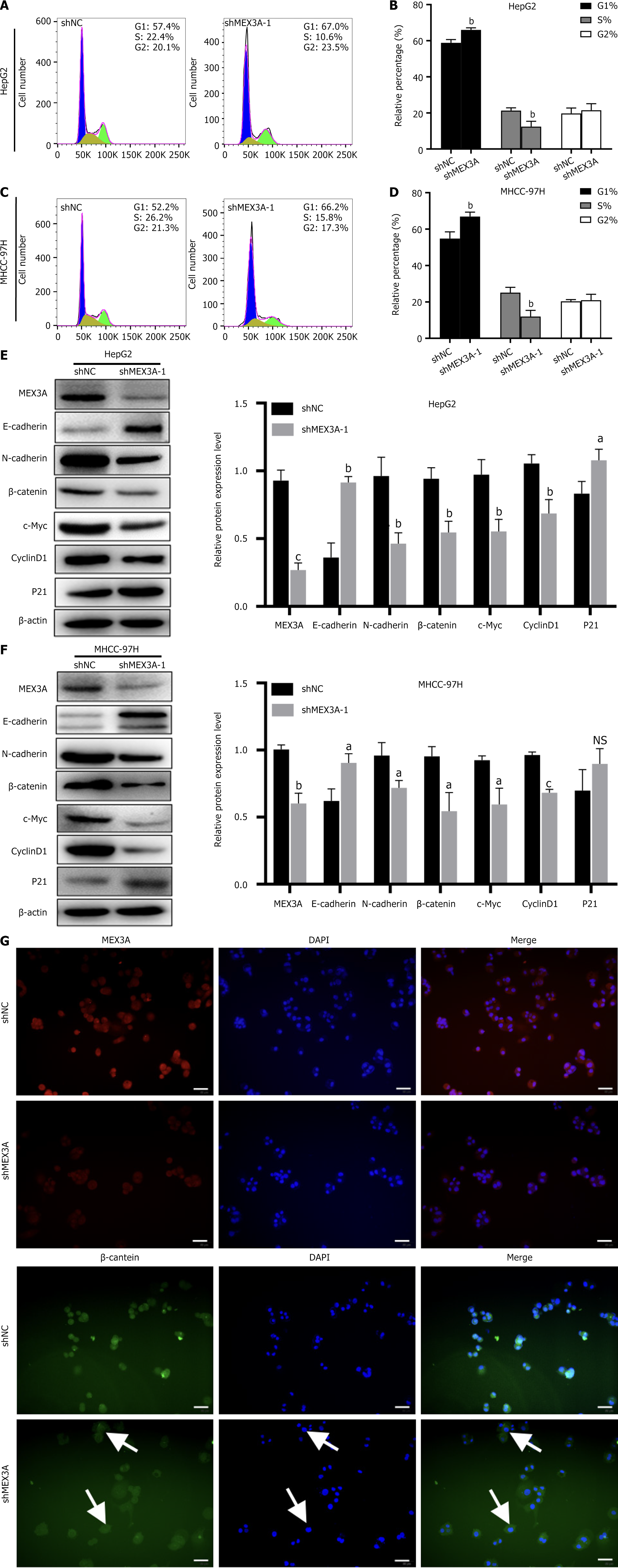
To explore the biological function of MEX3A in HCC, we designed two MEX3A shRNA sequences and constructed the stable transfected strains in HCC cell lines HepG2 and MHCC-97H, which were denoted as shMEX3A-1 and shMEX3A-2, respectively. The control group was denoted as shNC. Western blotting showed that both shRNA sequences downregulated expression of MEX3A (Figure 4A and B). We explored the effect of knockdown of MEX3A on cell proliferation in HepG2 and MHCC-97H cells. Using the previously constructed control group (shNC) and MEX3A knockdown group (shMEX3A-1, shMEX3A-2) cell lines, CCK-8 assay was used to detect cell growth. The proliferation of HepG2 and MHCC-97H cells was significantly inhibited after MEX3A knockdown, especially at days 3 and 4 (Figure 4C and D). Since the inhibitory role of shMEX3A-1 on HCC cell proliferation was more pronounced, we selected it for the subsequent study. The effect of MEX3A on clone formation was examined and showed that MEX3A silencing significantly inhibited clone formation compared with the control group (Figure 4E and F). These results indicated that MEX3A could promote cell proliferation in HCC cells and might play an important role in the occurrence and development of HCC.
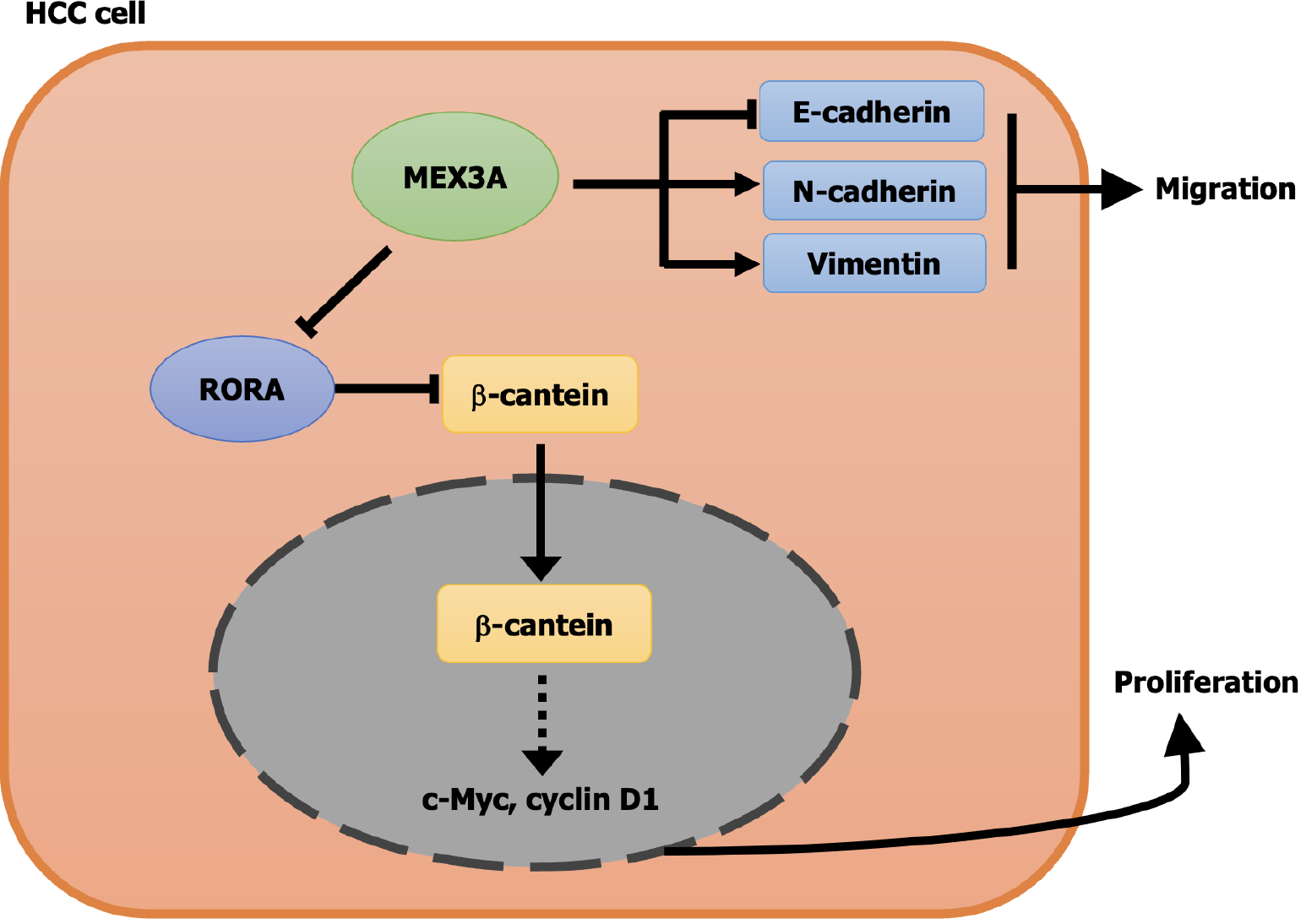
We explored the role of MEX3A in the cell cycle. Flow cytometry showed that inhibition of MEX3A expression resulted in an increase in G1 phase cells and a decrease percentage in S phase HepG2 cells (Figure 5A and B) and MHCC-97H cells (Figure 5C and D), which indicated that knockdown of MEX3A inhibited cell proliferation, resulting in cell cycle G1 phase arrest. The Wnt/β-catenin signaling pathway plays crucial roles in HCC progression, including proliferation and migration[23] and c-Myc and cyclin D1 are targets of the Wnt/β-catenin pathway[24]. P21 is a broad-spectrum cell cycle-dependent kinase inhibitor and a negative regulator of the cell cycle with the ability to prevent cells from passing through the G1/S phase[25]. Therefore, we examined the effect of MEX3A silencing on these proteins. Knockdown of MEX3A decreased expression of β-catenin, c-Myc and cyclin D1 but increased expression of p21 (Figure 5E and F). We evaluated the localization of β-catenin in the cells by immunofluorescence. The results indicated that knockdown of MEX3A leads to decreased expression of β-catenin in the nucleus, which meant that the activity of Wnt/β-catenin pathway was inhibited (Figure 5G). These results suggested that MEX3A promoted HCC proliferation through enhancing the activity of Wnt/β-catenin signaling pathway.
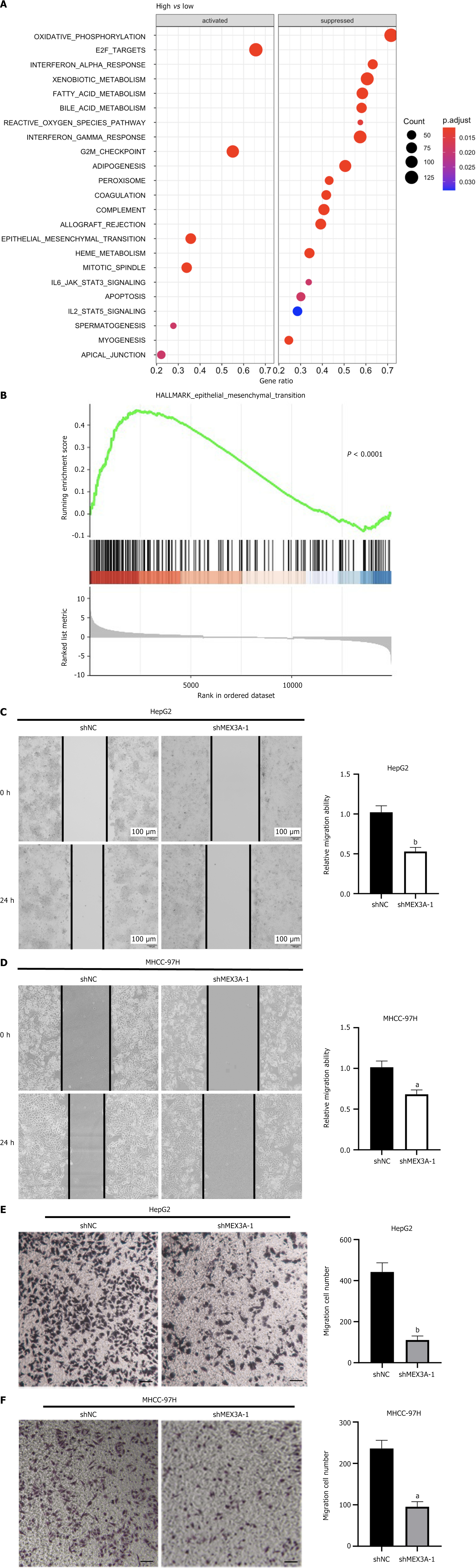
Gene set enrichment analysis (GSEA) found that MEX3A has important effects on genes that were mainly related to oxidative phosphorylation, G2/M checkpoint, EMT and the JAK/STAT pathway (Figure 6A). Also, some evidence has shown that MEX3A promotes the invasion and metastasis of pancreatic ductal adenocarcinoma by affecting expression of EMT-related proteins, such as N-cadherin[26]. Further GSEA revealed that high expression of MEX3A was enriched in the EMT pathway in HCC (Figure 6B), indicating that MEX3A influenced the cell migration of HCC through the EMT pathway. We explored the effect of MEX3A on the migration of HCC by cell scratch and Transwell experiments. Compared with the shNC groups, the shMEX3A groups of the two cell lines showed low migration (Figure 6C-F), indicating that knockdown of MEX3A impairs the migratory ability of HCC cells. The expression of E-cadherin was increased and N-cadherin was decreased by MEX3A silencing (Figure 5E and F), further supporting that MEX3A may facilitate cell migration through the EMT pathway in HCC.
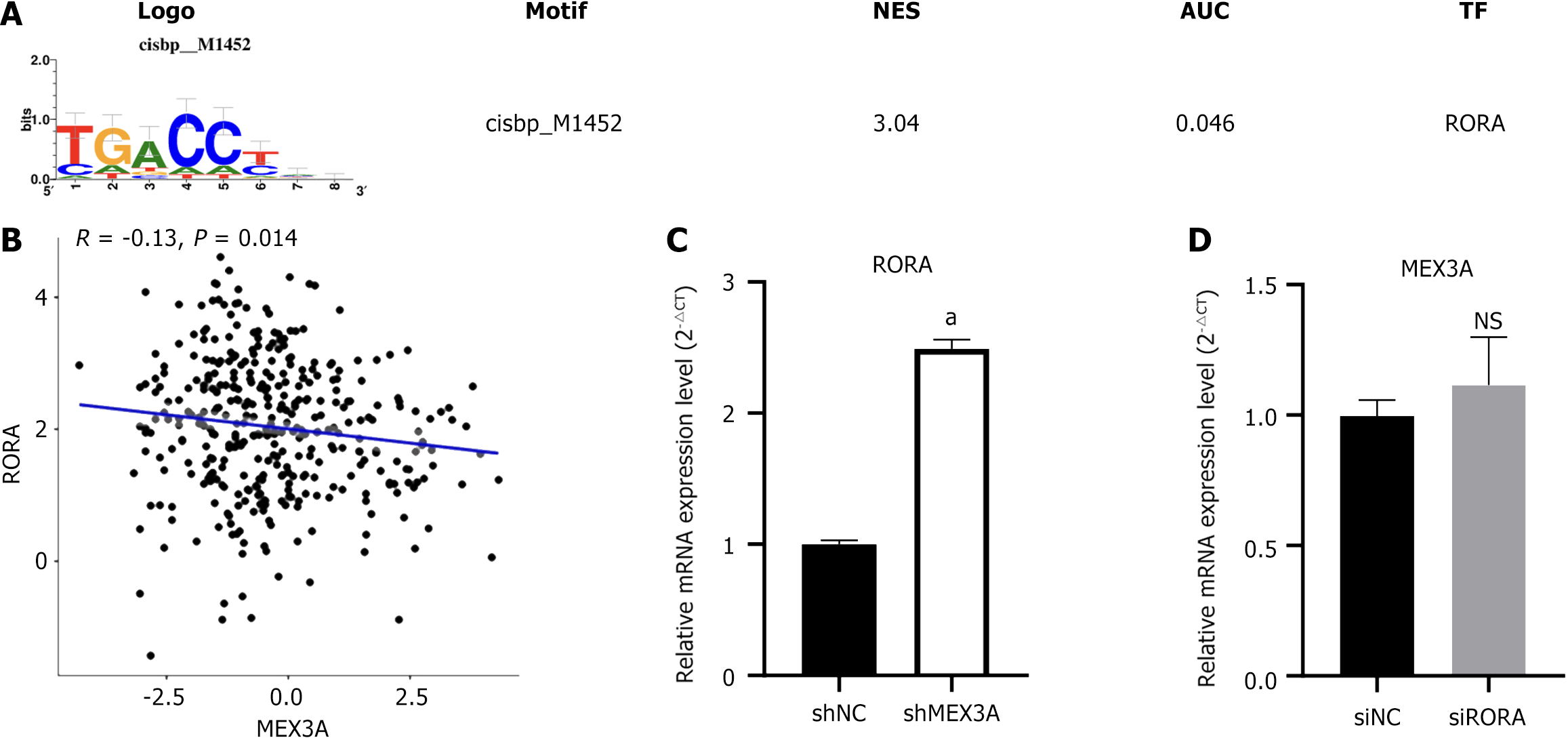
Through transcription factor analysis, we found there was a motif of transcription factor RORA in the promoter region of differential genes related to expression of MEX3A (Figure 7A), suggesting that RORA might regulate expression of differential genes at transcriptional level. To verify the correlation between MEX3A and RORA, we conducted correlation analysis and found that MEX3A and RORA were negatively correlated (Figure 7B). The upstream and downstream relationship between differential genes and MEX3A is not clear, and RORA may be upstream or downstream of MEX3A. We synthesized siRNA to inhibit expression of RORA and found that knockdown of MEX3A upregulated expression of RORA in HepG2 cells, while knockdown of RORA had no significant effect on MEX3A expression (Figure 7C and D), which suggests that RORA is a downstream gene of MEX3A.
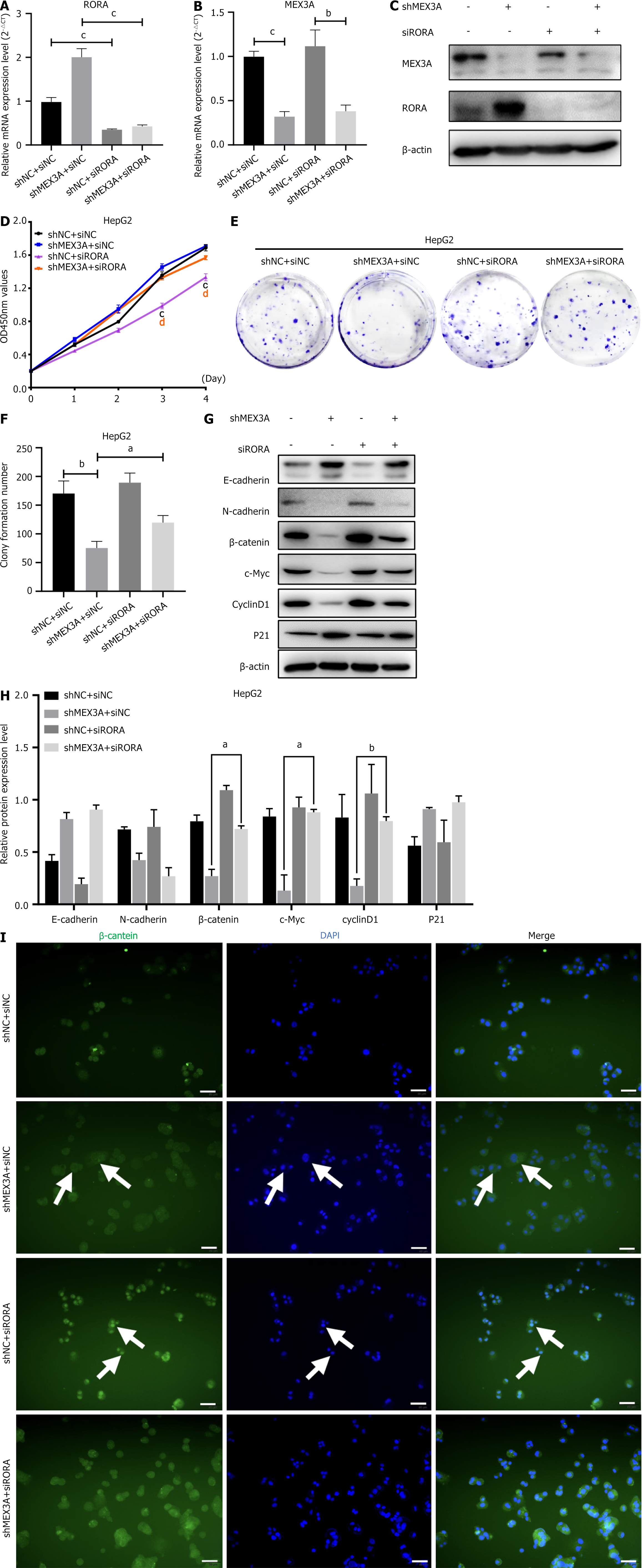
It has been reported that RORA plays a tumor-inhibitory role in several cancers by attenuating the Wnt/β-catenin pathway[27-29]. To test whether MEX3A stimulates cell growth in HCC through RORA/β-catenin, we transfected RORA siRNA in shMEX3A-HepG2 cell line to construct a double knockdown cell model. RT-qPCR and western blotting showed that expression of RORA obviously increased in shMEX3A group, while apparently decreased in double knockdown group (Figure 8A-C), which proved again that RORA is downstream of MEX3A. Next, we investigated the impact of three different genetic knockdown strategies on cell proliferation in HepG2 cells. Specifically, we compared the effects of the shMEX3A group, the RORA siRNA group, and the MEX3A/RORA double knockdown group. CCK-8 assay and clone formation showed that MEX3A knockdown inhibited cell growth, which is consistent with the previous results. Compared with the MEX3A knockdown group, cell growth was increased in the MEX3A/RORA double knockdown group (Figure 8D-F), indicating that interference of RORA could compensate for the proliferation inhibition in HepG2 cells induced by knockdown of MEX3A. Western blotting was used to examine the effect of RORA on protein levels in the Wnt/β-catenin pathway. Downregulation of β-catenin expression caused by MEX3A knockdown was partially reversed in the double knockdown group. Similar results were found for c-Myc and cyclin D1 (Figure 8G and H). Additionally, the immunofluorescence experiments revealed that silencing RORA can counteract the inhibitory effect of MEX3A knockdown on β-catenin nuclear translocation (Figure 8I). These data suggested that RORA was at least partially involved in the activation of the Wnt/β-catenin pathway induced by MEX3A. However, knockdown of RORA had little effect on expression of E-cadherin and N-cadherin (Figure 8G and H), suggesting that MEX3A regulates the EMT pathway not through RORA, but by an alternative mechanism.
In this study, the TCGA data were collected and subjected to bioinformatics analysis. The results showed that there was a very significant upregulation of MEX3A mRNA expression in HCC compared with adjacent tissues, and the expression of this gene was closely related to the age, clinical stage and the poor prognosis of liver cancer, which agrees with the report of Yang et al[30]. The TCGA database has been widely used in cancer research, encompassing studies on single genes in individual cancers[31], gene sets in individual cancers[32], single genes across multiple cancers (pan-cancer)[33], and gene sets across multiple cancers (pan-cancer)[34]. Although the TCGA database offers a wealth of resources for cancer research, it also has some potential biases. The generation and analysis of TCGA data may have technical limitations, and gene expression is complex and multifaceted. These factors need to be considered carefully[35].
Our initial bioinformatics analysis of TCGA data revealed significant insights into the role of MEX3A in HCC, highlighting its potential as a biomarker and therapeutic target. Building on these findings, we proceeded to validate these observations through experimental studies using clinical HCC specimens. This approach allowed us to confirm the bioinformatic predictions and delve deeper into the clinical significance of MEX3A expression in HCC.
Subsequently, 26 pairs of HCC specimens were used to verify the MEX3A expression and relationship between MEX3A and clinical features. The results showed that MEX3A was positively expressed in all HCC tissues, while negatively expressed in adjacent tissues. Ki-67 and vimentin was also detected and the result showed that both were highly expressed in samples with high expression of MEX3A, which hinted that MEX3A might play roles in cell proliferation and migration in HCC. The high expression of MEX3A was associated with tumor diameter and differentiation, indicating that MEX3A plays an important role in the development of HCC. We also found that the expression of MEX3A was correlated with the infection status of HBV, suggesting that MEX3A plays a key role in HBV-related HCC. However, the current study was limited by its small sample size. Future research with larger cohorts is needed to validate the relationship between MEX3A and HBV infection, as well as the specific mechanisms by which MEX3A contributes to the development and progression of HCC. Nevertheless, there was no relationship between MEX3A expression and age or clinical stage. Although the MEX3A mRNA in the TCGA database showed a significant difference with age, the difference in MEX3A mRNA was small. Our immunohistochemistry results indicated that MEX3A protein expression was not affected by age. It is hypothesized that the minor difference in mRNA level may not lead to a significant change in protein expression. In addition, the lack of association with age might have been related to our elatively small sample size. It is reported that SOX9 and c-Myc can act as differentiation markers because of their ability to sustain the stemness characteristics of HCC[36], and we found that expression of MEX3A was positively related to the expression of SOX9 and c-Myc by bioinformatics analysis. Our western blotting results showed that knockdown of MEX3A could decreased expression of c-Myc, which suggests that MEX3A expression is correlated with differentiation of HCC, indicating that MEX3A is a potential prognostic biomarker for HCC. Our CCK-8 assay and clone formation experiment showed that downregulation of MEX3A inhibited growth of HepG2 and MHCC-97H cell lines. Also, knockdown of MEX3A induced cell cycle G1 phase arrest by increasing the number in G1 phase and decreasing the number in S phase, which verifies the proliferative effect of MEX3A on HCC. Cyclins are a family of proteins that play important roles in regulating the cell cycle. Cyclin D1 drives G1/S phase transition[37]. P21 is a cell cycle inhibitor that can prevent cell cycle progression at G1 phase[38]. Fei et al[39] reported that cyclin D1 and p21 participated in G0/G1 cell cycle arrest in HepG2 cells. We found that MEX3A silencing decreased expression of cyclin D1 and increased the level of p21, which blocked the G1/S transition. There is accumulating evidence that the Wnt/β-catenin signaling pathway plays critical roles in progression of HCC[40]. C-Myc and cyclin D1 are downstream molecules of the Wnt/β-catenin pathway[24,41,42]. C-Myc is a well-studied proto-oncogene, and its abnormal expression can lead to 30%-50% of human malignant tumors[43]. Our results showed that knockdown of MEX3A decreased the nuclear translocation of β-catenin, and inhibited expression of c-Myc and cyclin D1. These data demonstrated that MEX3A could promote cell proliferation by activating the Wnt/β-catenin pathway.
HCC progresses rapidly, and its invasiveness is closely linked to its migratory and invasive capabilities. EMT induces cancer cell metastasis and invasion, playing a key role in migration of HCC[44]. E-cadherin, N-cadherin and vimentin are involved in EMT and they are important indicators of this pathway. During EMT, the expression of E-cadherin gradually decreased, while the expression of N-cadherin and vimentin increased[45]. GSEA found that high expression of MEX3A in HCC was closely related to the EMT pathway. The cell scratch and Transwell assays showed that downregulation of MEX3A inhibited migration of HCC. Immunohistochemistry showed that vimentin was highly expressed in HCC tissues. Western blotting found that expression of E-cadherin was upregulated and expression of N-cadherin was downregulated after MEX3A knockdown, indicating that MEX3A could promote HCC migration through the EMT pathway. The EMT pathway plays a complex and diverse role in cancer metastasis, particularly in HCC. Some studies have shown that signaling pathways within the EMT pathway, such as PI3K/PDK1/AKT and Ras-GTP/Raf/MEK/ERK/MAPK, are activated through interactions with various receptor tyrosine kinases of growth factors, leading to expression of EMT factors such as Twist and Snail Homolog 1/2. The ERK/MAPK pathway requires the combination with other signaling pathways, such as Notch, Wnt, and NF-κB, to more effectively induce EMT[46]. Although we have found that MEX3A affects gene expression changes related to EMT, validation of these findings can be considered in future studies using patient-derived xenograft models, which can simulate the heterogeneity and complexity of human cancers, thereby better understanding the metastatic mechanisms of HCC and developing more effective treatment methods[47].
GSEA obtained differential genes from MEX3A high expression and low expression groups. Transcription factor analysis found that a RORA motif in the promoter region of differential genes. Although MEX3A was weakly correlated with RORA, Lee et al[29] have reported that RORA can act as a transcription factor to regulate the Wnt/β-catenin pathway. Therefore, we wanted to observe whether MEX3A can regulate cell proliferation as well as the Wnt/β-catenin pathway through RORA. We found that RORA was located downstream of MEX3A and reversed the cell proliferation induced by MEX3A. Western blotting also revealed that RORA could partly antagonize the effects of MEX3A on the expression of β-catenin, c-Myc and cyclin D1, which suggested that RORA participated in regulation of MEX3A on the Wnt/β-catenin signaling pathway.
In conclusion, MEX3A is highly expressed and plays important roles in HCC, and promotes cell proliferation through the RORA/β-catenin signaling pathway. However, how MEX3A exerts its regulatory effect on RORA is still unknown. Since MEX3A is an RNA-binding protein, it may regulate the expression of RORA at the post-transcriptional level, which will be the subject of our next study. MEX3A can regulate the migration of HCC cells through the EMT pathway, but is not associated with RORA (Figure 9).
Our study demonstrates that MEX3A promotes cell proliferation in HCC by regulating the RORA/β-catenin pathway, highlighting its potential as a prognostic marker and therapeutic target.
We would like to thank all the patients who generously participated in this study.
| 1. | Zhang CH, Cheng Y, Zhang S, Fan J, Gao Q. Changing epidemiology of hepatocellular carcinoma in Asia. Liver Int. 2022;42:2029-2041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 192] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 2. | Ma C, Zhang Q, Greten TF. MDSCs in liver cancer: A critical tumor-promoting player and a potential therapeutic target. Cell Immunol. 2021;361:104295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 3. | . Erratum to "Cancer statistics, 2024". CA Cancer J Clin. 2024;74:203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 33] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 4. | Sonkin D, Thomas A, Teicher BA. Cancer treatments: Past, present, and future. Cancer Genet. 2024;286-287:18-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 173] [Article Influence: 173.0] [Reference Citation Analysis (0)] |
| 5. | Buchet-Poyau K, Courchet J, Le Hir H, Séraphin B, Scoazec JY, Duret L, Domon-Dell C, Freund JN, Billaud M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007;35:1289-1300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 120] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Huang Y, Fang C, Shi JW, Wen Y, Liu D. Identification of hMex-3A and its effect on human bladder cancer cell proliferation. Oncotarget. 2017;8:61215-61225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Li F, Zhao C, Diao Y, Wang Z, Peng J, Yang N, Qiu C, Kong B, Li Y. MEX3A promotes the malignant progression of ovarian cancer by regulating intron retention in TIMELESS. Cell Death Dis. 2022;13:553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Jiang H, Zhang X, Luo J, Dong C, Xue J, Wei W, Chen J, Zhou J, Gao Y, Yang C. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol Med Rep. 2012;6:575-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Hu B, Yang XB, Sang XT. Development and Verification of the Hypoxia-Related and Immune-Associated Prognosis Signature for Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2020;7:315-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 10. | Ding J, He X, Luo W, Zhou W, Chen R, Cao G, Chen B, Xiong M. Development and Validation of a Pyroptosis-Related Signature for Predicting Prognosis in Hepatocellular Carcinoma. Front Genet. 2022;13:801419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Chen G, Wang C, Wang J, Yin S, Gao H, Xiang LU, Liu H, Xiong Y, Wang P, Zhu X, Yang LI, Zhang R. Antiosteoporotic effect of icariin in ovariectomized rats is mediated via the Wnt/β-catenin pathway. Exp Ther Med. 2016;12:279-287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Patel M, Post Y, Hill N, Sura A, Ye J, Fisher T, Suen N, Zhang M, Cheng L, Pribluda A, Chen H, Yeh WC, Li Y, Baribault H, Fletcher RB. A WNT mimetic with broad spectrum FZD-specificity decreases fibrosis and improves function in a pulmonary damage model. Respir Res. 2024;25:153. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 13. | Ziki RA, Colnot S. Glutamine metabolism, a double agent combating or fuelling hepatocellular carcinoma. JHEP Rep. 2024;6:101077. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 14. | Liu L, Zhu XD, Wang WQ, Shen Y, Qin Y, Ren ZG, Sun HC, Tang ZY. Activation of beta-catenin by hypoxia in hepatocellular carcinoma contributes to enhanced metastatic potential and poor prognosis. Clin Cancer Res. 2010;16:2740-2750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 15. | Zhao Z, Cui T, Wei F, Zhou Z, Sun Y, Gao C, Xu X, Zhang H. Wnt/β-Catenin signaling pathway in hepatocellular carcinoma: pathogenic role and therapeutic target. Front Oncol. 2024;14:1367364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 16. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22632] [Cited by in RCA: 28993] [Article Influence: 1812.1] [Reference Citation Analysis (0)] |
| 17. | Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4238] [Cited by in RCA: 8317] [Article Influence: 831.7] [Reference Citation Analysis (0)] |
| 18. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11591] [Cited by in RCA: 22038] [Article Influence: 1695.2] [Reference Citation Analysis (0)] |
| 19. | Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 4694] [Article Influence: 1173.5] [Reference Citation Analysis (0)] |
| 20. | Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, Verbeiren T, De Maeyer D, Reumers J, Saeys Y, Aerts S. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247-2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 738] [Article Influence: 147.6] [Reference Citation Analysis (0)] |
| 21. | Kawai T, Yasuchika K, Ishii T, Miyauchi Y, Kojima H, Yamaoka R, Katayama H, Yoshitoshi EY, Ogiso S, Kita S, Yasuda K, Fukumitsu K, Komori J, Hatano E, Kawaguchi Y, Uemoto S. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci Rep. 2016;6:30489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 22. | Holczbauer Á, Factor VM, Andersen JB, Marquardt JU, Kleiner DE, Raggi C, Kitade M, Seo D, Akita H, Durkin ME, Thorgeirsson SS. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 2013;145:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 23. | Deldar Abad Paskeh M, Mirzaei S, Ashrafizadeh M, Zarrabi A, Sethi G. Wnt/β-Catenin Signaling as a Driver of Hepatocellular Carcinoma Progression: An Emphasis on Molecular Pathways. J Hepatocell Carcinoma. 2021;8:1415-1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 24. | Lecarpentier Y, Schussler O, Hébert JL, Vallée A. Multiple Targets of the Canonical WNT/β-Catenin Signaling in Cancers. Front Oncol. 2019;9:1248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 25. | Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 506] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 26. | Loh CY, Chai JY, Tang TF, Wong WF, Sethi G, Shanmugam MK, Chong PP, Looi CY. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 729] [Cited by in RCA: 852] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 27. | Sun X, Dongol S, Qiu C, Xu Y, Sun C, Zhang Z, Yang X, Zhang Q, Kong B. miR-652 Promotes Tumor Proliferation and Metastasis by Targeting RORA in Endometrial Cancer. Mol Cancer Res. 2018;16:1927-1939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 28. | Huang JL, Fu YP, Gan W, Liu G, Zhou PY, Zhou C, Sun BY, Guan RY, Zhou J, Fan J, Yi Y, Qiu SJ. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 2020;476:140-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 29. | Lee JM, Kim IS, Kim H, Lee JS, Kim K, Yim HY, Jeong J, Kim JH, Kim JY, Lee H, Seo SB, Kim H, Rosenfeld MG, Kim KI, Baek SH. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol Cell. 2010;37:183-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Yang D, Jiao Y, Li Y, Fang X. Clinical characteristics and prognostic value of MEX3A mRNA in liver cancer. PeerJ. 2020;8:e8252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | Liu H, Weng J, Huang CL, Jackson AP. Is the voltage-gated sodium channel β3 subunit (SCN3B) a biomarker for glioma? Funct Integr Genomics. 2024;24:162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Reference Citation Analysis (0)] |
| 32. | Liu H, Tang T. A bioinformatic study of IGFBPs in glioma regarding their diagnostic, prognostic, and therapeutic prediction value. Am J Transl Res. 2023;15:2140-2155. [PubMed] |
| 33. | Liu H, Dilger JP, Lin J. A pan-cancer-bioinformatic-based literature review of TRPM7 in cancers. Pharmacol Ther. 2022;240:108302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 34. | Liu H, Tang T. Pan-cancer genetic analysis of cuproptosis and copper metabolism-related gene set. Front Oncol. 2022;12:952290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 35. | Liu H, Guo Z, Wang P. Genetic expression in cancer research: Challenges and complexity. Gene Rep. 2024;37:102042. [RCA] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 36. | Buchynska LG, Brieieva OV, Iurchenko NP. Assessment of HER-2/neu, с-MYC and CCNE1 gene copy number variations and protein expression in endometrial carcinomas. Exp Oncol. 2019;41:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Park MS, Koff A. Overview of the cell cycle. Curr Protoc Cell Biol. 2001;Chapter 8:Unit 8.1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Huang GL, Zhang W, Ren HY, Zhou P, Chen Y, Chen QX, Shen DY. Oncogenic retinoic acid receptor α promotes human colorectal cancer growth through simultaneously regulating p21 transcription and GSK3β/β-catenin signaling. Cancer Lett. 2017;388:118-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 39. | Fei HR, Chen HL, Xiao T, Chen G, Wang FZ. Caudatin induces cell cycle arrest and caspase-dependent apoptosis in HepG2 cell. Mol Biol Rep. 2012;39:131-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 40. | Leung RWH, Lee TKW. Wnt/β-Catenin Signaling as a Driver of Stemness and Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 41. | He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3533] [Cited by in RCA: 3601] [Article Influence: 133.4] [Reference Citation Analysis (0)] |
| 42. | Jia XX, Zhu TT, Huang Y, Zeng XX, Zhang H, Zhang WX. Wnt/β-catenin signaling pathway regulates asthma airway remodeling by influencing the expression of c-Myc and cyclin D1 via the p38 MAPK-dependent pathway. Exp Ther Med. 2019;18:3431-3438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 43. | Zheng K, Cubero FJ, Nevzorova YA. c-MYC-Making Liver Sick: Role of c-MYC in Hepatic Cell Function, Homeostasis and Disease. Genes (Basel). 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 44. | Xu X, Zhang Y, Wang X, Li S, Tang L. Substrate Stiffness Drives Epithelial to Mesenchymal Transition and Proliferation through the NEAT1-Wnt/β-Catenin Pathway in Liver Cancer. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 45. | Kaszak I, Witkowska-Piłaszewicz O, Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F, Jurka P. Role of Cadherins in Cancer-A Review. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 214] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 46. | Liaghat M, Ferdousmakan S, Mortazavi SH, Yahyazadeh S, Irani A, Banihashemi S, Seyedi Asl FS, Akbari A, Farzam F, Aziziyan F, Bakhtiyari M, Arghavani MJ, Zalpoor H, Nabi-Afjadi M. The impact of epithelial-mesenchymal transition (EMT) induced by metabolic processes and intracellular signaling pathways on chemo-resistance, metastasis, and recurrence in solid tumors. Cell Commun Signal. 2024;22:575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 47. | Li R, Huang Y, Liu H, Dilger JP, Lin J. Abstract 2162: Comparing volatile and intravenous anesthetics in a mouse model of breast cancer metastasis. Cancer Res. 2018;78:2162-2162. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |