Published online Feb 15, 2025. doi: 10.4251/wjgo.v17.i2.100552
Revised: October 31, 2024
Accepted: November 20, 2024
Published online: February 15, 2025
Processing time: 151 Days and 11.2 Hours
In the following editorial, we discuss the article by Wu et al. In this contribution, we critically review the authors’ perspective and analyze the relevance of the results obtained in the original article of clinical research by Liu et al. We consider that additional factors associated with colon cancer progression have recently been described in extensive clinical research, and should be included in this analysis to achieve a more accurate prognosis. These factors include inflammation, gut microbiota composition, immune status and nutritional balance, as they influence the post-surgical survival profile of patients with stage II colorectal cancer. We also address the clinical implementation and limitations of these analyses. Evaluation of the patient´s entire context is essential for selection of the most appropriate therapy.
Core Tip: The treatment of patients with stage II colon cancer after surgical resection of the tumor is still under debate. Standard management based on the clinicopathological perspective is insufficient in cases not categorized as high-risk patients. Risk factors associated with the tumor and its microenvironment, including immune and inflammatory status, diet, and nutritional balance, should not be ruled out. The use of non-invasive molecular techniques to analyze these factors can provide relevant information for medical decisions in this group of patients.
- Citation: Novoa Díaz MB, Gentili C, Martín MJ, Carriere P. Prognosis in stage II colon cancer: Expanding the horizons of risk factors. World J Gastrointest Oncol 2025; 17(2): 100552
- URL: https://www.wjgnet.com/1948-5204/full/v17/i2/100552.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i2.100552
Colon cancer is the third most frequent oncological disease worldwide[1]. Sporadic intestinal tumors are the most frequent (70%-80% of cases), surpassing 20%-30% of cases of hereditary tumors[2]. This type of neoplasm has been related to dysbiosis and persistent inflammation, which impair gut permeability[3,4]. Furthermore, the heterogeneous presentation, clinical course, and prognosis of colon cancer[5], involve a high variability in the histology, primary location, and the expression of molecular markers. Therefore, the analysis of other key relapse factors such as the gut microbiota composition, and the nutritional and immune status, which strongly influence the tumor microenvironment (TME), can improve the understanding of these clinical cases. According to the Union for International Cancer Control and The American Joint Committee on Cancer and its tumor-node-metastasis (TNM) classification, T4N0M0 (stage II) colon cancer is defined as a tumor with early evolution, transmural bowel wall invasion and negative lymph nodes or metastasis[6]. In this group of patients, resection surgery alone is related to a high cure rate (80%). Therefore, the role of adjuvant chemotherapy remains under debate concerning its cost-benefit[7,8]. For stage II colon cancer patients, current guidelines advocate adjuvant chemotherapy especially if they have high-risk clinicopathological features[7]. In line with this, it is important to identify those patients who may have a real benefit from chemotherapy based on their risk factors for relapse. The purpose of the present editorial article is to highlight and discuss the risk factors of colon cancer and the validation of new models employed to analyze or predict its progression.
Recent evidence has highlighted an integral view of oncologic patients' status for prognosis evaluation and medical decisions. In this regard, the study and consideration of several molecular markers, the gut microbiota, immune cells and intestinal tissue interaction have received great attention[3,9]. In our research group, we have observed that factors present in the tumor and its microenvironment such as parathyroid hormone-related peptide (PTHrP), Secreted Protein Acidic and Cysteine Rich and other pro-tumor molecules are related to a more aggressive phenotype of colon cancer cells[1,10]. In addition, we analyzed the expression of molecular markers associated with malignant progression in human colon cancer samples, such as mesenchymal-epithelial transition receptor and parathyroid hormone receptor type 1, which directly correlates with PTHrP expression. We found that PTHrP could be relevant in the early stages of tumor development promoting molecular mechanisms associated with a worse prognosis[11]. These data, in line with those reported by other authors who propound the analysis of circulating tumor DNA (ctDNA) or stem cells/migration/invasion tumor markers[7,12,13] suggest expanding the spectrum of evaluation of molecular markers related to the progression of colon cancer in tissue and blood samples.
Diverse physiological and pathological processes are influenced by changes in intestinal barrier permeability, driven by microbial ecology and the products of fermentation derived from its metabolism[3]. Although the diagnostic/prognostic value of gut microbiota signatures is not well established to date, several authors have reported an association between colon cancer development and progression and gut enrichment in punctual species. For instance, Fusobacterium nucleatum (F. nucleatum) abundance has been associated with a shortened recurrence[14,15]. In addition, Bacterioides fragilis, Escherichia coli, and Helicobacter pylori, are other abundant species found in the tract of colon cancer patients[3,16]. In this regard, clinical evaluations of fecal composition, such as fecal occult blood and immunochemical tests, combined with microbiome analysis, can significantly enhance colon cancer diagnosis[15]. Fecal microbiome analysis is emerging as a promising diagnostic tool as specific microbial patterns are closely linked to tumor presence and progression. By examining bacterial diversity and abundance in stool samples, early-stage biomarkers of the disease can be identified, offering a non-invasive, cost-effective, and informative diagnostic approach[17].
On the other hand, the patient's lifestyle and nutritional status also have a significant influence on their microbiological and immune balance and should be considered as risk factors in both pre-operative and post-operative evaluations. Recent investigations have highlighted that the consumption of functional foods containing prebiotics and probiotics can strengthen the intestinal barrier, thus enhancing the immune status, and the production of beneficial bacterial metabolites. Together, these events can prevent events related to tumorigenesis[18]. On the contrary, the consumption of drugs that alter the microbiome such as antibiotics, can deplete beneficial bacteria, reducing the production of anti-inflammatory metabolites such as short-chain fatty acids. This microbial imbalance can favor tumorigenesis, due to a weakened intestinal barrier, immune dysfunction and inflammation. Other alarming effects associated with antibiotic-induced dysbiosis is interference with the efficacy of therapies with immune checkpoint inhibitors (ICIs), mainly due to an easier evasion of colon cancer cells to immune surveillance[19,20]. In this context, nutrigenomic approaches are emerging tools to assess diet-induced changes in the expression of genes related to tumorigenesis[21]. Moreover, it has been proposed to combine nutritional indicators with conventional risk factors analysis in scoring systems to predict the response of stage II colon cancer patients after surgery[22-24]. Therefore, when estimating overall survival (OS) and disease-free survival (DFS), it is essential to consider all these aspects. Recognizing this, the eighth edition of the TNM classification has incorporated inflammatory and nutritional scores as important prognostic factors related to the patient's condition in cases of liver, pancreatic, and esophageal cancer[25].
As mentioned by Liu et al[26] in the original article, enterostomy surgery, which was found to be an independent risk factor for OS, affects the patient's hydration and nutritional status. It is known that temporary or permanent surgical procedures can improve intestinal function, particularly in cases of obstructive colon cancer or inflammation. Despite this, there is evidence that these procedures can have a significant detrimental effect on health. This is mainly attributed to anatomical changes affecting the intestinal microbiota, which can potentially lead to dysbiosis - a potential risk factor that has been discussed in the previous paragraphs and, in this case, contribute to postoperative deterioration[27]. As discussed in the work by Wu et al[28] several risk factors were analyzed in the original clinical research work for the construction of a nomogram based on the standard management of patients in stage II to evaluate their prognosis and outcome. In their letter to the editor, they indicate that the nomogram model shows good consistency[28]. Despite this, we believe this analysis in terms of which patients are under consideration (T4N0M0 colon cancer) should be completed, focusing mainly on those risk factors that influence this stage of the disease. In medical practice, a cut-off point has commonly been established in those patients concerning certain high-risk features (cut-off of 20%) such as high T stage, lymphovascular invasion, bowel obstruction or perforation, and poorly differentiated histology[6]. However, patients are often classified as having an average risk, which complicates medical decisions regarding adjuvant chemotherapy administration[6]. Tie and colleagues recently highlighted the importance of detecting ctDNA in patients with stage II colon cancer; they analyzed the expression of this marker in a substantial number of patients. Detecting ctDNA after surgical resection enables the positive selection of those patients who could benefit from adjuvant chemotherapy[7]. This work suggests a new method for managing stage II patients that is noteworthy, primarily because ctDNA can be analyzed from blood samples, making it a real-time monitoring tool that is non-invasive[6,7,29]. Regarding clinical validation of the ctDNA detection tool, Jin et al[30] through a multiplex assay of several ctDNA detected in a cohort of 179 patients with colon cancer and healthy controls, found that this method has a sensitivity and specificity greater than 80%. Furthermore, they identified that this technique has high efficiency for the early detection of stage I and II colon tumors and the prognostic evaluation of these patients[30]. Notably, a recent study demonstrated that the cost of this type of analysis can have a very positive economic impact, since it allows adopting the criterion of not administering unnecessary chemotherapy without compromising survival[31].
Recent evidence highlights the influence of tumor-associated immune features in the clinical outcome of colon cancer. Several investigations have demonstrated that a high count of CD4+ T cells, regulatory T cells, and tumor-associated macrophages (TAMs) are indicators of poor clinical prognosis in colon tumors[32,33]. Nevertheless, Yang et al[34] observed that B cells and CD8+ T cells infiltration was related to low-risk subtypes of relapse in colon cancer patients. In particular, for stage II colon cancer patients, the focus of this editorial, Feng et al[35] postulate that TAMs are post-resection prognostic and predictive markers. In this work, CD68 and CD206 immunohistochemical analysis of two cohorts of patients who underwent TAMs detection was conducted, and the infiltration densities and CD206/CD68 ratio were then calculated. From this extensive analysis arises the criterion of TAMs evaluation as a complementary analysis to standard management (high-risk factor) for improving precision treatment[35]. Another publication reflects the prognostic value of the immunoscore for this group of patients based on a multicenter and international study according to the intratumor or invasive margins of CD3+ and CD8+ T cell densities. The authors found that a low immunoscore reliably identifies stage II patients at risk of relapse and who may benefit from adjuvant therapy[36].
Despite this, although the analysis of tumor infiltration cells can by itself allow the evaluation of low or high-risk patients, combining this test with other tools such as ctDNA detection would result in an improved risk estimation and improved criterion for adjuvant therapy administration[37]. In line with this, it is also important to highlight the relevance of assessing additional aspects of colon cancer such as DNA instability alongside the immunoscore[38]. For instance, some microsatellite-stable tumors show certain DNA mutations that confer an extensive infiltration by immune cells making them susceptible to ICIs[39]. However, this is not always applicable. A clear example are those colon cancer patients with low microsatellite instability or high mismatch repair since this group appears not to benefit from ICIs such as programmed cell death protein 1 (PD1)/PD1-ligand 1 (PD1-L1) or cytotoxic T-lymphocyte-associated protein 4 inhibitors[39–41]. Also, overexpression of genetic signatures such as PD-L1, lemur tyrosine kinase-3 and lymphocyte activation gene-3, immune escape-related signals, has been detected in colon cancer cells and can predict patient outcome[40].
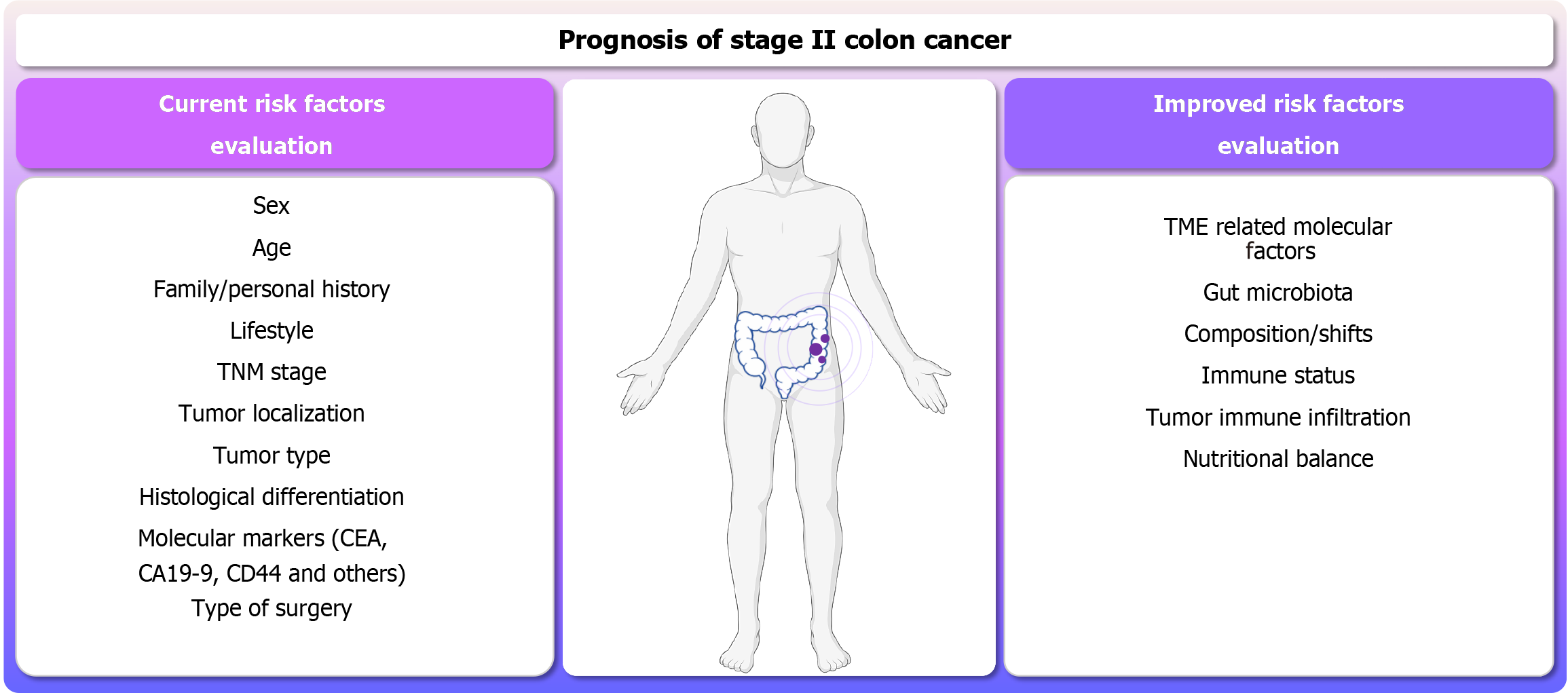
Currently, new single-cell RNA sequencing techniques make it possible to determine the contribution of some tumor cells to the overall tumor response and to identify early transcriptomic responses to tumor cell differentiation. This allows not only the identification of new genes and epigenetic mechanisms but also cell populations that participate in the progression and spatiotemporal characteristics of the preinvasive niche for different stages of primary colorectal cancer[42]. The single-cell resolution, together with advances in multi-omic analysis, has allowed the development of a large amount of information for projects such as the Human Tumor Atlas Network which will enable accurate information to be provided for clinical decisions. In terms of the implementation of models including non-conventional parameters for colon cancer prognosis, Mazaki et al[23] constructed novel nomograms to predict DFS in stage II and III colon patients based on nutritional and inflammatory markers during a postoperative period of 1 year to 5 years. Wang et al[43] also emphasized the importance of these risk factors; in their recent publication, which focused on the construction of prospective/predictive nomograms through artificial intelligence. In this case, machine learning on pathophysiology, radiomics features, immunoscore, and clinical factors served to predict postoperative outcomes of colon cancer patients[43]. The application of these methods/models could benefit from more accurate disclosure, enhancing the prognostic prediction performance for stage II patients, especially with non-invasive techniques. A summary of the risk factors typically considered in standard management, along with additional risk factors extensively discussed in this work that should be considered to achieve a more accurate prognosis is shown in Figure 1.
The analysis of inflammatory and nutritional markers has been shown to predict cancer recurrence after curative surgery in patients with stage II colon cancer[23]. The original research by Liu et al[26] evaluates, through a retrospective analysis, the clinical outcomes and prognostic factors in T4N0M0 colon cancer patients (stage II) after resection surgery without microscopic or macroscopic residual tumors. Several risk factors associated with this oncological disease were investigated in a training cohort (n = 127) and a validation cohort (n = 88) applying statistical techniques of univariate and multivariate analysis concerning 3 years OS. A nomogram model was constructed with these data[26]. It is highlighted that this article analyzes risk factors like sex, age, and relevant molecular markers in colon cancer such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9), pathological type, status, tumor size and location; but in addition includes the study of variables such as the type of surgery achieved (laparotomy/laparoscopy), whether anastomosis/enterostomy was performed, preoperative or postoperative complications and patients/clinicians compromised with regular reviews. These aspects, which only a few studies consider, are extremely relevant when evaluating OS and DFS in patients with colon cancer. However, it would be beneficial to expand the study and to include in an extensive cohort other reliable factors in predicting relapses such as nutritional balance and immune status. In this regard, we consider that the examination of gut microbiota composition and nutritional and immune status is also compelling. The rationale for focusing on these studies lies in the fact that an imbalance in the microbiota is closely associated with not healthy diets and lifestyles, but disruptions in the integrity of the gut barrier and alterations in the immune response, including the infiltration and activation of immune cells in intestinal tissue[16]. Consequently, a pro-inflammatory intestinal environment and dysbiosis, probably associated with poor diet, may contribute to tumor growth, progression, and resistance to therapy in colon cancer patients[3].
Although various nomograms with genetic and immune risk factors are being proposed today[39,40], their implementation is still challenging. The great number of analyzed genes, their interpretation in the tumor context, and the heterogeneity of the disease hinder the establishment of a single genetic signature or study panel. However, this analysis provides fundamental data in pursuit of personalized medicine and the early detection of those patients who may relapse or benefit from certain drugs and therapies. Hence, it is crucial to validate the clinical impact and relevance of all these new risk factors and nomograms proposed by studying several populations, subjecting the results to rigorous statistical methods, and comparing them with those obtained by standard management. In addition, to prevent the overfitting of these models to a data set, a large-scale study must be accomplished. Since this exploration requires considerable effort, artificial intelligence, and current data analysis, web tools could greatly contribute to this work[44,45].
It is also relevant to highlight that the detection of gene signatures or microbiome analysis by metagenomic sequencing or metabolome exploration has economic limitations in clinical practice, particularly in resource-limited environments. Genetic markers associated with the immune system[40], or the microbiota[46] can be studied in these cases using cheaper techniques than microarray or next-generation sequencing such as PCR, immunohistochemistry, or immunofluorescence. An exhaustive anamnesis with analysis of the patient's diet and lifestyle can provide significant information regarding the abundance of beneficial gut microorganisms. Finally, a risk factor and microbial biomarker such as F. nucleatum, is enriched on both stools and tumor tissue and can then be detected for the early prognostic prediction of colon cancer[47].
On the other hand, considering that the original research work is a retrospective study, access to the patient's information is limited. However, with the advent of artificial intelligence for the analysis of computed tomography images, a deeper examination of available scans could predict the progression of the primary tumor. Regarding the study of CEA and CA19-9 markers, the authors of the original article do not specify the measurement method. They highlight that right hemicolon and CA19-9 levels are independent risk factors for OS. The arrival of tissue microarray technology and a large amount of publicly available data for the analysis of patient samples could show whether the expression of these tumor markers can vary according to the location of the tumor and thus reinforce the researchers' conclusion contributing to validating this method on a large scale.
Finally, despite the opinion expressed by Wu et al[28] concerning the model achieved in the original work, the internal and external validation of nomograms should always be carefully reviewed before clinicians apply these to their patients. The relationship between the risk factors and the outcome measurements, the relationships among covariates, and synergistic effects should be assessed. To reinforce this point, the model should be subjected to cross-validation of patient data sets and how it would respond to expansion of the study population.
Prognostic evaluation of stage II colon cancer patients and chemotherapy recommendation based on their clinicopathological features results are insufficient. The development of nomogram models based solely on standard management is therefore imprecise, possibly exposing patients to unnecessary treatments or enabling relapse of patients who did not receive the correct treatment. Other aspects in the management of this group of patients should also be considered, such as the molecular analysis of biomarkers in both the pre-operative and post-operative situations, the prognostic nutritional index, immunoscore, TME cell composition and an integral review of clinical conditions to categorize with more accuracy low, average or high-risk patients. However, the development of new nomograms for colon cancer patient management requires not only internal validation to evaluate the accuracy and precision of the results, but also external validation to prevent overfitting and ensure the model can adapt to variations when expanding the population studied. Furthermore, specifying molecular signatures that show dependence on the variables analyzed in the original study could enhance medical decision-making.
| 1. | Carriere P, Calvo N, Novoa Díaz MB, Lopez-Moncada F, Herrera A, Torres MJ, Alonso E, Gandini NA, Gigola G, Contreras HR, Gentili C. Role of SPARC in the epithelial-mesenchymal transition induced by PTHrP in human colon cancer cells. Mol Cell Endocrinol. 2021;530:111253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 2. | Stastna M, Janeckova L, Hrckulak D, Kriz V, Korinek V. Human Colorectal Cancer from the Perspective of Mouse Models. Genes (Basel). 2019;10:788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | Novoa Díaz MB, Carriere P, Gentili C. How the interplay among the tumor microenvironment and the gut microbiota influences the stemness of colorectal cancer cells. World J Stem Cells. 2023;15:281-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Yuan J, Wei Z, Xu X, Ocansey DKW, Cai X, Mao F. The Effects of Mesenchymal Stem Cell on Colorectal Cancer. Stem Cells Int. 2021;2021:9136583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley CM, Wu CH, Beggs AD, McDermott U, Dunne PD, Meade A, Walker SM, Murray GI, Samuel L, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH; S:CORT consortium. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70:544-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 6. | Rebuzzi SE, Pesola G, Martelli V, Sobrero AF. Adjuvant Chemotherapy for Stage II Colon Cancer. Cancers (Basel). 2020;12:2584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 7. | Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, Wong R, Shapiro J, Lee M, Harris S, Khattak A, Burge M, Harris M, Lynam J, Nott L, Day F, Hayes T, McLachlan SA, Lee B, Ptak J, Silliman N, Dobbyn L, Popoli M, Hruban R, Lennon AM, Papadopoulos N, Kinzler KW, Vogelstein B, Tomasetti C, Gibbs P; DYNAMIC Investigators. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med. 2022;386:2261-2272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 559] [Article Influence: 186.3] [Reference Citation Analysis (0)] |
| 8. | Baxter NN, Kennedy EB, Bergsland E, Berlin J, George TJ, Gill S, Gold PJ, Hantel A, Jones L, Lieu C, Mahmoud N, Morris AM, Ruiz-Garcia E, You YN, Meyerhardt JA. Adjuvant Therapy for Stage II Colon Cancer: ASCO Guideline Update. J Clin Oncol. 2022;40:892-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 161] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 9. | Tanca A, Abbondio M, Fiorito G, Pira G, Sau R, Manca A, Muroni MR, Porcu A, Scanu AM, Cossu-Rocca P, De Miglio MR, Uzzau S. Metaproteomic Profile of the Colonic Luminal Microbiota From Patients With Colon Cancer. Front Microbiol. 2022;13:869523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Novoa Díaz MB, Carriere PM, Martín MJ, Calvo N, Gentili C. Involvement of parathyroid hormone-related peptide in the aggressive phenotype of colorectal cancer cells. World J Gastroenterol. 2021;27:7025-7040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Novoa Díaz MB, Carriere P, Gigola G, Zwenger AO, Calvo N, Gentili C. Involvement of Met receptor pathway in aggressive behavior of colorectal cancer cells induced by parathyroid hormone-related peptide. World J Gastroenterol. 2022;28:3177-3200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (1)] |
| 12. | Ehteram H, Aslanbeigi F, Ghoochani Khorasani E, Tolouee M, Haddad Kashani H. Expression and Prognostic Significance of Stem Cell Marker CD133 in Survival Rate of Patients with Colon Cancer. Oncol Ther. 2022;10:451-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 13. | Garouniatis A, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. FAK, CD44v6, c-Met and EGFR in colorectal cancer parameters: tumour progression, metastasis, patient survival and receptor crosstalk. Int J Colorectal Dis. 2013;28:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1473] [Article Influence: 184.1] [Reference Citation Analysis (0)] |
| 15. | Liu Y, Lau HC, Cheng WY, Yu J. Gut Microbiome in Colorectal Cancer: Clinical Diagnosis and Treatment. Genomics Proteomics Bioinformatics. 2023;21:84-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 16. | Lee SA, Liu F, Riordan SM, Lee CS, Zhang L. Global Investigations of Fusobacterium nucleatum in Human Colorectal Cancer. Front Oncol. 2019;9:566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 17. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, Wu WK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BA, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TY, Ng SC, Cheng AS, Wong VW, Chan FK, Xu X, Yang H, Madsen L, Datz C, Tilg H, Wang J, Brünner N, Kristiansen K, Arumugam M, Sung JJ, Wang J. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 781] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 18. | Wang L, Yu KC, Hou YQ, Guo M, Yao F, Chen ZX. Gut microbiome in tumorigenesis and therapy of colorectal cancer. J Cell Physiol. 2023;238:94-108. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 19. | Patel P, Poudel A, Kafle S, Thapa Magar M, Cancarevic I. Influence of Microbiome and Antibiotics on the Efficacy of Immune Checkpoint Inhibitors. Cureus. 2021;13:e16829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Sambi M, Bagheri L, Szewczuk MR. Current Challenges in Cancer Immunotherapy: Multimodal Approaches to Improve Efficacy and Patient Response Rates. J Oncol. 2019;2019:4508794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 178] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 21. | Ho J, Puoplo N, Pokharel N, Hirdaramani A, Hanyaloglu AC, Cheng CW. Nutrigenomic underpinnings of intestinal stem cells in inflammatory bowel disease and colorectal cancer development. Front Genet. 2024;15:1349717. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 22. | Ma Z, Liu R, Liu H, Zheng L, Zheng X, Li Y, Cui H, Qin C, Hu J. New scoring system combining computed tomography body composition analysis and inflammatory-nutritional indicators to predict postoperative complications in stage II-III colon cancer. J Gastroenterol Hepatol. 2023;38:1520-1529. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 23. | Mazaki J, Katsumata K, Tago T, Kasahara K, Enomoto M, Ishizaki T, Nagakawa Y, Tsuchida A. Novel and Simple Nomograms Using Inflammation and Nutritional Biomarkers for Stage II-III Colon Cancer, Taking "Time after Curative Surgery" into Consideration. Nutr Cancer. 2022;74:2875-2886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Xu YS, Liu G, Zhao C, Lu SL, Long CY, Zhong HG, Chen Y, Huang LX, Liang Z. Prognostic Value of Combined Preoperative Carcinoembryonic Antigen and Prognostic Nutritional Index in Patients With Stage II-III Colon Cancer. Front Surg. 2021;8:667154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Ahiko Y, Shida D, Nakamura Y, Imaizumi J, Takamizawa Y, Moritani K, Tsukamoto S, Kanemitsu Y. Preoperative Nutritional Scores as Host-Related Prognostic Factors for Both Overall Survival and Postoperative Complications in Patients With Stage II to III Colorectal Cancer. Dis Colon Rectum. 2021;64:1222-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Liu B, Zhang ZX, Nie XY, Sun WL, Yan YJ, Fu WH. Clinical outcome and prognostic factors of T4N0M0 colon cancer after R0 resection: A retrospective study. World J Gastrointest Oncol. 2024;16:1869-1877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (5)] |
| 27. | Lin XH, Jiang JK, Luo JC, Lin CC, Ting PH, Yang UC, Lan YT, Huang YH, Hou MC, Lee FY. The long term microbiota and metabolic status in patients with colorectal cancer after curative colon surgery. PLoS One. 2019;14:e0218436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Wu CY, Ye K. Risk factors for the prognosis of colon cancer. World J Gastrointest Oncol. 2024;16:3738-3740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 29. | Petrelli F, Labianca R, Zaniboni A, Lonardi S, Galli F, Rulli E, Rosati G, Corallo S, Ronzoni M, Cardellino GG, Mattioli R, Mambrini A, Ciuffreda L, Banzi M, Pusceddu V, Maiello E, Zampino M, Zagonel V, Marchetti P, Corsi D, Rimassa L, Cinieri S, Sobrero A. Assessment of Duration and Effects of 3 vs 6 Months of Adjuvant Chemotherapy in High-Risk Stage II Colorectal Cancer: A Subgroup Analysis of the TOSCA Randomized Clinical Trial. JAMA Oncol. 2020;6:547-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Jin S, Zhu D, Shao F, Chen S, Guo Y, Li K, Wang Y, Ding R, Gao L, Ma W, Lu T, Li D, Zhang Z, Cai S, Liang X, Song H, Ji L, Li J, Zheng Z, Jiang F, Wu X, Luan J, Zhang H, Yang Z, Cantor CR, Xu C, Ding C. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci USA. 2021;118:e2017421118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 31. | Li Y, Heer AK, Sloane HS, Edelstein DL, Tie J, Gibbs P, Barzi A. Budget Impact Analysis of Circulating Tumor DNA Testing for Colon Cancer in Commercial Health and Medicare Advantage Plans. JAMA Health Forum. 2024;5:e241270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Zhou H, Wang Y, Zhang Z, Xiong L, Liu Z, Wen Y. A novel prognostic gene set for colon adenocarcinoma relative to the tumor microenvironment, chemotherapy, and immune therapy. Front Genet. 2022;13:975404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 33. | Labanieh L, Majzner RG, Mackall CL. Programming CAR-T cells to kill cancer. Nat Biomed Eng. 2018;2:377-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 295] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 34. | Yang S, Liu T, Cheng Y, Bai Y, Liang G. Immune cell infiltration as a biomarker for the diagnosis and prognosis of digestive system cancer. Cancer Sci. 2019;110:3639-3649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 35. | Feng Q, Chang W, Mao Y, He G, Zheng P, Tang W, Wei Y, Ren L, Zhu D, Ji M, Tu Y, Qin X, Xu J. Tumor-associated Macrophages as Prognostic and Predictive Biomarkers for Postoperative Adjuvant Chemotherapy in Patients with Stage II Colon Cancer. Clin Cancer Res. 2019;25:3896-3907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 36. | Mlecnik B, Lugli A, Bindea G, Marliot F, Bifulco C, Lee JJ, Zlobec I, Rau TT, Berger MD, Nagtegaal ID, Vink-Börger E, Hartmann A, Geppert CI, Kolwelter J, Merkel S, Grützmann R, Van den Eynde M, Jouret-Mourin A, Kartheuser A, Léonard D, Remue C, Wang J, Bavi P, Roehrl MHA, Ohashi PS, Nguyen LT, Han S, MacGregor HL, Hafezi-Bakhtiari S, Wouters BG, Masucci GV, Andersson EK, Zavadova E, Vocka M, Spacek J, Petruzelka L, Konopasek B, Dundr P, Skalova H, Nemejcova K, Botti G, Tatangelo F, Delrio P, Ciliberto G, Maio M, Laghi L, Grizzi F, Fredriksen T, Buttard B, Lafontaine L, Maby P, Majdi A, Hijazi A, El Sissy C, Kirilovsky A, Berger A, Lagorce C, Paustian C, Ballesteros-Merino C, Dijkstra J, van de Water C, Vliet SVL, Knijn N, Mușină AM, Scripcariu DV, Popivanova B, Xu M, Fujita T, Hazama S, Suzuki N, Nagano H, Okuno K, Torigoe T, Sato N, Furuhata T, Takemasa I, Patel P, Vora HH, Shah B, Patel JB, Rajvik KN, Pandya SJ, Shukla SN, Wang Y, Zhang G, Kawakami Y, Marincola FM, Ascierto PA, Fox BA, Pagès F, Galon J. Multicenter International Study of the Consensus Immunoscore for the Prediction of Relapse and Survival in Early-Stage Colon Cancer. Cancers (Basel). 2023;15:418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 37. | Taieb J, Karoui M, Basile D. How I treat stage II colon cancer patients. ESMO Open. 2021;6:100184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Yu G, Wu Y, Wang W, Xu J, Lv X, Cao X, Wan T. Correction to: Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell Mol Immunol. 2020;17:111-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 39. | Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front Immunol. 2020;11:369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 346] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 40. | Wei L, Xu J, Hu X, Lyu G. Development of a risk model based on immune genes in patients with colon adenocarcinoma. Cancer Rep (Hoboken). 2023;6:e1712. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 41. | Stein A, Folprecht G. Immunotherapy of Colon Cancer. Oncol Res Treat. 2018;41:282-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 42. | Wang M, Deng C, Yang C, Yan M, Lu H, Zhang Y, Liu H, Tong Z, Ma J, Wang J, Zhang Y, Wang J, Xuan Y, Cheng H, Zhao K, Zhang J, Chai C, Li M, Yu Z. Unraveling temporal and spatial biomarkers of epithelial-mesenchymal transition in colorectal cancer: insights into the crucial role of immunosuppressive cells. J Transl Med. 2023;21:794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 43. | Wang R, Dai W, Gong J, Huang M, Hu T, Li H, Lin K, Tan C, Hu H, Tong T, Cai G. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. 2022;15:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 142] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 44. | Chen T, Jiang Q, Wang Z, Zhang H, Fu Z. The roles of lncRNA AP001469.3 in clinical implications, immune landscape and carcinogenesis of colorectal cancer. Transl Cancer Res. 2024;13:3465-3481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 45. | Kothalawala WJ, Győrffy B. Transcriptomic and Cellular Content Analysis of Colorectal Cancer by Combining Multiple Independent Cohorts. Clin Transl Gastroenterol. 2023;14:e00517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 46. | Yang Y, Du L, Shi D, Kong C, Liu J, Liu G, Li X, Ma Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun. 2021;12:6757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 148] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 47. | Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023;31:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 162] [Article Influence: 81.0] [Reference Citation Analysis (0)] |