Published online Aug 15, 2024. doi: 10.4251/wjgo.v16.i8.3651
Revised: May 27, 2024
Accepted: June 18, 2024
Published online: August 15, 2024
Processing time: 105 Days and 20.8 Hours
Hepatocellular carcinoma (HCC) is a major cause of cancer mortality worldwide, and metastasis is the main cause of early recurrence and poor prognosis. How
To determine the possible mechanism affecting HCC metastasis and provide a possible theoretical basis for HCC treatment.
The candidate molecule lecithin-cholesterol acyltransferase (LCAT) was screened by gene microarray and bioinformatics analysis. The expression levels of LCAT in clinical cohort samples was detected by quantitative real-time polymerase chain reaction and western blotting. The proliferation, migration, invasion and tumor-forming ability were measured by Cell Counting Kit-8, Transwell cell migration, invasion, and clonal formation assays, respectively. Tumor formation was detected in nude mice after LCAT gene knockdown or overexpression. The immunohistochemistry for Ki67, E-cadherin, N-cadherin, matrix metalloproteinase 9 and vascular endothelial growth factor were performed in liver tissues to assess the effect of LCAT on HCC. Gene set enrichment analysis (GSEA) on various gene signatures were analyzed with GSEA version 3.0. Three machine-learning algorithms (random forest, support vector machine, and logistic regression) were applied to predict HCC metastasis in The Cancer Genome Atlas and GEO databases.
LCAT was identified as a novel gene relating to HCC metastasis by using gene microarray in HCC tissues. LCAT was significantly downregulated in HCC tissues, which is correlated with recurrence, metastasis and poor outcome of HCC patients. Functional analysis indicated that LCAT inhibited HCC cell proliferation, migration and invasion both in vitro and in vivo. Clinicopathological data showed that LCAT was negatively associated with HCC size and metastasis (HCC size ≤ 3 cm vs 3-9 cm, P < 0.001; 3-9 cm vs > 9 cm, P < 0.01; metastatic-free HCC vs extrahepatic metastatic HCC, P < 0.05). LCAT suppressed the growth, migration and invasion of HCC cell lines via PI3K/AKT/mTOR signaling. Our results indicated that the logistic regression model based on LCAT, TNM stage and the serum level of α-fetoprotein in HCC patients could effectively predict high metastatic risk HCC patients.
LCAT is downregulated at translational and protein levels in HCC and might inhibit tumor metastasis via attenuating PI3K/AKT/mTOR signaling. LCAT is a prognostic marker and potential therapeutic target for HCC.
Core Tip: For the first time, we found that lecithin-cholesterol acyltransferase, a molecule related to lipid metabolism, is closely related to the metastasis of hepatocellular carcinoma (HCC), which provides a new prospective for studying the recurrence and metastasis mechanism of HCC, and also finds strong evidence for revealing the connection between lipid metabolism and HCC metastasis.
- Citation: Li Y, Jiang LN, Zhao BK, Li ML, Jiang YY, Liu YS, Liu SH, Zhu L, Ye X, Zhao JM. Lecithin-cholesterol acyltransferase is a potential tumor suppressor and predictive marker for hepatocellular carcinoma metastasis. World J Gastrointest Oncol 2024; 16(8): 3651-3671
- URL: https://www.wjgnet.com/1948-5204/full/v16/i8/3651.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i8.3651
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related deaths worldwide[1]. Despite the significant progress in the treatment of liver cancer that has been made in the past few decades[2], the prognosis is still dismal due to the high probability of metastasis even after potentially curative treatment[3]. Metastasis is a complex multistep process that is regulated by a complex network of intra- and intercellular signal transduction cascades[4]. Studies showed that lipid metabolism disorders were associated with poor prognosis after HCC radical resection[5,6] and initiate tumor metastasis[7]. However, little is known about lipid-metabolism-related molecules that regulate HCC metastasis or their possible molecular mechanisms in HCC recurrence.
Nowadays, discovering new potential biomarkers and functional molecules from massive omics data and translating them into clinical practice to improve the outcome of HCC patients are challenging[8]. In this study, by using HCC microarrays analysis, we found that LCAT was the top-ranked downregulated gene, and its lower expression was correlated with poorer recurrence-free survival (RFS) and overall survival (OS) of HCC patients.
Lecithin-cholesterol acyltransferase (LCAT) is a crucial enzyme in the extracellular metabolism of plasma lipoproteins, mainly synthesized and secreted by the liver into plasma[9-11]. Recently, an association between LCAT expression and tumor malignancy has been found in some cancers, including HCC[12,13]. However, the molecular role and regulatory mechanism of LCAT in HCC metastasis or recurrence are still largely undetermined.
Here, we found that the expression level of LCAT in HCC tissue is lower than that in non-cancerous liver tissue, and the low expression of LCAT is closely related to HCC metastasis and recurrence. We demonstrated that LCAT attenuated the proliferation, migration and invasion of HCC cells. In addition, we revealed that LCAT inhibited the PI3K/AKT/mTOR signaling pathway. Our finding indicated that LCAT may serve as a potential prognostic biomarker and therapeutic target for HCC.
The microarray dataset of gene expression profiles and related clinical features analyzed in this study was downloaded from the publicly available GEO database (httPs://www.ncbi.nlm.nih.gov/geo/). GSE14520 from GEO database was conducted by GPL3921 (Affymetrix HT Human Genome U133A Array), including 225 HCC samples and 220 paired nontumor tissue samples. Eleven high metastasis risk HCC tissue samples, 18 low metastasis risk HCC tissue samples and 11 paired nontumor tissue samples were selected to unveil the universally metastatic-related differential genes in early high metastatic risk HCC. The inclusion criteria were as follows: HCC tissue confirmed by pathology; solitary tumors with diameters < 5 cm; BCLC stage 0 or A; TNM stage I or II; ALT(Alanine aminotransferase) ≤ 50 U/L and α-fetoprotein (AFP) ≤ 300 ng/mL. GSE54236 from GEO database was conducted by GPL6480, including 81 HCC cases with different tumor doubling times.
From The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/; as of September 1, 2018), a total of 421 RNA-sequencing profiles (371 HCC samples and 50 nontumor samples), 430 DNA methylation profiles (380 HCC samples and 50 nontumor samples) and corresponding clinical information of HCC patients were obtained. Among the 380 HCC samples that yielded DNA methylation data, 371 HCC samples included both RNA-sequencing data and paired DNA methylation data. Illumina HiSeq 2000 RNA sequencing platform was used to obtain HCC gene expression data form TCGA. Illumina Infinium Human methylation 450 platform was used to obtain DNA methylation data. DNA methylation value in a gene was calculated with the average DNA methylation value of all CpG sites.
To search for genes critical for HCC metastasis, we identified differentially expressed genes between 11 high metastasis risk HCC samples and 29 non-high metastasis risk HCC samples (18 low metastasis risk HCC tissue samples and 11 paired nontumor tissue samples) from GSE14520 utilizing the “limma” R package. To select lipid-metabolism-related genes for further analysis, we curated the gene set of lipid metabolic pathway (766 genes) (Supplementary Table 1) based on the latest Reactome annotations[14], and then restricted the list to genes with available RNA expression data in the GSE14520 dataset. Finally, the P value < 0.0001 and |log2 fold change (FC)| > 2 were utilized as cutoff criteria[15].
HCC samples including early recurrence (recurrence within 2 years after curative resection) or recurrence free within 3 years after curative resection were paired into 15 groups. Extrahepatic metastatic HCCs [defined as extrahepatic metastasis (EHMH), classic imaging features or pathological diagnosis appeared within 1-year follow-up after curative resection], metastatic-free HCCs (defined as MFH), cirrhotic liver (derived from portoazygous devascularization) and normal liver (derived from surgery for hepatic hemangioma) were also prepared. All samples were obtained from the Department of Hepatobiliary Surgery, Fifth Medical Center of Chinese PLA General Hospital between January 2018 and December 2019. Surgically removed tissues were quickly frozen in liquid nitrogen until analysis. All specimens were evaluated by three independent pathologists.
Total RNA from frozen tissues and cultured cells was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-stranded cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen). Real time fluorescence quantitative polymerase chain reaction (PCR) using SYBR Green (Thermo, Waltham, MA, USA) was performed in StepOne Real time PCR system (Applied Biosystems). Data of the relative expression level of RNA were presented using the comparative Ct method. Gene-specific primers are shown in Supplementary Table 2 with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control.
Tissue samples were fixed in 4% formalin for 24-36 h and subsequently embedded in paraffin. For tissue analysis, 3-5-μm thick sections were cut and deparaffinized. Antigen retrieval was performed using high pressure antigen retrieval with citrate at pH 6.0 for 3 min. The slides with sections were washed with phosphate-buffered saline, and then subjected to immunostaining with the primary antibody LCAT (Abcam, ab109417, 1:150), Ki67 (ZSbio, China, ZM-0166, 1:200), E-cadherin (ZSbio, ZA-0565, 1:200), N-cadherin (ZSbio, ZM-0094, 1:200), matrix metalloproteinase (MMP)9 (ZSbio, ZA-0562, 1:200), and vascular endothelial growth factor (VEGF) (ZSbio, ZM-0265, 1:200) respectively, followed by incubating with horseradish-peroxidase-conjugated secondary antibodies.
Total cell and tissues lysates were prepared in RIPA protein extraction reagent (80004, QIAGEN, Germany) supplemented with a protease inhibitor cocktail (Roche, CA, USA) and phenymenthylsulfonyl fluoride (Roche). Total proteins (100 μg) were separated by 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes. After incubation with antibodies specific for LCAT (Abcam, ab109417, 1:1000), AKT (Cell Signaling Technology, 9272s, 1:1000), phospho (p)-AKT (Thr308) (Cell Signaling Technology, 13038s, 1:1000), mammalian target of rapamycin (mTOR) (Cell Signaling Technology, 2972s, 1:1000), p-mTOR (Ser2448) (Cell Signaling Technology, 2971s, 1:1000), E-cadherin (Cell Signaling Technology, 49398T, 1:1000), vimentin (Cell Signaling Technology, 49398T, 1:1000), transcription factor (TCF)-8) (Cell Signaling Technology, 49398T, 1:1000), Snail (Cell Signaling Technology, 49398T, 1:1000) or β-actin (ZSbio, TA-09, 1:5000). IRdye 800-conjugated goat anti-rabbit IgG or IRdye 700-conjugated goat anti-mouse IgG were used to incubate the blots and then detected with Odyssey infrared scanner (Li-Cor). β-actin was used as an internal control.
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in an atmosphere containing 5% CO2. HepG2, Hep3B, PLC/PRF/5, Huh7, SNU-182, SNU-387, Li-7, and 293FT cells were obtained from the Chinese Academy of Sciences Cell Bank and authenticated. Mycoplasma contamination was excluded via a PCR-based method. Cells were treated with different concentration decitabine (0-6 μM) (Selleck.cn) for 72 h.
The pLKO.1 Lentiviral shRNA clones (TRCN0000034694 and TRCN0000034698) were applied to knockdown LCAT. For lentiviral packaging, 6 μg shRNA plasmid and packaging plasmids (pMD2.G and psPAX2) were cotransfected into 293FT cells with Lipofectamine 2000 reagent. Virus was collected twice after 48 and 72 h of transfection, and used to infect HepG2 cells in the presence of 8 μg/mL polybrene (Sigma, Catalog # H9268). The stably transduced cells were further selected with puromycin (2 μg/mL) for 7 d.
The reported three plasmid system was applied to CRISPR activation (CRISPRa)[12]: lenti dCAS-VP64_Blast (Addgene, cat. No. 61425), lenti MS2-P65-HSF1_Hygro (Addgene, cat. No. 61426) and lenti_sgRNA (MS2)_zeo backbone (Addgene, cat. No. 61427). For activating the transcription of LCAT, two single guide RNAs (sgRNAs) targeting the first 200 bp upstream of LCAT were synthesized (guide#1 GGGAGGGACGGCCTGGCCAG, guide#2 CCTGAGGCTGTGCCCCTTTC), annealed and ligated to the lenti_sgRNA (MS2)_zeo backbone plasmid with the BsmBI restriction site. The lentiviral particles carrying lenti_dCAS-VP64_Blast, lenti_MS2-P65-HSF1_Hygro and lenti_sgRNA (MS2)_zeo backbone including sgRNA sequences or control sequence were made as previously described[12], and coinfected Huh7 cells to generate a stable cell line. After selection with zeocin, hygromycin and blasticidin for 7 d, the samples were collected and examined the expression of LCAT.
To conduct cell proliferation experiments, 3000 cells were cultured in a 96-well plate. Cell proliferation was detected with the Cell Counting Kit-8 (Dojindo Laboratories) after 12 h cells culture. For colony formation assays, 3000 cells were incubated in a six-well plate for 10 d with normal culture medium. Clones were fixed and stained with 0.50% crystal violet, and the number of colonies was counted. For soft agar colony formation assay, ~104 cells were suspended in a top layer of 0.33% soft agar (Agarose, Amresco) and plated on a bottom layer of 0.65% soft agar (Agar, Amresco) containing complete MEM (HepG2) or DMEM (Huh-7) supplemented with 10% FBS in six-well plates. After 3 wk of culture, the colonies (diameter > 50 µm) were photographed using a microscope.
A 24-well Transwell plate (8 μm pore size, Corning, NY, USA) was used to measure cell migration and invasion. For migration assays, 7.5 × 104 cells in 250 μL DMEM without FBS were placed into the upper chamber, and 500 μL medium containing 10% FBS was added to the lower chambers. Chemicals (if used) such as rapamycin (100 ng/mL) and MK-2206 (1 μM) were also added to the upper chambers. For invasion assays, chamber inserts were precoated with 50 μL 1:5 mixture of BD Matrigel (BD Biosciences, San Jose, CA, USA) and DMEM overnight under sterile conditions. Then, 5 × 105 cells with or without MK-2206 (1 μM) or rapamycin (100 ng/mL) were seeded in the upper chamber. After 24 h (migration assays) or 72 h (invasion assays), cells on the top side of each insert were scraped off and the wells were fixed in 4% phosphate-buffered neutral formalin (pH 7.4), and stained by crystal violet. Three random microscopic view fields were calculated for each group. The experiments were repeated three times independently at least.
Gene set enrichment analysis (GSEA) on various gene signatures were perform with GSEA version 3.0. Gene sets were acquired from published gene signatures. The enrichment score to enrichment results generated from 1000 random permutations of the gene set was compared statistically to obtain FDR q values.
The animal studies were approved by the Committee on Ethics of Chinese PLA General Hospital. Male athymic BALB/c nude mice (age 5 wk) were used for animal studies. Cells (107) in 100 μL PBS were injected subcutaneously into the left or the right flanks of randomly grouped mice (6 per group). Tumor growth was recorded every 2 d with a caliper, and the tumor volume was calculated as a × b2 × 0.5 (a, longest diameter; b, shortest diameter). Tumors were allowed to grow for 3-4 wk, and then the mice were killed, followed by photography. The tumors were dissected out, and the tumor volume and weight were measured. The samples were fixed with paraformaldehyde (4%) followed by dehydration and embedding in paraffin. Hematoxylin and eosin (H&E) staining or immunohistochemistry (IHC) were performed according to standard protocols.
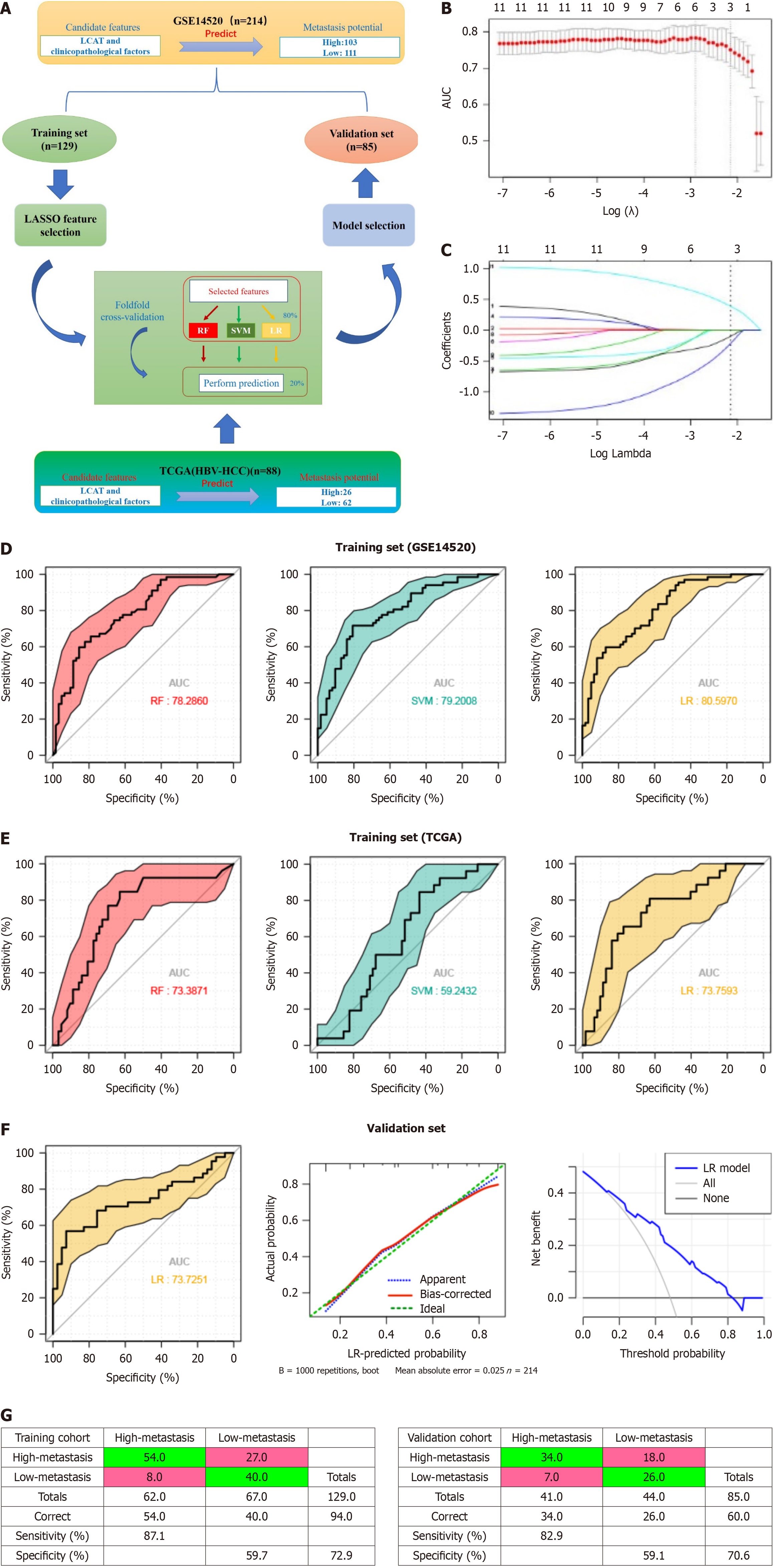
To assess the predictive power of LCAT combined with clinicopathological characteristics for two HCC metastatic risk subtypes (high vs low), we randomly divided the GSE14520 cohort into training and test sets. Adapted from Yuan et al[16], we applied three widely used machine-learning algorithms [random forest (RF), support vector machine (SVM), and logistic regression (LR)] to predict the subtype (as a binary variable). Logarithmic transformation expression levels of LCAT and pathological data were used as candidate features. We evaluated the performance of classifiers through fivefold cross-validation within the training set. Specifically, we randomly divided the training set into five equal parts. In each of the five iterations, the R package glmnet[17] was used to apply the minimum absolute shrinkage and selection operator as feature selection methods to four fifths of the training data, and use the selected features to train the classifier (RF is 1000 trees, SVM is radial kernel, and other parameters are set by default). We applied the trained classifiers to the remaining fifth of the training data for prediction. We merged and compared the predicted results of five iterations with the true values, drew the receiver operating characteristic curve[15] and calculated the area under the curve (AUC). Finally, we used the best performing algorithm identified through cross validation (with the highest AUC) to select features from the entire training set and construct a classifier, which is then applied to the test set to independently verify the predictive ability.
All data are shown as means and standard errors of the mean. Prism 6 (GraphPad Software, La Jolla, CA, USA) and R 3.5.2 were used for the Wilcoxon matched-pairs test, Student’s t-test, χ2 test, Fisher's exact test, or Pearson's correlation test. Kaplan-Meier analysis and Cox proportional hazards regression models were used for survival analysis. P < 0.05 was considered statistically significant.
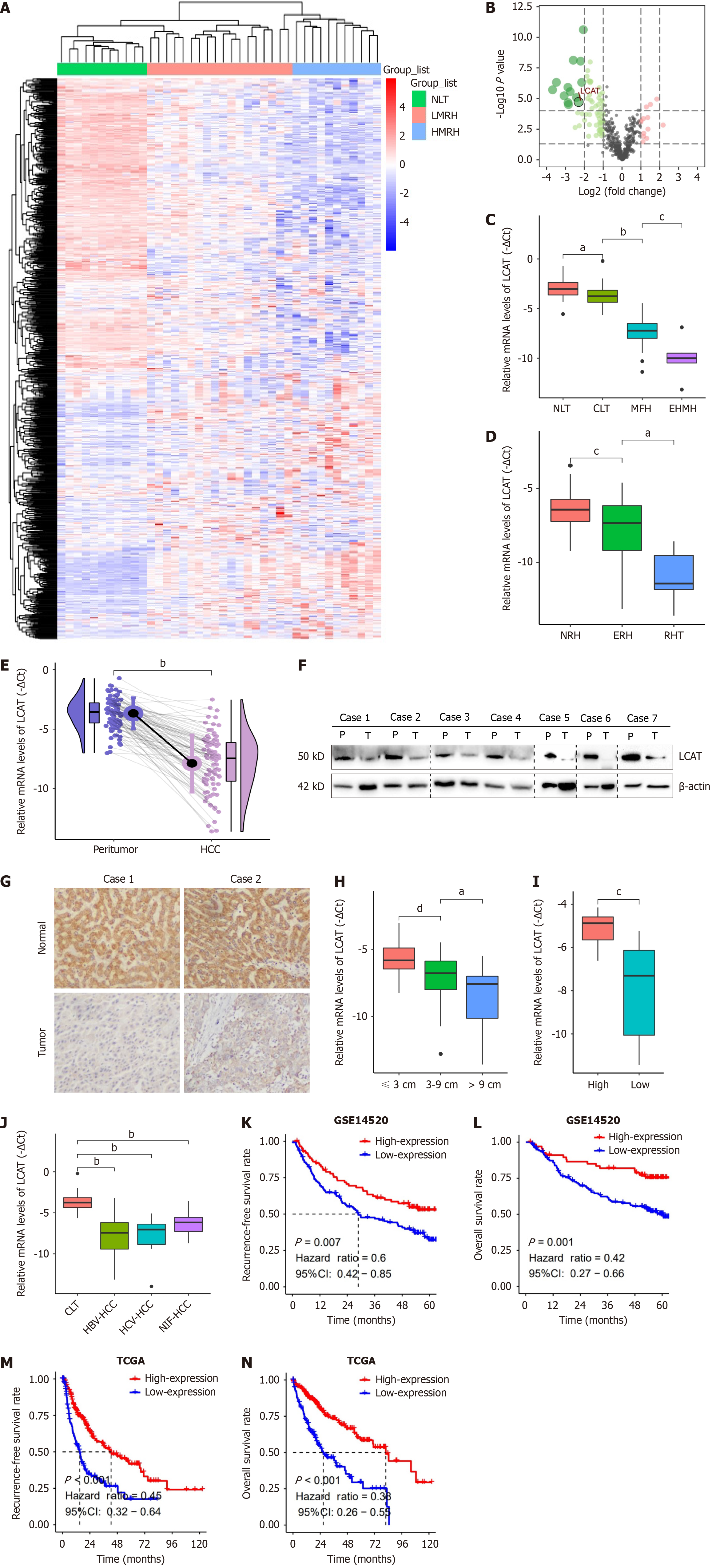
To identify potential steroid metabolic genes involved in HCC metastasis, we analyzed the microarray data downloaded from the GEO database (GSE 14520) to define candidate genes. We performed hierarchical clustering and differential analysis on genes related to lipid metabolism. Results showed that consistent with the actual group, the tissues were significantly clustered into three groups (Figure 1A), and lipid-metabolism-related gene expression profiles were significantly different between high metastasis risk HCC (HMRH) and low metastasis risk HCC (LMRH) as well as normal liver tissues (NLT). It is worth noting that, LCAT, which encodes a pivotal steroid metabolic enzyme, was identified as the leading lipid metabolic gene among the significantly downregulated genes in HMRH compared with LMRH and NLT (Figure 1B). Due to the disruption of cholesterol homeostasis is associated with the lowest OS and the greatest risk of a poor prognosis after first-line surgery in HCC patients[4], we focused on the functional role and molecular mechanisms of LCAT in promoting HCC metastasis.
First, we analyzed the clinical relevance of LCAT expression using quantitative real-time PCR (qRT-PCR). The data showed that mRNA levels of LCAT significantly decreased in HCC samples, especially in HCC with EHMH, compared with NLT and cirrhotic liver tissues (CLT) (Figure 1C). Early HCC recurrence within 2 years after tumor resection is believed to be mainly related to the dissemination of metastatic HCC tissues. Our data indicated that LCAT expression levels were significantly lower in early recurrence HCC than that in RF HCC after curative resection. In addition, LCAT was further downregulated in recurrent HCC tissues (Figure 1D). These data suggest that there is a close association between LCAT downregulation and HCC metastasis.
We detected the mRNA levels of LCAT in 90 human HCC tissues and their matched nontumor liver samples using qRT-PCR. The results showed significant downregulation of LCAT mRNA levels in HCC tissues (Figure 1E). Decreased protein levels of LCAT in HCC tissues were confirmed in seven paired samples using western blotting (Figure 1F) and in two paired samples using IHC randomly selected from 90 paired samples (Figure 1G).
To investigate further the relationship between LCAT and HCC metastasis, especially the prognostic value of LCAT in HCC patients, we conducted prognostic analysis. RT-qPCR indicated that LCAT downregulated expression was closely correlated with tumor size and differentiation status (Figure 1H and I). Notably, compared to CLTs, downregulation of LCAT was independent of etiology (Figure 1J). Using Wang’s cohort (GSE14520, n = 225), Kaplan-Meier survival analysis showed that HCC patients with low LCAT mRNA level had shorter RFS (P = 0.007) (Figure 1K) and OS (P = 0.001) (Figure 1L). We obtained consistent results from TCGA (TCGA-LIHC dataset, n = 364) (Figure 1M and N). In summary, these results indicated a significant correlation between downregulation of LCAT and poor prognosis in HCC patients.
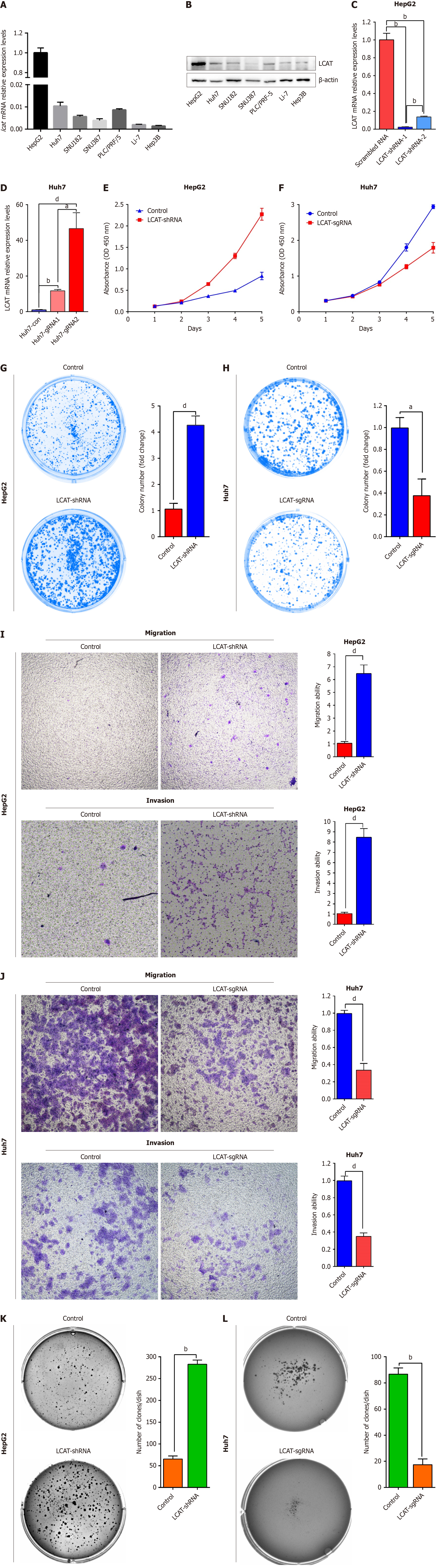
To examine the function of LCAT in HCC metastasis, we first checked the expression levels of LCAT in different HCC cell lines. The data showed that both mRNA and protein levels of LCAT were low in HCC cell lines, except HepG2 cells (Figure 2A and B). HepG2 and Huh7 cell lines were chosen for subsequent studies. We took the approach of RNA interference to knockdown LCAT in HepG2 cells (Figure 2C). Cell proliferation, colony formation, migration and invasion were detected. The data showed that the knockdown of LCAT resulted in an increase in HCC cell proliferation and colony formation (Figure 2E and G) and to enhanced migration and invasion (Figure 2I). In addition, LCAT-knockdown HepG2 cells formed more colonies in soft agar than the control cells (Figure 2K), implying that downregulation of LCAT increased the tumorigenicity of HepG2 cells.
To investigate further the function of LCAT in HCC cells, we activated the expression of endogenous LCAT in Huh7 cells using a CRISPR-Cas9 based transcriptional activation system[12]. As shown in Figure 2D, sgRNA2 significantly activated expression of LCAT and was thus chosen for subsequent studies. The data showed that overexpression of LCAT in Huh7 cells exhibited impaired cell proliferation (Figure 2F), colony formation (Figure 2H), migration and invasion (Figure 2J) compared with the controls. LCAT-overexpressed Huh7 cells formed fewer colonies in soft agar than the control cells (Figure 2L), suggesting LCAT impaired in vitro tumorigenicity. These results suggest that LCAT is a tumor suppressor gene, which regulates HCC metastasis via affecting proliferation, migration, invasion and tumorigenicity.
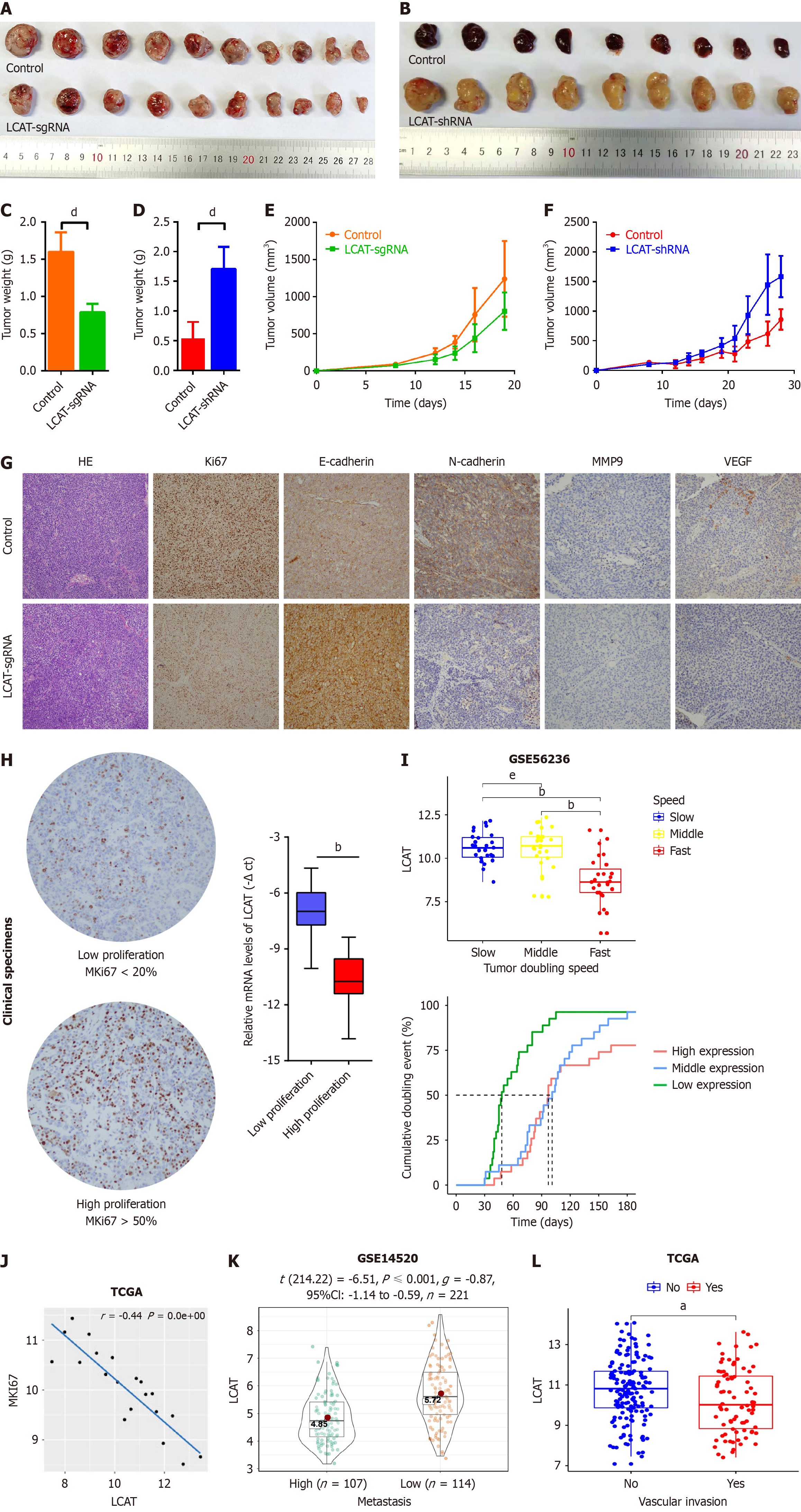
We investigated the role of LCAT in regulating HCC growth and metastasis in vivo. LCAT-overexpressed Huh7 cells (Huh-7-LCAT-sgRNA) or control Huh7 cells were injected subcutaneously into athymic nude mice (Figure 3A-F). The mice were monitored once every 2 d, and killed on day 19. The growth rate of tumors derived from Huh-7-LCAT-sgRNA cells was slower than that of control cells (Figure 3E). In nude mice inoculated with Huh-7-LCAT-sgRNA cells in week 3, the tumor size and average weight were significantly lower than those of the control group (Figure 3A and C). We dissected xenograft tumors and performed immunostaining with indicator antibodies. As shown in Figure 3G, the levels of Ki67, N-cadherin, MMP9, and VEGF in tumors formed from Huh-7-LCAT-sgRNA cells were lower than these in tumors from control cells, while the level of E-cadherin in tumors formed from Huh-7-LCAT-sgRNA cells was lower than that in tumors from control cells. This verified the role of LCAT in affecting HCC cell proliferation, migration and vascular invasion. To examine the effect of LCAT attenuation on HCC progression, we subcutaneously injected HepG2 knockdown cells (HepG2-LCAT-shRNA) or control cells into thymic nude mice The tumors from HepG2-LCAT-shRNA cells grew faster than that from control cells (Figure 3F), and the tumor size and average weight from HepG2-LCAT-shRNA cells were significantly greater compared with the controls (Figure 3B and D). We then analyzed the data from our HCC cohort combined with other HCC patients’ cohorts (TCGA-LICH, GSE54236, and GSE14520). The HCC samples with lower LCAT level had the lower proliferation index using Ki67 as the indicator (Figure 3H), and HCC with the lower levels of LCAT grew faster and had higher metastasis potential and stronger vascular invasion (Figure 3I-L). These results indicate that LCAT suppresses cancer growth and metastasis, which confers antitumor potency in HCC.
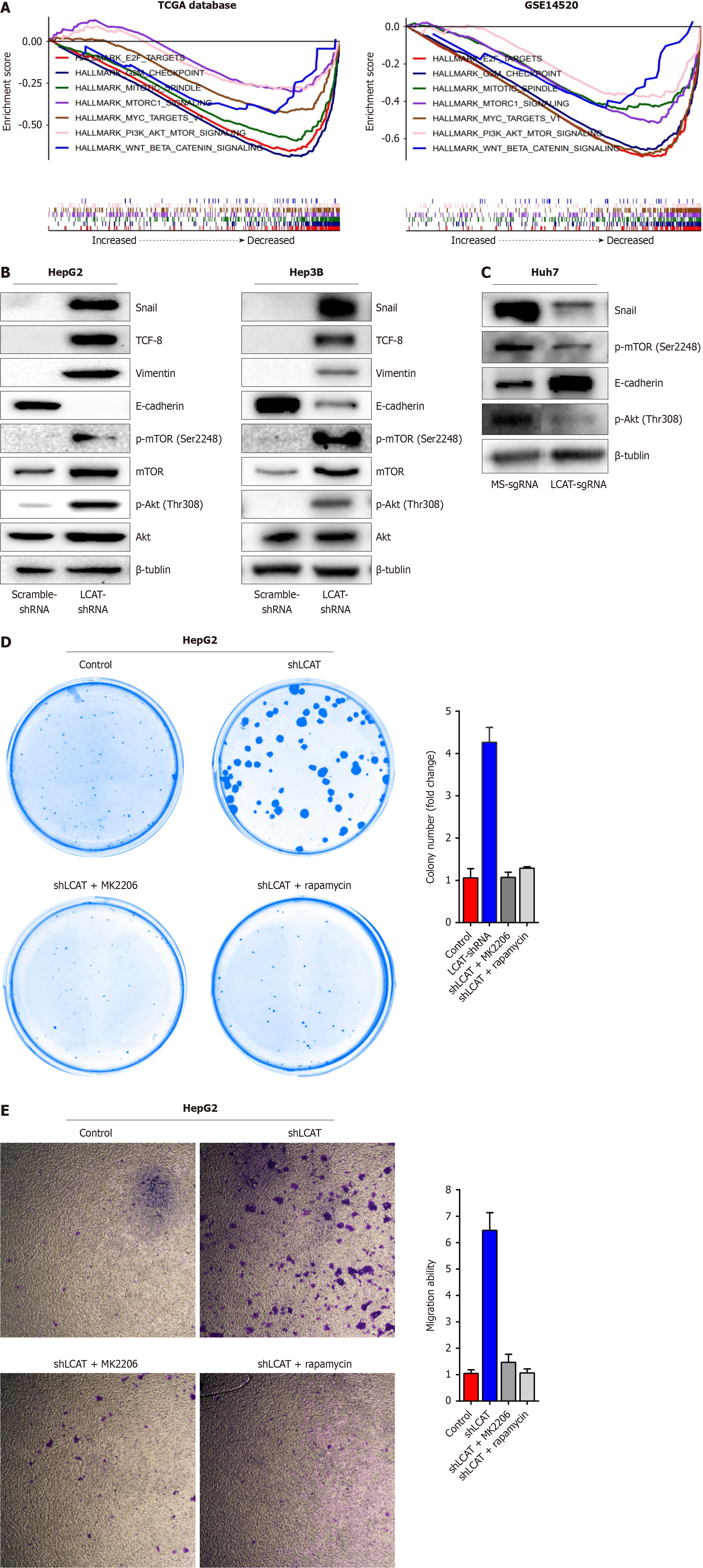
To elucidate the molecular mechanism of LCAT in attenuating HCC metastasis, we performed GSEA in two different HCC cohorts (TCGA and GSE14520). PI3K/Akt/mTOR, mTOR, and proliferation-related signaling (such as E2F target, G2M checkpoint) were significantly enriched in the LCAT low-expression group (Figure 4A). Given the crucial role of the PI3K/Akt/mTOR signaling in tumor progression, we focused on the effect of LCAT on this pivotal signaling pathway. We found that knockdown of LCAT in HepG2 and Hep3B cells led to increased phosphorylation and expression of AKT and mTOR (Figure 4B), indicative of activation of PI3K/Akt/mTOR signaling. We also examined the expression patterns of essential key regulatory factors in epithelial-mesenchymal transition (EMT) and tumor metastasis. The expression level of E-cadherin was reduced, while vimentin, TCF-8, and Snail levels were upregulated in LCAT-knockdown cells (Figure 4B). On the contrary, in Huh-7 cells, overexpression of LCAT reduced the activity of PI3K/Akt/mTOR signaling and altered the expression patterns of E-cadherin and Snail (Figure 4C). Inhibition of Akt or mTOR with their specific inhibitors abolished the increased colony formation and migration due to LCAT knockdown in HepG2 cells (Figure 4D and E). In summary, these results demonstrate that LCAT inhibits HCC growth and metastasis through blocking PI3K/AKT/mTOR signaling.
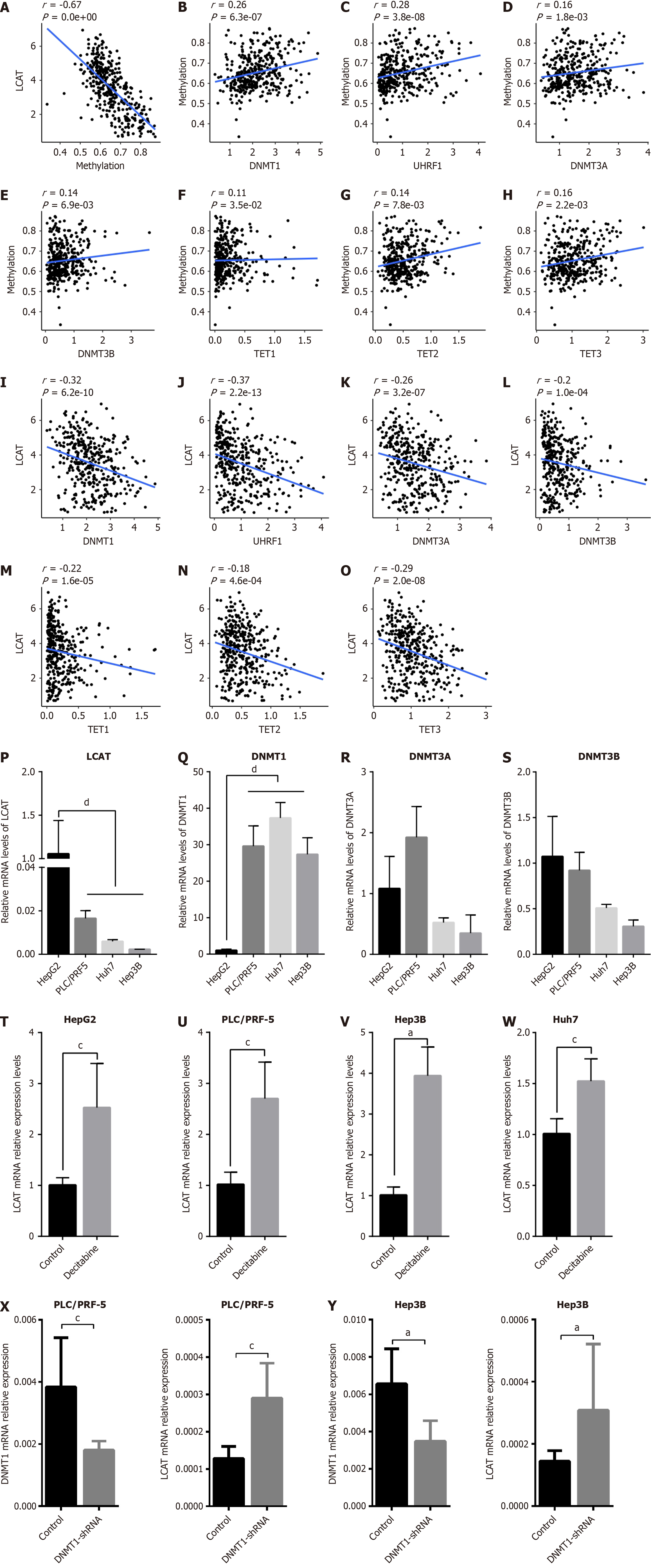
There is increasing evidence that the expression of pivotal tumor-related genes is regulated by aberrant DNA methylation, which is crucial for the progression of tumors. To investigate whether LCAT was regulated by DNA methylation, we performed correlation analysis between the mRNA expression and DNA methylation of LCAT, utilizing the TCGA database. There was a significant negative correlation between LCAT mRNA expression and LCAT gene DNA methylation (Figure 5A). We also explored which DNA methylation-related molecule affected the DNA methylation of LCAT. The strongest positive correlation was between DNMT1 and its mode subunit UHRF1 (Figure 5B-H). We postulated that DNMT1-mediated DNA methylation was the main cause of aberrant LCAT DNA methylation. Subsequently, correlation analysis was conducted between LCAT mRNA and different DNA-methylation-related enzymes. The mRNA expression level of LCAT was negatively correlated with DNA methyltransferases DNMT1, UHRF1, DNMT3A and DNMT3B (Figure 5I-L) and DNA demethylases TET1, TET2 and TET3 (Figure 5M-O), further verifying our hypotheses. We also examined the expression level of LCAT, DNMT1, DNMT3A and DNMT3B in different HCC cell lines using qRT-PCR. Only DNMT1 had a significantly negative correlation with LCAT (Figure 5P-S). This was due to the strongest positive correlation between DNMT1 and DNA methylation, and the strongest negative correlation with gene expression of LCAT. Therefore, we analyzed DNA methyltransferase inhibitor desitabine (1 μM), which can significantly downregulate the protein level of DNMT1[13]. As shown in Figure 5T-W, Decitabine significantly upregulated LCAT expression in different HCC cell lines after treatment for 72 h. To exclude the nonspecificity of decitabine, we knocked down DNMT1 with shRNA, and found that LCAT mRNA expression level was significantly increased as DNMT1 was downregulated in PLC/PRF5 and Hep3B cells (Figure 5X and Y). Our results suggest that abnormal DNA methylation mediated by DNMT1 contributes to the downregulation of LCAT mRNA expression levels during development of HCC.
Metastasis leads to 90% of cancer-related deaths, highlighting the importance of predicting metastasis risk[18]. Moreover, the improved ability to distinguish between high-risk and low-risk HCC patients with metastasis may lead to the development of more effective treatment options and personalized treatment, which hold the promise to prolong survival and improve overall outcome. Given the role of LCAT in metastasis, we first used dataset GSE14520 and applied multivariate logistic regression analysis to evaluate the classification ability of LCAT for both high and low subtypes of metastasis risk. The results indicated that LCAT could be an independent risk factor for clinicopathological characteristics (Supplementary Table 3). Subsequently, we explored a new liver cancer metastasis risk grading prediction model based on LCAT. We used rigorous machine learning methods to evaluate the ability of LCAT expression combined with clinicopathological factors to classify these two subtypes. First, we divided Wang’s cohort (GSE14520) HCC samples into training and test sets according to their metastatic risk. Second, in the training set, we use the least absolute shrinkage and selection operator (LASSO) method to select the most useful predictive features. We applied the selected features in three machine learning algorithms including RF, SVM and LR. The area under the receiver operating characteristics curve (AUC SCORE) indicated the performances through fivefold cross-validation. Using the same pattern, the TCGA database was used to train the best predictive model (Figure 6A). In LCAT and clinicopathological characteristics, 11 features were reduced to three potential predictors on the basis of 129 HCC patients in the training set (43:1 ratio; Figure 6B and C), and LCAT, TNM stage and serum AFP were selected with nonzero coefficients in the LASSO logistic regression model. Combination of the expression level of LCAT, TNM stage and serum AFP could accurately distinguish patients with HCC at high risk of metastasis from those at low risk (RF, AUC score = 0.783, SVM, AUC score = 0.792, LR, AUC score = 0.806, Figure 6D). In the TCGA-LICH cohort, the above combination achieved a similar distinguishing capability (Figure 6E). The best-performing LR algorithm also achieved a high AUC of 0.737 and demonstrated good risk estimation calibration curves on an independent test set (Figure 6F). The decision curve analysis for the LR model in the test set was also presented in Figure 6F. The decision curve showed that if the threshold probability of a patient or doctor is 16%, using the LR model to predict HCC metastasis risk is beneficial for patients to determine whether treatment is needed in the future. We generated Kaplan-Meier curves in the training and validation data sets using the LCAT marker. The high-risk group had 62 observations with 54 events in the training data set and 41 observations with 34 events in the validation data set; and the low-risk group has 67 observations with 40 events in the training dataset and 44 observations with 26 events in the validation data set (Figure 6G). These results indicate that the model based on LCAT can effectively and accurately classify these two subtypes of metastasis and has clinical application value. The LR model based on LCAT may provide a potential approach for predicting the risk of HCC metastasis.
HCC is one of the deadliest malignant tumors worldwide, due to its uncontrolled growth and metastasis, resulting in high mortality and recurrence rates[18,19]. Identifying the factors that lead to uncontrolled expansion and transfer of HCC is important. Increasing evidence suggests that lipid metabolism disorders are associated with poor prognosis after first-line surgery for HCC, and affect the proliferation and migration of HCC[20-22].
Metabolic disorders and immune evasion are commonly present as two important indicators of cancer. LCAT gene encodes an extracellular cholesterol esterifying enzyme in which cholesterol esterification is required for cholesterol transportation[23]. The LCAT gene defect may cause hypercholesterolemia, leading to accumulation of cholesterol in immune cells, promoting inflammatory responses[24]. Many metabolic genes are involved in the occurrence and development of HCC. Due to the crucial role of the liver in many biological processes, we speculated that LCAT dysregulation may have multiple roles in the occurrence and development of HCC. The positive correlation of LCAT activity with high-density lipoprotein indicated the dysregulation of cholesterol in HCC[25]. We observed a decrease in LCAT and demonstrated its prognostic role. Previous studies have demonstrated that LCAT is mainly involved in extracellular metabolism of plasma lipoproteins. Mutations in human LCAT may cause fish-eye disease as well as LCAT deficiency[26]. Our findings firstly demonstrated that LCAT plays an anticancer role in HCC by regulating the PI3K/AKT/mTOR signaling pathway. This pathway is important in various diseases and plays a crucial role in HCC[27]. The PI3K/AKT/mTOR signaling pathway is reported in 40%-50% of HCC[28]. Activation of the PI3K/AKT/mTOR pathway is associated with lower tumor differentiation and early recurrence in HCC[29]. A significant metabolic change in HCC is an increase in lipid uptake and synthesis to support cell growth, proliferation, and tumorigenesis[30]. Our study found that LCAT was hypermethylated in HCC tissues. LCAT could be a good biomarker at predicting HCC diagnosis, prognosis and recurrence. Currently, there is little understanding of the expression and role of LCAT in HCC. LCAT may enhance HCC progression through different mechanisms. Here, through LCAT-associated gene analysis in HCC, we identified the PI3K/Akt/mTOR pathway, in which, LCAT might be involved. To examine the impact of the decreased expression of LCAT, our study investigated the interaction between LCAT and the PI3K/Akt/mTOR pathway. We observed a modest increase in PI3K, Akt, and phosphorylation of Akt and mTOR. The PI3K/Akt/mTOR pathway is an important signaling pathway that regulates cell proliferation, metabolism, tumor cell differentiation, autophagy, apoptosis, oxidative stress, and EMT. The PI3K/Akt/mTOR pathway is overexpressed in almost 50% of HCCs and dysregulated activation of the pathway leading to altered protein expression, DNA damage and carcinogenesis is common in HCC. Increasing evidence demonstrates that the PI3K/Akt/mTOR signaling pathway profoundly affects tumorigenesis and HCC progression. PI3K, AKT, mTOR and phosphatase are key targets in the PI3K/AKT/mTOR pathway. Cellular cholesterol depletion may lead to sustained phosphorylation of various signaling molecules and transcription factors, including Akt. In addition to lipid metabolism (cholesterol metabolism and fatty acid metabolism), immune processes (DNA methylation, allograft, oncogenic signaling and inflammatory response) were also included, indicating the multiple potential of LCAT in HCC. The key role of tumor microenvironment in tumor progression has been extensively studied and confirmed[20]. The downregulation, prognostic effects and their correlation indicated that LCAT might be involved in the PI3K/Akt/mTOR pathway during HCC development and progression. LCAT, as an important component of the tumor microenvironment, plays a crucial role in tumor progression and is a cause of poor prognosis in many malignant tumors, including HCC[21,31]. Inhibiting the activity of the PI3K/AKT/mTOR signaling pathway can prevent abnormal cell proliferation, cellular metabolism, and tumor angiogenesis, thus providing potential molecular targeted therapeutics[32]. Our in vitro and in vivo data showed that LCAT can affect cell proliferation, migration and tumor angiogenesis. Knockdown of LCAT can contribute to activation of the PI3K/AKT/mTOR pathway and inhibition of this specific signaling pathway reverses the related phenotypes. These results imply that LCAT plays a role as a tumor suppressor gene in the development of HCC by regulating the PI3K/AKT/mTOR signaling pathway. The relationship between LCAT and the PI3K/Akt/mTOR pathway may have uncovered a novel signaling pathway in HCC pathogenesis that targets metabolism and forms a metabolic network during HCC development. The underlying mechanism for LCAT regulation of HCC through inhibition of the PI3K/AKT/mTOR pathway has not been entirely elucidated. Future studies are needed to evaluate the detailed molecular mechanisms of LCAT in regulating the PI3K/AKT/mTOR signaling pathway.
Current research has shown that HCC, similar to other tumors, is caused by genetic changes as well as epigenetic abnormalities[33]. The overall DNA methylation pattern changes during tumor development lead to high methylation of CpG islands and low methylation of non-CpG islands[34]. In most types of cancer, DNA hypermethylation can lead to dysregulation and silencing of several tumor suppressor genes[35,36]. Therefore, it is necessary to clarify whether specific tumor-associated genes are regulated by DNA methylation, which can contribute to the development of demethylating drugs with fewer adverse reactions, optimize early diagnosis of HCC, improve the prognosis of HCC, and strengthen HCC treatment[37]. Our study shows that expression of LCAT is significantly negatively correlated with the average DNA methylation of CpGs, and the DNMT1 specific inhibitor (desitabine) restores downregulated LCAT expression in HCC cell lines, indicating that DNMT1-mediated DNA methylation is the main mechanism of LCAT silencing. We explored the possibility of constructing a predictive model based on LCAT using three well-established machine-learning algorithms (RF, SVM and LR) to predict individual metastatic risk. The performance of the LR model including LCAT, TNM stage and serum AFP level was the best and verified in an independent test set. Thus, it may provide a potential predictive method for HCC metastatic risk. Although the predictive model was well constructed based on GEO database and TCGA data and was validated using an independent test set, prospective studies in different populations are required to validate the model. In the future, we plan to collect HCC tissues, TNM stage, serum AFP level and follow-up information of HCC patients and examine the LCAT mRNA expression to validate the predictive ability of the potential model for HCC metastatic risk.
Downregulation of LCAT in HCC was observed in our study. It demonstrated that LCAT played a pivotal role in HCC by regulating the activity of the PI3K/AKT/mTOR signaling pathway. In HCC patients’ survival analysis, low expression of LCAT was associated with RFS and OS. In addition, the predictive model based on LCAT effectively predicted the risk of HCC progression. LCAT is a potential biomarker for the prognosis of HCC and a prospective therapeutic target for HCC. The function of LCAT needs further study.
The authors wish to thank all the patients and family members that participated in the study. We also thank Dr. Xiao-Wen Huang and technician Peng-Fei Xu for their support and assistance during the experimental process.
| 1. | Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, Pikarsky E, Zhu AX, Finn RS. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 1016] [Article Influence: 338.7] [Reference Citation Analysis (2)] |
| 2. | Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 781] [Article Influence: 156.2] [Reference Citation Analysis (0)] |
| 3. | Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3059] [Cited by in RCA: 3622] [Article Influence: 258.7] [Reference Citation Analysis (0)] |
| 4. | Jiang Y, Sun A, Zhao Y, Ying W, Sun H, Yang X, Xing B, Sun W, Ren L, Hu B, Li C, Zhang L, Qin G, Zhang M, Chen N, Zhang M, Huang Y, Zhou J, Zhao Y, Liu M, Zhu X, Qiu Y, Sun Y, Huang C, Yan M, Wang M, Liu W, Tian F, Xu H, Zhou J, Wu Z, Shi T, Zhu W, Qin J, Xie L, Fan J, Qian X, He F; Chinese Human Proteome Project (CNHPP) Consortium. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567:257-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 623] [Article Influence: 103.8] [Reference Citation Analysis (0)] |
| 5. | Peng X, Chen Z, Farshidfar F, Xu X, Lorenzi PL, Wang Y, Cheng F, Tan L, Mojumdar K, Du D, Ge Z, Li J, Thomas GV, Birsoy K, Liu L, Zhang H, Zhao Z, Marchand C, Weinstein JN; Cancer Genome Atlas Research Network, Bathe OF, Liang H. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018;23:255-269.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 211] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 6. | Pascual G, Avgustinova A, Mejetta S, Martín M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, Bescós C, Di Croce L, Benitah SA. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 668] [Cited by in RCA: 1035] [Article Influence: 115.0] [Reference Citation Analysis (0)] |
| 7. | Piper DE, Romanow WG, Gunawardane RN, Fordstrom P, Masterman S, Pan O, Thibault ST, Zhang R, Meininger D, Schwarz M, Wang Z, King C, Zhou M, Walker NP. The high-resolution crystal structure of human LCAT. J Lipid Res. 2015;56:1711-1719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 8. | Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19:670-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 168] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 9. | Kosek AB, Durbin D, Jonas A. Binding affinity and reactivity of lecithin cholesterol acyltransferase with native lipoproteins. Biochem Biophys Res Commun. 1999;258:548-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Clay MA, Pyle DH, Rye KA, Barter PJ. Formation of spherical, reconstituted high density lipoproteins containing both apolipoproteins A-I and A-II is mediated by lecithin:cholesterol acyltransferase. J Biol Chem. 2000;275:9019-9025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 11. | Yang K, Wang J, Xiang H, Ding P, Wu T, Ji G. LCAT- targeted therapies: Progress, failures and future. Biomed Pharmacother. 2022;147:112677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 12. | Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 2091] [Article Influence: 190.1] [Reference Citation Analysis (0)] |
| 13. | Negrotto S, Hu Z, Alcazar O, Ng KP, Triozzi P, Lindner D, Rini B, Saunthararajah Y. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res. 2011;71:1431-1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 14. | Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, Matthews L, May B, Milacic M, Rothfels K, Shamovsky V, Webber M, Weiser J, Williams M, Wu G, Stein L, Hermjakob H, D’Eustachio P. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481-D487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 988] [Cited by in RCA: 1003] [Article Influence: 100.3] [Reference Citation Analysis (0)] |
| 15. | Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940-3941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2054] [Cited by in RCA: 2020] [Article Influence: 101.0] [Reference Citation Analysis (0)] |
| 16. | Yuan Y, Xu Y, Xu J, Ball RL, Liang H. Predicting the lethal phenotype of the knockout mouse by integrating comprehensive genomic data. Bioinformatics. 2012;28:1246-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7447] [Cited by in RCA: 8425] [Article Influence: 561.7] [Reference Citation Analysis (0)] |
| 18. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19834] [Cited by in RCA: 19515] [Article Influence: 780.6] [Reference Citation Analysis (0)] |
| 19. | Bouek JW. Navigating Networks: How Nonprofit Network Membership Shapes Response to Resource Scarcity. Soc Probl. 2018;65:11-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 20. | Lamarca A, Mendiola M, Barriuso J. Hepatocellular carcinoma: Exploring the impact of ethnicity on molecular biology. Crit Rev Oncol Hematol. 2016;105:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Samarin J, Laketa V, Malz M, Roessler S, Stein I, Horwitz E, Singer S, Dimou E, Cigliano A, Bissinger M, Falk CS, Chen X, Dooley S, Pikarsky E, Calvisi DF, Schultz C, Schirmacher P, Breuhahn K. PI3K/AKT/mTOR-dependent stabilization of oncogenic far-upstream element binding proteins in hepatocellular carcinoma cells. Hepatology. 2016;63:813-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 22. | Song TJ, Ip EW, Fong Y. Hepatocellular carcinoma: current surgical management. Gastroenterology. 2004;127:S248-S260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 170] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 23. | Lu K, Shi TS, Shen SY, Shi Y, Gao HL, Wu J, Lu X, Gao X, Ju HX, Wang W, Cao Y, Chen D, Li CJ, Xue B, Jiang Q. Defects in a liver-bone axis contribute to hepatic osteodystrophy disease progression. Cell Metab. 2022;34:441-457.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 65] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 24. | Santamarina-Fojo S, Lambert G, Hoeg JM, Brewer HB Jr. Lecithin-cholesterol acyltransferase: role in lipoprotein metabolism, reverse cholesterol transport and atherosclerosis. Curr Opin Lipidol. 2000;11:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 25. | Bakiri L, Hamacher R, Graña O, Guío-Carrión A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK, Hasenfuss SC, Wagner EF. Liver carcinogenesis by FOS-dependent inflammation and cholesterol dysregulation. J Exp Med. 2017;214:1387-1409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 87] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 26. | Selitsky SR, Dinh TA, Toth CL, Kurtz CL, Honda M, Struck BR, Kaneko S, Vickers KC, Lemon SM, Sethupathy P. Transcriptomic Analysis of Chronic Hepatitis B and C and Liver Cancer Reveals MicroRNA-Mediated Control of Cholesterol Synthesis Programs. mBio. 2015;6:e01500-e01515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 27. | Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: current state and future trends. J Hepatol. 2014;60:855-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 261] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 28. | Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, Battiston C, Van Laarhoven S, Fiel MI, Di Feo A, Hoshida Y, Yea S, Toffanin S, Ramos A, Martignetti JA, Mazzaferro V, Bruix J, Waxman S, Schwartz M, Meyerson M, Friedman SL, Llovet JM. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972-1983, 1983.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 591] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 29. | Luo YD, Fang L, Yu HQ, Zhang J, Lin XT, Liu XY, Wu D, Li GX, Huang D, Zhang YJ, Chen S, Jiang Y, Shuai L, He Y, Zhang LD, Bie P, Xie CM. p53 haploinsufficiency and increased mTOR signalling define a subset of aggressive hepatocellular carcinoma. J Hepatol. 2021;74:96-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 30. | Ning Z, Guo X, Liu X, Lu C, Wang A, Wang X, Wang W, Chen H, Qin W, Liu X, Zhou L, Ma C, Du J, Lin Z, Luo H, Otkur W, Qi H, Chen D, Xia T, Liu J, Tan G, Xu G, Piao HL. USP22 regulates lipidome accumulation by stabilizing PPARγ in hepatocellular carcinoma. Nat Commun. 2022;13:2187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 115] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 31. | Xing S, Yu W, Zhang X, Luo Y, Lei Z, Huang D, Lin J, Huang Y, Huang S, Nong F, Zhou C, Wei G. Isoviolanthin Extracted from Dendrobium officinale Reverses TGF-β1-Mediated Epithelial⁻Mesenchymal Transition in Hepatocellular Carcinoma Cells via Deactivating the TGF-β/Smad and PI3K/Akt/mTOR Signaling Pathways. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Wang J, Zhou Y, Li D, Sun X, Deng Y, Zhao Q. TSPAN31 is a critical regulator on transduction of survival and apoptotic signals in hepatocellular carcinoma cells. FEBS Lett. 2017;591:2905-2918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Feitelson MA. Parallel epigenetic and genetic changes in the pathogenesis of hepatitis virus-associated hepatocellular carcinoma. Cancer Lett. 2006;239:10-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1689] [Cited by in RCA: 1864] [Article Influence: 116.5] [Reference Citation Analysis (0)] |
| 35. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3397] [Cited by in RCA: 3722] [Article Influence: 161.8] [Reference Citation Analysis (0)] |
| 36. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3299] [Cited by in RCA: 3406] [Article Influence: 189.2] [Reference Citation Analysis (0)] |
| 37. | Hernandez-Meza G, von Felden J, Gonzalez-Kozlova EE, Garcia-Lezana T, Peix J, Portela A, Craig AJ, Sayols S, Schwartz M, Losic B, Mazzaferro V, Esteller M, Llovet JM, Villanueva A. DNA Methylation Profiling of Human Hepatocarcinogenesis. Hepatology. 2021;74:183-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |