Published online Aug 15, 2024. doi: 10.4251/wjgo.v16.i8.3585
Revised: June 6, 2024
Accepted: June 25, 2024
Published online: August 15, 2024
Processing time: 215 Days and 21.7 Hours
Gastrointestinal stromal tumors (GISTs) are typical gastrointestinal tract neo
To further investigate the mechanism of IGF2 specific to GISTs.
IGF2 was screened and analyzed using Gene Expression Omnibus (GEO: GSE225819) data. After IGF2 knockdown or overexpression by transfection, the phenotypes (proliferation, migration, invasion, apoptosis) of GIST cells were characterized by cell counting kit 8, Transwell, and flow cytometry assays. We used western blotting to evaluate pathway-associated and epithelial-mesen
Data from the GEO indicated that IGF2 expression is high in GISTs, associated with liver metastasis, and closely related to drug resistance. GIST cells with high expression of IGF2 had increased proliferation and migration, invasiveness and EMT. Knockdown of IGF2 significantly inhibited those activities. In addition, OE-IGF2 promoted GIST metastasis in vivo in nude mice. IGF2 activated IGF1R signaling in GIST cells, and IGF2/IGF1R-mediated glycolysis was required for GIST with liver metastasis. GIST cells with IGF2 knockdown were sensitive to imatinib treatment when IGF2 overexpression significantly raised imatinib resistance. Moreover, 2-deoxy-D-glucose (a glycolysis inhibitor) treatment reversed IGF2 overexpression-mediated imatinib resistance in GISTs.
IGF2 targeting of IGF1R signaling inhibited metastasis and decreased imatinib resistance by driving glycolysis in GISTs.
Core Tip: Our study found that insulin-like growth factor 2 (IGF2) regulated metastasis and imatinib resistance in gastrointestinal stromal tumors (GISTs). IGF2 interacted with IGF1R to regulate glycolysis. Our results confirm that IGF2 targeting of IGF1R signaling inhibited metastasis and improved imatinib chemosensitivity by driving glycolysis in GISTs and indicated that IGF2 might be used to reverse imatinib resistance in GIST patients.
- Citation: Li DG, Jiang JP, Chen FY, Wu W, Fu J, Wang GH, Li YB. Insulin-like growth factor 2 targets IGF1R signaling transduction to facilitate metastasis and imatinib resistance in gastrointestinal stromal tumors. World J Gastrointest Oncol 2024; 16(8): 3585-3599
- URL: https://www.wjgnet.com/1948-5204/full/v16/i8/3585.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i8.3585
Primary gastrointestinal stromal tumors (GISTs) account for 2% of gastrointestinal tumors[1,2]. GISTs are encoded by the receptor tyrosine kinase gene KIT or PDGFRA[3]. These mutations cause ligand-dependent activation and constitutive activation of signal transduction mediated by PDGFRA or KIT[4]. The downstream molecular pathways of the KIT mutation include PI3K/AKT, JAK-STAT, Src family kinases, and Ras-ERK)[5,6]. Activation of molecular pathways follows KIT activation and leads to the occurrence of GISTs tumors by activation of cell proliferation and inhibition of apoptosis signals [7].
Imatinib remains the primary treatment of GIST patients with advanced or metastatic tumors[8,9]. Imatinib significantly improves the prognosis of patients in the advanced stages of the disease, but those undergoing imatinib treatment often encounter challenges associated with both primary and secondary drug resistance, which, unfortunately, restricts long-term efficacy[10].
Insulin-like growth factor 2 (IGF2) is a genomic imprinting gene in growth on the chromosome 11 short arm[11]. IGF2 overexpression is observed in a variety. of cancers and is related to chemotherapy resistance and a worse prognosis[12-14]. Studies of IGF1R have increased recently. Insulin-like growth factor (IGF) is comprised of the two ligands IGF1 and IGF2, their target tyrosine kinase receptors, IGF1 receptor (IGF1R) and the insulin receptor, as well as the IGF2 receptor (IGF2R) and IGF-binding proteins that regulate IGF ligand availability[15]. IGF1R, is a tyrosine kinase receptor with binding affinity for both IGF1 and IGF2 ligands[16]. Upon ligand binding, the activated tyrosine kinase domain initiates signaling cascades that specifically activate the GPTase Ras-Raf-ERK/MAPK and PI3K-AKT/mTOR pathways. These pathways, regulate the proliferation rate and apoptosis of cancer cells[17,18]. The IGF pathway family gene expression (such as IGF1, IGF2, and IGF1R) has been reported to distinguish subsets of GISTs wild type for KIT and PDGFRA[19]. Although data on IGF1R in GISTs have been reported[20-22], further research on the mechanisms of IGF2 and IGF1R in GISTs is needed.
Sequencing data from the Gene Expression Omnibus (GEO) database (GSE225819 and GSE155880) were examined by bioinformatics. We found that IGF2 acted as a cancer-promoting factor and was involved in cell proliferation, apoptosis, liver metastasis, and epithelial-mesenchymal transition (EMT) in GISTs. Moreover, the role of IGF2 in GIST cells and the IGF2-IGF1R regulatory axis contributed to imatinib resistance of GISTs by regulating glycolysis and represents a target for GISTs therapy.
Gene expression data based on RNA sequencing were obtained from the GEO. Two eligible datasets (GSE225819, GSE155880) were combined. The aligned reads were calculated by FeatureCounts (subread/2.0, http://subread.sourceforge.net/) and differentially expressed genes (DEGs) were analyzed by the R package DESeq2/3.1.0 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)[23]. A total of 2578 DEGs (1398 downregulated, and 1188 upregulated) were identified by screening GSE225819, including 20 normal samples and 20 GISTs samples with liver metastasis (|log2FC| > 1; P < 0.05) (Supplementary Table 1). Based on Deseq2, 1386 DEGs (939 downregulated, and 447 upregulated) were identified by screened GSE155880 including seven Imatinib-sensitive samples and seven imatinib-resistant GIST patients (|log2FC| > 1; P < 0.05) (Supplementary Table 2).
RGM-1 normal human gastric mucosal cells, GIST882, and GIST-T1 cells were cultured in Iscove's modified Dulbecco's medium containing10% fetal bovine serum and 1% antibiotics, The culture temperature was 37 °C with 5% CO2. The imatinib concentration was increased from 1 nM to 100 nM over 10 mon and repeated to obtain imatinib-resistant GIST882 (GIST882-R) and GISTT1 (GISTT1R) cells. GIST882 and GIST-T1 cells were transfected with OE-IGF2, sh-IGF2 plasmids and sh-NC, OE-NC negative controls (RiboBio, Beijing, China) using Lipofectamine 3000 (Invitrogen, Waltham, MA, United States) and cultured for 2 d. Transfection efficiency was determined by western blotting. Imatinib mesylate was purchased from Selleckchem (Houston, TX, United States). GIST-T1 and GIST-882 cells were treated with serial dilutions of 1 μM imatinib in dimethyl sulfoxide for 4 h.
We lysed transfected cells with RIPA buffer, the total protein was purified, and the protein concentration was determined with bicinchoninic kits (ThermoFisher Scientific, Waltham, MA, United States). The proteins were resolved by 10% SDS-PAGE and transferred to PVDF membranes for incubation with anti-IGF2 (1:1000, ab177467; Abcam, Cambridge, United Kingdom), anti-vimentin (1:1000, ab92547; Abcam), anti-N-cadherin (1:1000, ab76011; Abcam), anti-E-cadherin (1:1000, ab40772; Abcam), anti-Twist1 (1:1000, ab50887; Abcam), anti-IGF1R (1:1000, ab182408; Abcam), anti-p-IGF1R (1:1000, ab39398; Abcam), anti-PI3K (1:1000, ab302958; Abcam), anti-AKT (1:1000, MA5-14916; Invitrogen), anti-phospho-AKT (1:1000, PA5-95669; Invitrogen), and anti-β-actin (1:1000, ab8227; Abcam) primary antibodies overnight at 4 °C after blocking with skimmed milk (5%). After washing the primary antibodies away, the proteins were incubated with the anti-rabbit secondary antibody (1:5000; SA00001-2; SanYing Biotechnology Inc, Wuhan, China) for 1 h. The protein bands were visualized using an ECL chemiluminescence system, and the protein blots were quantified with Image J.
The concentration of IGF2 was measured using ELISA kits (Abcam) according to the manufacturer′s instructions. The samples were prepared from cell culture supernatants and the IGF2 concentration was measured at 450 nm using a microplate reader.
We determined GIST cell proliferation by cell counting kit-8 (CCK-8) assay. OE-IGF2- or sh-IGF2-transfected GIST882 and GIST-T1 cells were added to 96-well plates (1 × 103/well). After 1 d, we added CCK-8 reagent (10 μL, Catalog No. AD10; Dojindo Molecular Technologies, Kumamoto, Japan) to each well at room temperature. Absorbance was monitored at 0, 24, 48, 72, and 96 h and the half inhibitory concentration of imatinib was determined at 450 nm. After overnight incubation, the cells were treated with imatinib at 0, 20, 40, 60, and 80 μmol/L for 48 h. CompuSyn software was used to calculate the combination index using the Chou-Talalay method[24] to determine the antagonistic influence.
For the migration assay, GIST cells were seeded into 8 µm well Transwell chambers (Corning; Corning, NY, United States). The upper chamber was filled with 200 µL serum-free medium containing 2 × 104 cells and the lower chamber was filled with 500 μL complete medium (10% FBS). After 48 h, the cells were fixed with formaldehyde and stained with 0.2% crystal violet for 10 min. To assay cell invasion, 500 μL culture supernatant was collected from transfected cells and added to the upper Transwell chamber. GIST cells (2 × 104 cells) in about 200 μL serum-free medium were added to the lower chamber. The cells were cultured for 2 d at 37 °C with 5% CO2. After culturing, cells remaining in the lower chamber were removed with cotton swabs and those in the upper chamber were stained with 0.2% crystal violet for 5 min. We used an inverted microscope to count the cells that had migrated through the membrane and invaded the upper chamber.
We bought 5-wk-old; male BALB/c nude mice from Vital River Laboratories (Beijing, China) and housed them for 1 wk to adapt to the environment. GIST-T1 cells (5 × 106) transfected with OE-IGF2/OE-NC, sh-IGF2/sh-NC were injected into the inguinal skin and the mice were monitored for growth of the tumor for 7 d before being randomized to four groups and treated with imatinib 50 mg/kg daily. After 4 wk, we killed the mice with an overdose of pentobarbital. All animal experiments were approved by the Animal Ethics Committee of Beijing Viewsolid Biotechnology Co. LTD (Protocol No. VS2126A00170) and all methods followed the ARRIVE guidelines. We fixed the liver tissue of mice in neutral formalin (10%), embedded it in paraffin, cut the tissue into 4 µm sections, and stained it with hematoxylin and eosin (HE). The sections were observed with a microscope.
Cells were incubated in commercial seahorse XF assay medium plus pyruvate (1 mmol/L), glucose (10 mmol/L) and glutamine (2 mmol/L) 37 °C for 1 h in a CO2-free incubator. The rate of extracellular acidification was measured before and after addition of oligomycin, glucose, and 2-deoxy-D-glucose (2-DG). FCCP, a mitochondrial uncoupling agent; oligomycin, an ATP synthase inhibitor; 2-DG, a glycolysis inhibitor; rotenone; and antimycin A were added and metabolic energy consumption was assayed with a Seahorse XF96 Analyzer (Agilent, Santa Clara, CA, United States).
The concentration of lactate in transfected cells was determined by ELISA with lactate assay kits (MAK064; Sigma-Aldrich, St Louis, MO, United States) according to the manufacturer’s protocol. The optical density of each well was determined at 570 nm (Plate Reader AF2000; Eppendorf, Waltham, MA, United States).
GIST cell apoptosis was assayed by flow cytometry (LSRII; BD Biosciences, Franklin Lakes, NJ, United States). using annexin V-FITC apoptosis detection kits. The apoptosis rate was determined by analysis of Q2 and Q3 quadrant cells.
We used GraphPad Prism 7.0 for data analysis. Data were reported as mean ± standard deviation of three independent experiments. Single-group comparisons were done with Student’s t-tests. Multiple group differences were compared by analysis of variance. P < 0.05 indicated significance.
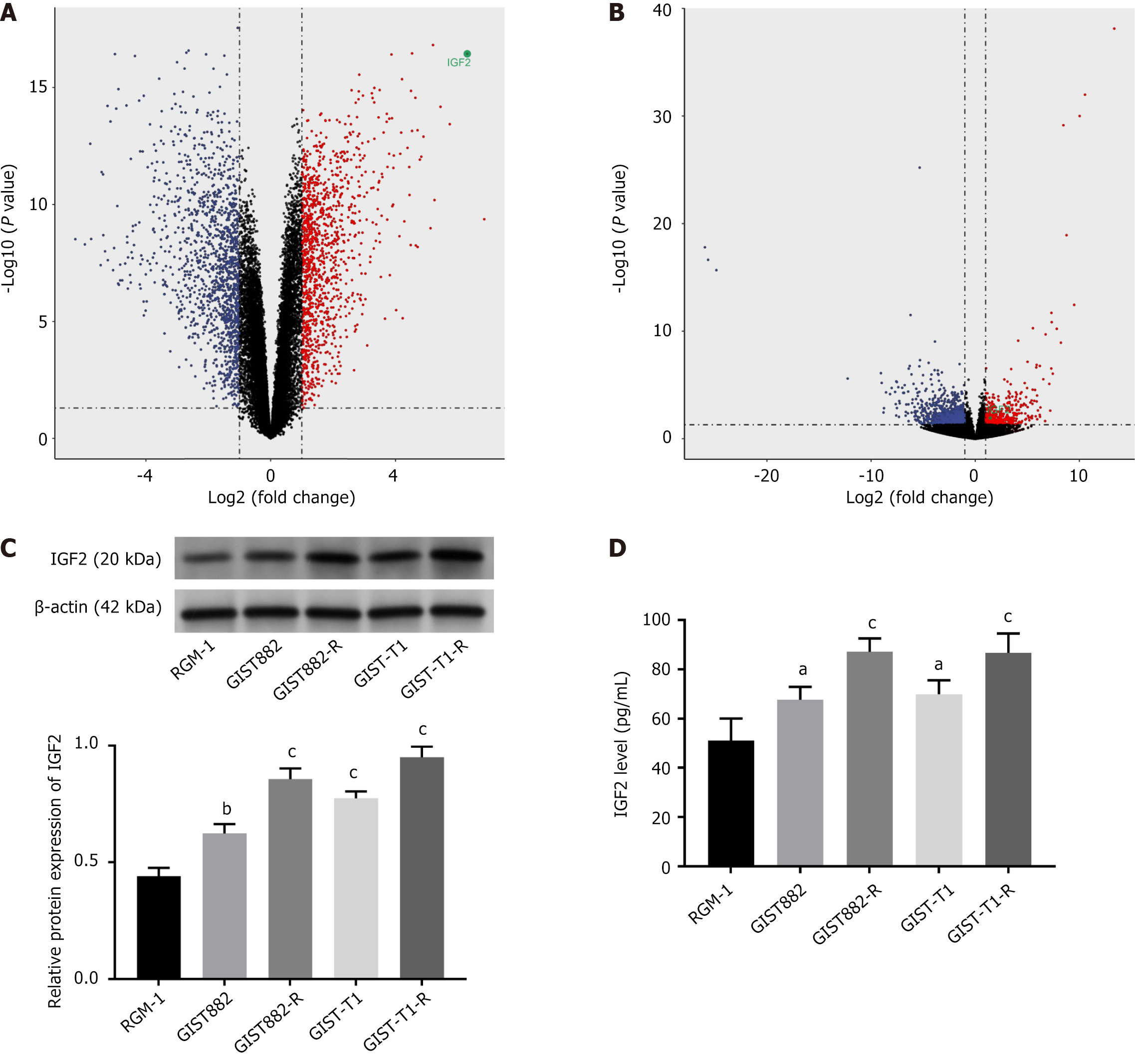
Based on the limma R package, a total of 2578 (DEGs 1398 downregulated and 1188 upregulated) were screened out from GEO: GSE225819 data, including 20 normal samples and 20 GIST samples with liver metastasis (|log2FC| > 1; P < 0.05), suggesting that these DEGs may be involved in liver metastasis in GIST patients (Figure 1A). The top 10 upregulated genes were PENK, IGF2, GPR20, CTSL, SCRG1, PNMAL1, NKX3-2, ANO1, PLAT, and BCHE. The top 10 downregulated genes were ATP4B, GKN1, MT1G, GKN2, ATP4A, SPINK1, TSPAN8, TFF1, KCNE2, and REG1A (Supplementary Table 1). Based on the Deseq2, 1386 DEGs (939 downregulated and 447 upregulated) were screened out in GSE155880, including seven Imatinib-sensitive samples and seven imatinib-resistant GIST patients (|log2FC| > 1; P < 0.05, Figure 1B). The intersection of the two analyses indicated that only IGF2 was involved in the drug resistance regulation and GIST metastasis in these DEGs (Supplementary Table 2). Moreover, we evaluated IGF2 expression in the GIST cell line. By western blotting, expression levels of IGF2 in GIST882, GIST882-R, GIST-T1, and GIST-T1-R were higher than those in normal RGM-1. Furthermore, IGF2 was significantly over expressed in GIST882-R/GIST-T1-R compared with other cell lines GIST882/GIST-T1 (P < 0.01, P < 0.001; Figure 1C). In addition, the expression levels of IGF2 in culture supernatants were measured using ELISA and compared (Figure 1D). We found that the ELISA and western blot results (P < 0.05, P < 0.001) were similar. IGF2 expression was high in drug-resistant GIST cell lines, suggesting that IGF2 overexpression may be closely related to drug resistance.
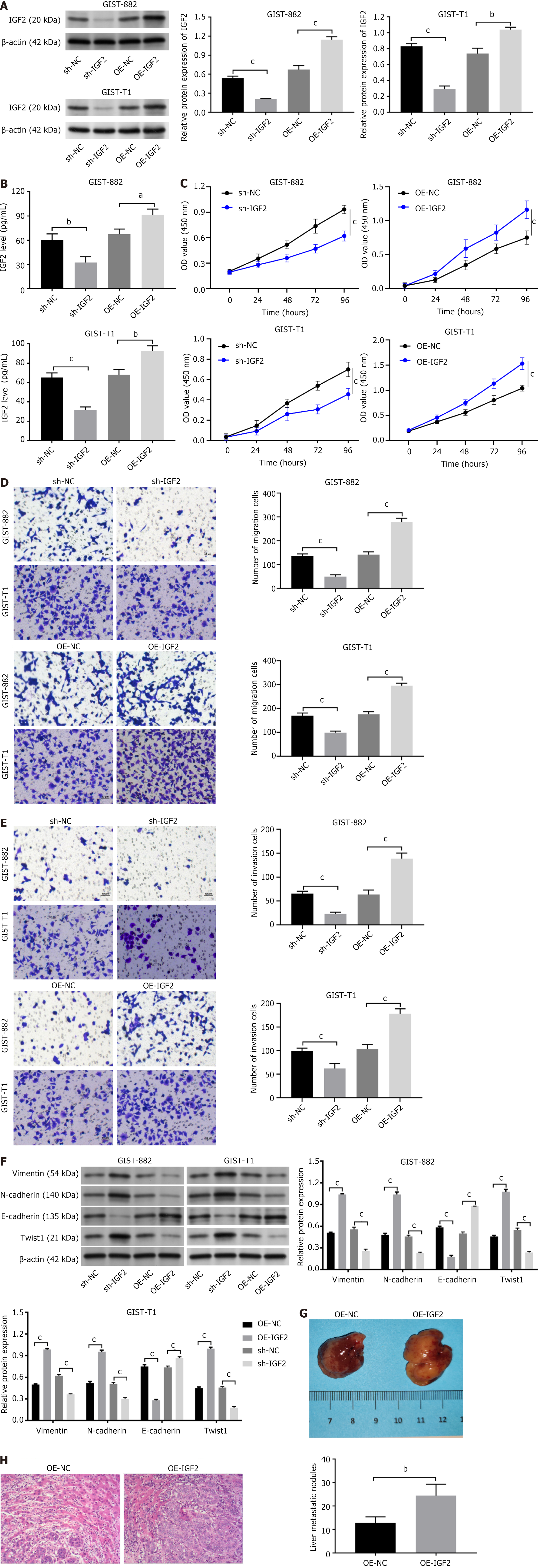
We transfected GIST882 and GIST-T1 cells with an IGF2 overexpressing plasmid (OE-IGF2) or a shRNA to knock down IGF2 (sh-IGF2). Western blotting detected the efficiency of cell transfection (Figure 2A). IGF2 was highly expressed in OE-IGF2-transfected cells compared with OE-NC cells, while IGF2 expression was low in sh-IGF2-transfected cells (P < 0.001). ELISA also found that IGF2 expression high in OE-IGF2 group compared with OE-NC-GIST882 and GIST-T1 cells and IGF2 was low expressed in sh-IGF2-transfected cells (Figure 2B, P < 0.05, P < 0.01, P < 0.001). The CCK-8 results showed that cell viability was significantly increased after exogenous expression of IGF2, sh-IGF2 transfection inhibited GIST882 and GIST-T1 cell viability (Figure 2C, P < 0.001). Likewise, the Transwell assays found more migrating and invading OE-IGF2-GIST882 and GIST-T1 cells compared with their respective control cells (Figure 2D and E, P < 0.001). We also found that sh-IGF2 transfection inhibited cell viability, migration and invasion. In addition, western blotting detect EMT-related proteins (E-cadherin, vimentin, Twist1, and N-cadherin) expression in cells. Silencing IGF2 increased E-cadherin expression, and inhibited vimentin, Twist1, and N-cadherin expression, but IGF2 overexpression had the opposite experimental findings (Figure 2F, P < 0.001). To further verify the functional role of IGF2 on the growth of GISTs, we performed nude mouse tumorigenesis experiments. OE-IGF2 transfected-GIST-T1 cell lines were injected into the spleen. We found that OE-IGF2 promoted the GIST-T1 cell metastasis in vivo, showing a significant decline in the number of liver metastatic nodules (Figure 2G and H, P < 0.01).
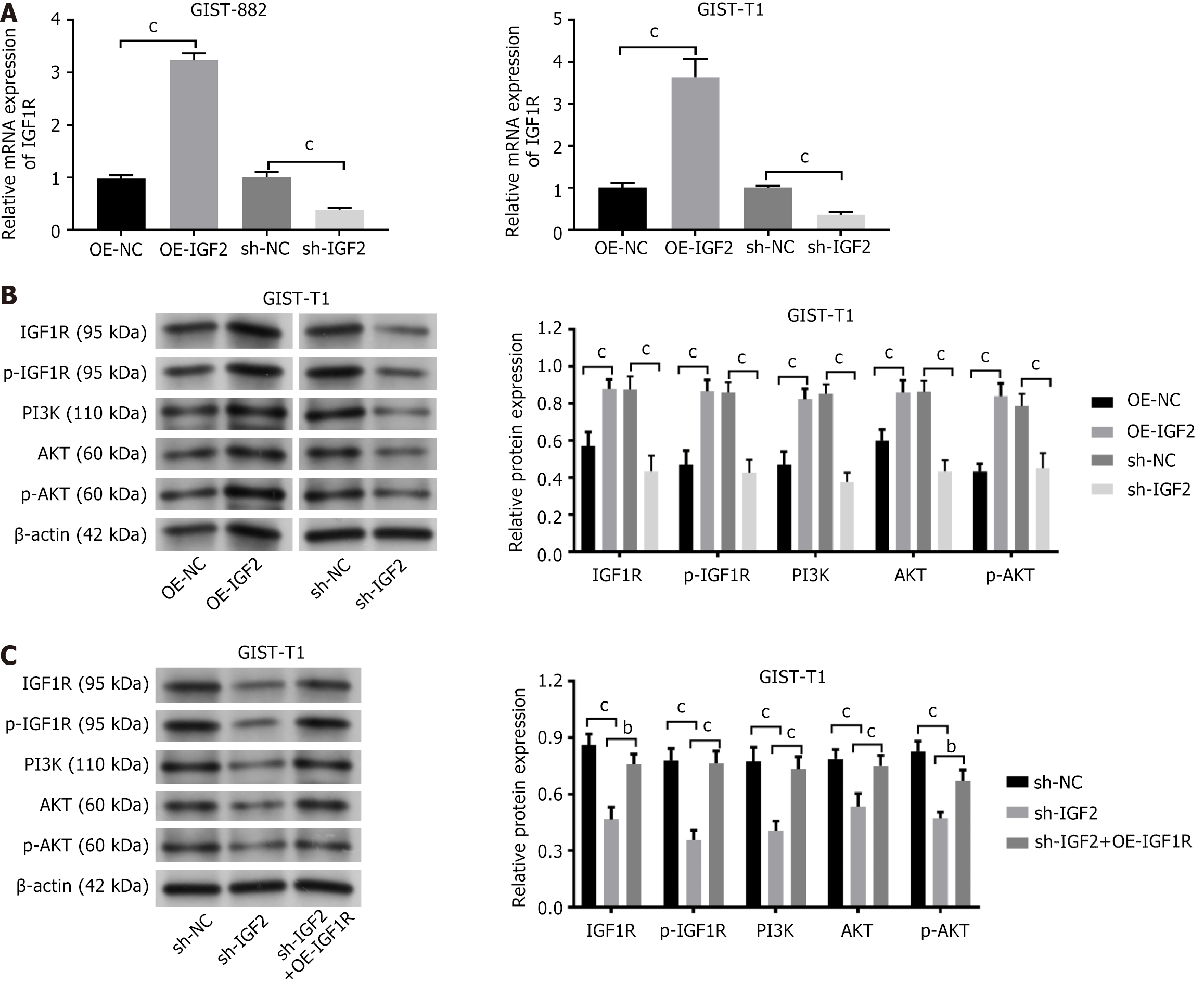
IGF1R mRNA expression was increased in GIST-T1 and GIST882 cells transfected with OE-IGF2, and IGF1R mRNA expression was decreased after sh-IGF2 transfection (Figure 3A, P < 0.001). PI3K-Akt signaling is the IGF2-IGF1R signal principal downstream target[25]. Expression of IGF2-IGF1R pathway-associated proteins (IGF1R, p-IGF1R, PI3K, AKT, p-AKT) in GIST-T1 cells was measured by western blotting. IGF2 overexpression increased the expression of IGF1R, p-IGF1R, PI3K, AKT, and p-AKT in GIST-T1 cells. The opposite result was noted after IGF2 knockdown (Figure 3B, P < 0.01, P < 0.001). Although sh-IGF2 reduced IGF1R, p-IGF1R, PI3K, AKT, and p-AKT expression in GIST-T1 cells, it was partially restored by overexpression of IGF2R (Figure 3C, P < 0.01, P < 0.001).
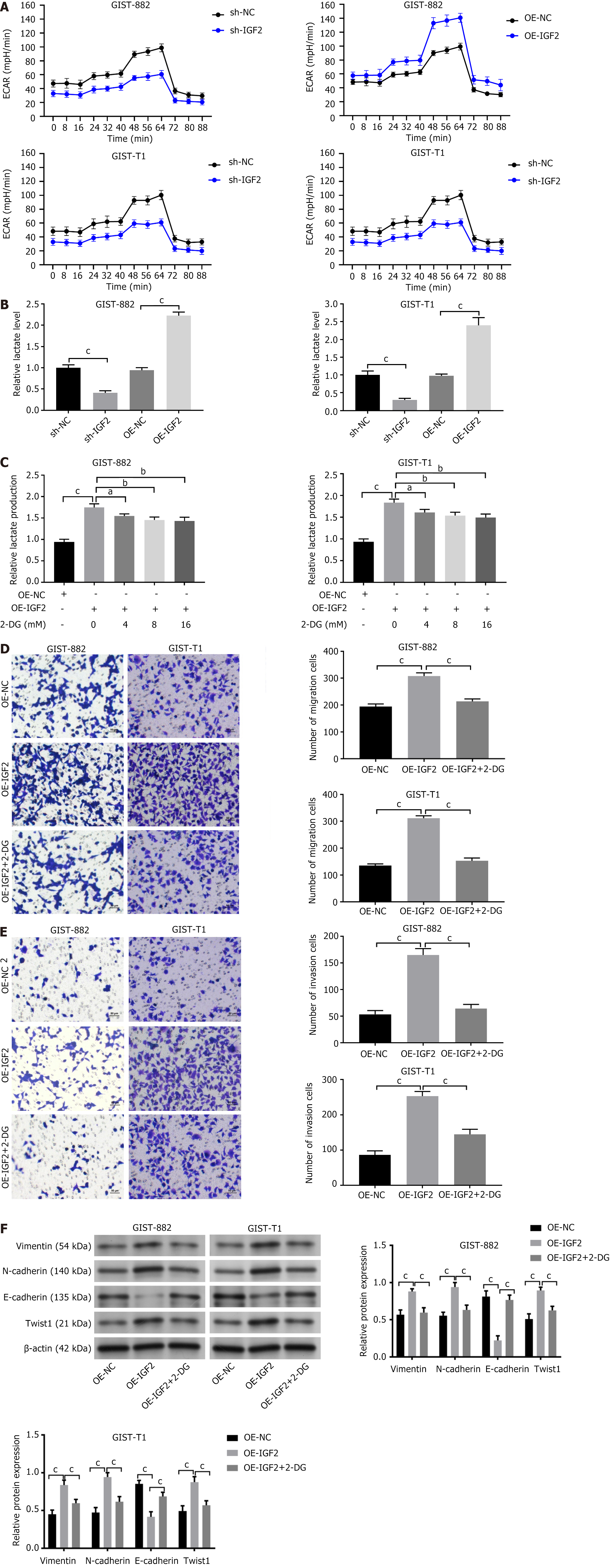
We analyzed glucose consumption and lactate production in GIST cells. Sh-IGF2 inhibited glucose consumption (Figure 4A), and lactate production in GIST882 and GIST-T1 cells (Figure 4B), but IGF2 overexpression had the opposite experimental findings (P < 0.001). To examine the role of the Warburg effect in liver metastasis of GISTs, we treated OE-NC-GIST882 and OE-IGF2-GIST882 cells with 2-deoxyglucose (2-DG, a glycolysis inhibitor) for 24 hat 0, 4, 8, and 16 mmol/L. 2-DG significantly inhibited glycolysis (Figure 4C, P < 0.05, P < 0.01, P < 0.001) and Transwell assays found that 2-DG treatment inhibited the promoting effect of OE-IGF2 on GIST882 and GIST-T1 cell invasion and migration (Figure 4D and E, P < 0.001). Similarly, OE-IGF2 increased vimentin, Twist1, and N-cadherin expression and inhibited E-cadherin expression in cells, but the expression was partially restored by 2-DG treatment (Figure 4F, P < 0.001).
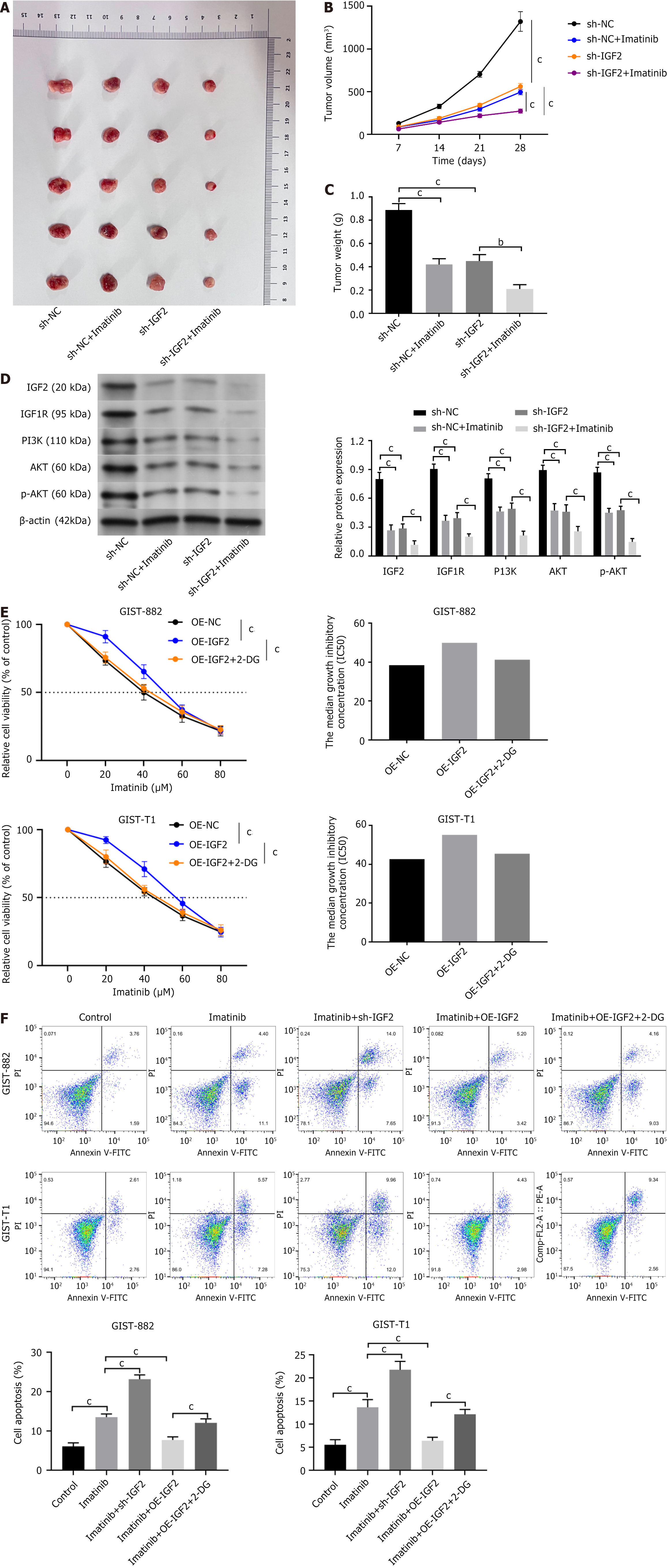
Figure 1 shows that IGF2 was involved in regulating drug resistance. Next, we will further verify. To test whether IGF2 also regulated drug resistance in GISTs in vivo, we established a xenograft model by inoculating sh-NC or sh-IGF2-GIST-T1 cells into nude mice. In the sh-IGF2-GIST-T1 mouse xenograft model, tumor volume and growth were inhibited by sh-IGF2, and imatinib had the same influence on tumor growth and volume. Combined treatment with imatinib and sh-IGF2 was more effective for reducing tumor progression than single treatment (Figure 5A-C, P < 0.001). The western blot results revealed that expression of IGF1R, p-IGF1R, AKT, PI3K, and p-AKT in tumor tissue was suppressed in both sh-IGF2-transfected cells and after imatinib treatment. Moreover, combined imatinib and sh-IGF2 were more effective than single therapy (Figure 5D, P < 0.001). The above data suggest that IGF2/IGF1R regulate imatinib resistance.
In addition, previous data shows that IGF2 regulates glycolysis in GIST cells. IGF2 regulates cell sensitivity to imatinib through its influence on glycolysis. We used 2-DG to inhibit glycolysis in GIST cells. OE-IGF2 increased drug sensitivity in GIST882 and GIST-T1 cells, but after treatment with 2-DG, transfection with OE-IGF2 no longer changed drug sensitivity in GIST cells (Figure 5E, P < 0.001). Flow cytometric analysis showed that sh-IGF2 suppressed imatinib-induced apoptosis and OE-IGF2 reduced imatinib-induced apoptosis in GIST cells. Treatment with 2-DG and transfection with OE-IGF2 no longer influenced imatinib-induced apoptosis in GIST cells (Figure 5F, P < 0.001). Therefore, the results show that IGF2 regulated imatinib sensitivity in GIST cells by affecting glycolysis.
GISTs is the most frequent malignant gastrointestinal sarcoma and causes significant patient harm[26,27]. Recently, anticancer drug resistance has become a significant challenge to the treatment of GISTs[28]. Treatment with tyrosine kinase inhibitors (TKIs) has led to substantial improvement of survival, both for patients with localized GISTs and those with advanced disease[29]. As the first-line TKI, imatinib offers treatment for advanced and metastatic GISTs, adjuvant therapy in high-risk GISTs and neoadjuvant treatment to downsize large tumors prior to resection[8]. We explored the mechanism of IGF2 in imatinib resistance in GISTs and whether IGF2 enhanced metastasis and imatinib resistance by driving glycolysis by targeting IGF1R signaling transduction.
IGF2, identified as an imprinted gene, exhibits a significant impact on cancer progression when its imprinting is lost or relaxed, leading to heightened autocrine IGF2 levels and increased secretion in malignant cells[30,31]. Numerous studies have revealed the upregulation of IGF2 in various cancers such as hepatocellular carcinoma, correlating with resistance to chemotherapy and a poorer prognosis[12-14]. Our investigation, which focused on DEGs associated with liver metastasis and drug resistance in GISTs, we observed elevated levels of IGF2 in GISTs cases linked to liver metastasis and drug resistance. Our comprehensive analysis included assessment of cell proliferation, viability, migration, and invasiveness. The findings strongly suggest that overexpression of IGF2 induce the proliferation, metastasis, and EMT of GIST cells.
IGF1R, is a tyrosine kinase receptor that can be triggered by IGF2 and has a pivotal role in regulating mammalian development, metabolism, and growth[32]. IGF1R is known to be upregulated in various human solid tumors[19]. Its involvement in cell promoting cell proliferation and inhibiting programmed cell death is facilitated by activation of its tyrosine kinase and the subsequent engagement of the Ras/Raf/MEK and PI3K/AKT/mTOR signaling pathways[23]. The IGF2-IGF1R signaling axis assumes critical significance in governing cell proliferation, differentiation, EMT, migration, drug resistance, and maintaining stemness in malignancies[33]. This investigation further demonstrated the activation of IGF1R signaling by IGF2 in GIST cells. It highlights the role of IGF2 as a pivotal chromatin factor that controls the expression level of IGF1R and modulates downstream signaling by the PI3K/AKT pathway. IGF2 also upregulated the expression of glycolytic and mitochondrial respiration markers. IGF2 overexpression has also been shown to cause metabolic reprogramming in breast cancer[31]. As expected, we also that IGF2 mediated the glycolysis in GISTs by targeting IGF1R signaling.
Increased expression of IGF2 is a common occurrence in various cancers and has been associated with increased resistance to chemotherapy, leading to a poorer prognosis[12,13]. Regarding GISTs, the standard first-line therapeutic approach involves the use of imatinib[34]. Imatinib, a potent TKI, is the primary treatment for GISTs, and significantly contributes to the progression-free survival of GIST patients[35,36]. Our investigation revealed a noteworthy correlation of increased IGF2 expression with the induction of GISTs resistance to imatinib concurrently with a reduction of imatinib-induced apoptosis in GIST cells. These findings underscore IGF2 as a potential regulator of GISTs imatinib resistance, and a promising target for interventions aimed at reversing such resistance. Intriguingly, our study further showed that IGF2 regulates cellular sensitivity to imatinib by modulating glycolysis.
The study had some limitations of this study. First, except for GIST cells, the role of IGF2 on GIST patient samples needs verification. Even though we found that IGF-2 overexpression increased the resistance of GIST cells to imatinib in cell culture, the clinical effect needs to be verified. Secondly, our results allows speculation that IGF2 was involved in the resistance to chemotherapy and a worse GISTs prognosis. However, the molecular mechanism of IGF2 specific to GISTs requires further investigation. We will consider these issues in future studies. In addition, studies have found that hypoglycemia in patients with non-islet cell tumor-induced GISTs may be aggravated by imatinib[37]. A recent case study reported that a GISTs that produced big-IGF2 also caused severe hypoglycemia[38]. We also hope to investigate that in future experiments.
This study investigated IGF2 regulation of metastasis and imatinib resistance in GISTs. IGF2 interacted with IGF1R to regulate glycolysis. Our results found that IGF2 targeting of IGF1R signaling improved metastasis and imatinib chemosensitivity via driving glycolysis in GISTs and support potential use of IGF2 to reverse imatinib resistance in GISTs patients.
| 1. | Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 827] [Cited by in RCA: 918] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 2. | Blay JY, Kang YK, Nishida T, von Mehren M. Gastrointestinal stromal tumours. Nat Rev Dis Primers. 2021;7:22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 249] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 3. | Nguyen V, Banerjee S, Sicklick JK. Moving gastrointestinal stromal tumours towards truly personalised precision therapy. Lancet Oncol. 2020;21:865-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Chen W, Li Z, Liu H, Jiang S, Wang G, Sun L, Li J, Wang X, Yu S, Huang J, Dong Y. MicroRNA-30a targets BECLIN-1 to inactivate autophagy and sensitizes gastrointestinal stromal tumor cells to imatinib. Cell Death Dis. 2020;11:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Lennartsson J, Rönnstrand L. The stem cell factor receptor/c-Kit as a drug target in cancer. Curr Cancer Drug Targets. 2006;6:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 6. | Cao CL, Niu HJ, Kang SP, Cong CL, Kang SR. miRNA-21 sensitizes gastrointestinal stromal tumors (GISTs) cells to Imatinib via targeting B-cell lymphoma 2 (Bcl-2). Eur Rev Med Pharmacol Sci. 2016;20:3574-3581. [PubMed] |
| 7. | Duensing A, Medeiros F, McConarty B, Joseph NE, Panigrahy D, Singer S, Fletcher CD, Demetri GD, Fletcher JA. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs). Oncogene. 2004;23:3999-4006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 248] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 8. | Lim KT, Tan KY. Current research and treatment for gastrointestinal stromal tumors. World J Gastroenterol. 2017;23:4856-4866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 9. | Nishida T, Blay JY, Hirota S, Kitagawa Y, Kang YK. The standard diagnosis, treatment, and follow-up of gastrointestinal stromal tumors based on guidelines. Gastric Cancer. 2016;19:3-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 323] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 10. | Reichardt P, Demetri GD, Gelderblom H, Rutkowski P, Im SA, Gupta S, Kang YK, Schöffski P, Schuette J, Soulières D, Blay JY, Goldstein D, Fly K, Huang X, Corsaro M, Lechuga MJ, Martini JF, Heinrich MC. Correlation of KIT and PDGFRA mutational status with clinical benefit in patients with gastrointestinal stromal tumor treated with sunitinib in a worldwide treatment-use trial. BMC Cancer. 2016;16:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 11. | Hsu CM, Lin PM, Lin HC, Lai CC, Yang CH, Lin SF, Yang MY. Altered Expression of Imprinted Genes in Squamous Cell Carcinoma of the Head and Neck. Anticancer Res. 2016;36:2251-2258. [PubMed] |
| 12. | Livingstone C. IGF2 and cancer. Endocr Relat Cancer. 2013;20:R321-R339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 13. | Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26:371-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 14. | Yang J, Li Y, Yu Z, Zhou Y, Tu J, Lou J, Wang Y. Circular RNA Circ100084 functions as sponge of miR‑23a‑5p to regulate IGF2 expression in hepatocellular carcinoma. Mol Med Rep. 2020;21:2395-2404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Andersson MK, Åman P, Stenman G. IGF2/IGF1R Signaling as a Therapeutic Target in MYB-Positive Adenoid Cystic Carcinomas and Other Fusion Gene-Driven Tumors. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1039] [Cited by in RCA: 1043] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 17. | Tao Y, Pinzi V, Bourhis J, Deutsch E. Mechanisms of disease: signaling of the insulin-like growth factor 1 receptor pathway--therapeutic perspectives in cancer. Nat Clin Pract Oncol. 2007;4:591-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 184] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 18. | Pantaleo MA, Astolfi A, Nannini M, Biasco G. The emerging role of insulin-like growth factor 1 receptor (IGF1r) in gastrointestinal stromal tumors (GISTs). J Transl Med. 2010;8:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Beadling C, Patterson J, Justusson E, Nelson D, Pantaleo MA, Hornick JL, Chacón M, Corless CL, Heinrich MC. Gene expression of the IGF pathway family distinguishes subsets of gastrointestinal stromal tumors wild type for KIT and PDGFRA. Cancer Med. 2013;2:21-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Braconi C, Bracci R, Bearzi I, Bianchi F, Sabato S, Mandolesi A, Belvederesi L, Cascinu S, Valeri N, Cellerino R. Insulin-like growth factor (IGF) 1 and 2 help to predict disease outcome in GIST patients. Ann Oncol. 2008;19:1293-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Tarn C, Rink L, Merkel E, Flieder D, Pathak H, Koumbi D, Testa JR, Eisenberg B, von Mehren M, Godwin AK. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci U S A. 2008;105:8387-8392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 185] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 22. | Janeway KA, Zhu MJ, Barretina J, Perez-Atayde A, Demetri GD, Fletcher JA. Strong expression of IGF1R in pediatric gastrointestinal stromal tumors without IGF1R genomic amplification. Int J Cancer. 2010;127:2718-2722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 23. | Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 932] [Cited by in RCA: 1795] [Article Influence: 179.5] [Reference Citation Analysis (0)] |
| 24. | Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res.. 2010;70 (2):440-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3361] [Cited by in RCA: 4285] [Article Influence: 285.7] [Reference Citation Analysis (0)] |
| 25. | Adamek A, Kasprzak A. Insulin-Like Growth Factor (IGF) System in Liver Diseases. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 26. | Miettinen M, Lasota J. Gastrointestinal stromal tumors--definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1178] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 27. | Yan J, Chen D, Chen X, Sun X, Dong Q, Hu C, Zhou F, Chen W. Downregulation of lncRNA CCDC26 contributes to imatinib resistance in human gastrointestinal stromal tumors through IGF-1R upregulation. Braz J Med Biol Res. 2019;52:e8399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Serrano C, George S. Gastrointestinal Stromal Tumor: Challenges and Opportunities for a New Decade. Clin Cancer Res. 2020;26:5078-5085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (1)] |
| 29. | von Mehren M, Joensuu H. Gastrointestinal Stromal Tumors. J Clin Oncol. 2018;36:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 30. | Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S, Woodfine K, Jagodic M, Nativio R, Dunning A, Moore G, Klenova E, Bingham S, Pharoah PD, Brenton JD, Beck S, Sandhu MS, Murrell A. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633-2643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 92] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Vella V, Nicolosi ML, Giuliano M, Morrione A, Malaguarnera R, Belfiore A. Insulin Receptor Isoform A Modulates Metabolic Reprogramming of Breast Cancer Cells in Response to IGF2 and Insulin Stimulation. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Forbes BE, Blyth AJ, Wit JM. Disorders of IGFs and IGF-1R signaling pathways. Mol Cell Endocrinol. 2020;518:111035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 33. | Hua H, Kong Q, Yin J, Zhang J, Jiang Y. Insulin-like growth factor receptor signaling in tumorigenesis and drug resistance: a challenge for cancer therapy. J Hematol Oncol. 2020;13:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 34. | Essat M, Cooper K. Imatinib as adjuvant therapy for gastrointestinal stromal tumors: a systematic review. Int J Cancer. 2011;128:2202-2214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Lopes LF, Bacchi CE. Imatinib treatment for gastrointestinal stromal tumour (GIST). J Cell Mol Med. 2010;14:42-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Li GZ, Raut CP. Targeted therapy and personalized medicine in gastrointestinal stromal tumors: drug resistance, mechanisms, and treatment strategies. Onco Targets Ther. 2019;12:5123-5133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 37. | Hamberg P, de Jong FA, Boonstra JG, van Doorn J, Verweij J, Sleijfer S. Non-islet-cell tumor induced hypoglycemia in patients with advanced gastrointestinal stromal tumor possibly worsened by imatinib. J Clin Oncol. 2006;24:e30-e31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 38. | Yamasaki H, Itawaki A, Morita M, Miyake H, Yamamoto M, Sonoyama H, Tanaka S, Notsu M, Yamauchi M, Fujii Y, Ishikawa N, Fukuda I, Ishihara S, Kanasaki K. A case of insulin-like growth factor 2-producing gastrointestinal stromal tumor with severe hypoglycemia. BMC Endocr Disord. 2020;20:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |