Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.3118
Peer-review started: September 27, 2023
First decision: December 7, 2023
Revised: January 2, 2024
Accepted: March 20, 2024
Article in press: March 20, 2024
Published online: July 15, 2024
Processing time: 289 Days and 4.6 Hours
In the quest to manage hepatocellular carcinoma (HCC), the focus has shifted to a more holistic approach encompassing both data analytics and innovative tre
To elucidate the immunological genes and the underlying mechanism of the combined Kombo knife and sorafenib regimen for HCC by analyzing data from TCGA and machine learning data.
Immune attributes were evaluated via TCGA's postablation HCC RNA seque
Immune genes–specifically, peptidylprolyl isomerase A and solute carrier family 29 member 3–emerged as significant prognostic markers. Enhanced therapeutic outcomes, such as prolonged progression-free survival and an elevated overall response rate, characterize the combined approach, with peripheral blood mononuclear cells displaying potent effects on HCC dynamics.
The combination of Kombo knife with sorafenib is an innovative HCC treatment modality anchored in immune-centric strategies.
Core Tip: In this groundbreaking study, we explore the synergistic effects of Kombo Knife and sorafenib in hepatocellular carcinoma (HCC) treatment, utilizing insights from the cancer genome atlas. Employing advanced data analytics, including weighted gene coexpression network analysis and machine learning, we identify key immune genes-particularly peptidylprolyl isomerase A and solute carrier family 29 member 3-as significant prognostic markers. Our findings highlight the enhanced therapeutic efficacy of the combined regimen, marked by improved progression-free survival and response rates. This research represents a significant stride in HCC therapy, pivoting towards innovative, immune-centric treatment strategies that promise to transform patient outcomes.
- Citation: Cao Y, Li PP, Qiao BL, Li QW. Kombo knife combined with sorafenib in liver cancer treatment: Efficacy and safety under immune function influence. World J Gastrointest Oncol 2024; 16(7): 3118-3157
- URL: https://www.wjgnet.com/1948-5204/full/v16/i7/3118.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i7.3118
Cryoablation technology plays a significant role in the treatment of liver cancer[1]. This technique employs either low or high temperatures to target and eradicate tumor cells within tumor tissue[2]. Cryoablation technology can be used to treat small liver cancer effectively, to manage residual lesions post-surgery and to control intraoperative bleeding[1]. Hence, the use of cold and heat ablation technology for the treatment of liver or other cancers has a substantial impact on patients and can enhance treatment effectiveness[1].
Liver cancer is a prevalent malignancy worldwide that imposes significant burdens on patients' lives[3]. Liver cancer has high incidence and mortality rates worldwide, which makes it a significant public health problem[4]. The distinct growth characteristics and specific location of this type of cancer pose significant challenges to its treatment[5]. In the majority of cases, patients are diagnosed when the tumor has already progressed to an advanced stage, resulting in limited treatment options[6]. Correspondingly, the prognosis of these patients is usually poor[7]. Therefore, it is crucial to explore new treatment strategies and enhance early detection capabilities[8]. Recent research has shown a strong correlation between the occurrence of liver cancer and the body's immune response[9]. The development of liver cancer can disrupt the body's immune surveillance mechanism[10]. This interference enables cancer cells to evade immune system attacks[11]. Hence, the restoration or enhancement of immune function could be indispensable in future liver cancer treatments[12].
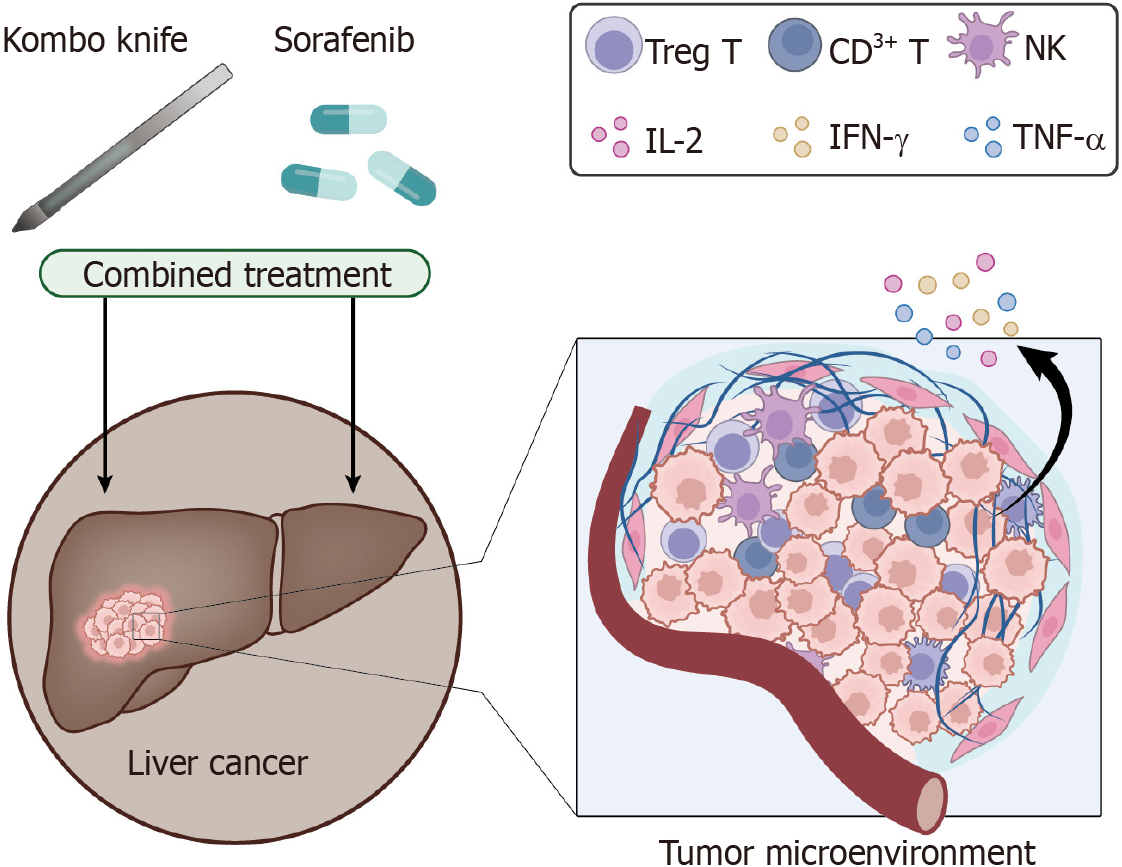
The combination of cryoablation technology, such as CyberKnife, with sorafenib has led to new hope for cancer treatment[13]. Multiple studies have demonstrated that this combination therapy not only significantly ablates tumors but also enhances the body's immune function[14]. Precise ablation using the Kombo knife combined with the pharmacological effects of sorafenib may activate and strengthen the immune system, consequently enhancing its ability to target cancer cells[15]. Moreover, this therapy exerts minimal impact on healthy tissues and enhances the stable functioning of the immune system[16]. Hence, this combination therapy has the potential to offer a novel approach for enhancing treatment efficacy and preserving immune function in the body[17].
The aim of this study was to assess the effectiveness and safety of combining Kombo knife with sorafenib for treating liver cancer. Furthermore, we also examined how this treatment method achieves its therapeutic effect by modulating patient immune function. This comprehensive investigation holds immense value in terms of revealing novel mechanisms for the treatment of liver cancer and enhancing patient prognosis.
The cancer genome atlas (TCGA)-liver hepatocellular carcinoma (HCC)-counts dataset was downloaded from the TCGA database (https://cancergenome.nih.gov/). The data included mRNA sequencing (mRNA-seq) expression profiles and corresponding clinical information. There were 371 tumor samples in the mRNA data, but the clinical sample information downloaded from the TCGA database showed that 6 patients had a survival status of 0 or not available (indicating missing survival information and excluding them from subsequent survival analysis). Therefore, 13 patients who were confirmed to have undergone ablation therapy and 235 patients who were confirmed to have not undergone ablation therapy were selected based on clinical information.
The expression data of HCC patients from TCGA were analyzed using the "ESTIMATE" R package to evaluate the number of stromal cells and immune cells in 13 HCC tumor tissues that received ablation therapy. The stromal and immune scores were measured, and the purity of the tumor was calculated based on this algorithm. We selected 13 genes from patients who underwent ablation therapy and obtained 9338 genes by calculating the mean and standard deviation. A coexpression network of 9338 genes from 13 patients who received ablation therapy was subsequently constructed using the "WGCNA v1.68" package in the R platform. WGCNA is a comprehensive biological algorithm with significant advantages in analyzing correlated gene expression patterns. The advantages of WGCNA include gene modular clustering and analysis of the correlation between modules and clinical features. We used the immune, stromal, and tumor purity scores obtained from the ESTIMATE algorithm as clinical features and analyzed the correlation between modules and traits using the Pearson correlation analysis (P < 0.05). The most important module contained immune-related genes (IRGs), which we used for subsequent analysis.
Based on the immune-associated genes obtained from WGCNA, the coexpressed genes were subjected to Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using DAVID 6.8 (https://david.ncifcrf.gov/) and the clusterProfiler package in R. Data visualization analysis was performed using the ggplot2 package. Among them, the three enriched terms in the GO enrichment analysis were cell component, biological process, and molecular function, which reflect the functional annotations of key genes.
Gene set enrichment analysis (GSEA) was performed on the high-risk and low-risk groups of HCC patients in the TCGA database using the clusterprofiler package in R software. All patients were classified as either high risk or low risk based on the optimal cutoff value of the risk score (RS) determined by least absolute shrinkage and selection operator analysis (LASSO) regression. Moreover, the enrichplot package in R was used to generate enrichment plots of the gene sets with annotations. A P value < 0.05 was considered to indicate statistical significance.
According to the univariate Cox regression analysis, independent prognostic genes with significant overall survival (OS) correlations were selected (P < 0.05). These independent prognostic genes were subsequently included in the Lasso Cox analysis to establish a risk model. The purpose of LASSO is to use L1 regularization to penalize model parameters and avoid overfitting the model. The LASSO model employs an L1 norm penalty mechanism to avoid overfitting. LASSO regularization can be defined as follows:
L (x, y) = [∑ni= 1(yi-hθ(xi))2] + λ∑ki= 1|θi|)
Model complexity is controlled by λ. The larger λ is, the greater the penalty for the linear model with more variables. LASSO can also be expressed as a constraint on the objective function.
An important feature of LASSO regularization is that it can force parameter values to be 0. The purpose of LASSO is to generate a sparse parameter space, which is an ideal feature selection characteristic.
We performed Kaplan–Meier (KM) curve and receiver operating characteristic (ROC) curve analyses on the genes obtained from LASSO to select genes with excellent prognostic performance and diagnostic ability to construct prognostic features. Using a risk-scoring formula based on prognostic characteristics, the risk level for each patient can be determined. The formula for the RS is defined as follows:
RS = ∑ni= 1Coe fi × Expi
Here, n represents the number of genes included in the prognostic feature set. The LASSO coefficients for gene i. Expi represents the expression value of gene i. A RS was assigned for each patient based on the expression of the feature genes in different samples. All patients were divided into high-risk or low-risk groups based on the optimal cutoff value determined by risk scoring. The prognostic significance and diagnostic ability of the RS were evaluated through K-M curves and ROC curve analysis.
The prognostic independence of the RS was analyzed under the interference of multiple clinical characteristics through univariate and multivariate Cox regression analyses. The independent prognostic features were subsequently incorporated into the construction of the column chart. Moreover, the consistency index between the actual survival rate and the predicted survival rate was used to evaluate the ability of the column chart to predict survival. The predicted and observed results were visualized in the calibration curve to measure the predictive performance of the line plot.
We utilized single-sample GSEA and Spearman's correlation analysis (r) from the R package to investigate the potential associations between the expression levels of peptidylprolyl isomerase A (PPIA) and solute carrier family 29 member 3
This study obtained approval from the ethics committee and required every patient to sign an informed consent form following the Helsinki Declaration.
By collecting data from patients diagnosed with primary HCC in our department between July 2022 and December 2022, we obtained the following relevant information: Personal information, medical history, routine blood and urine tests, liver and kidney function, coagulation function, blood biochemical liver and gallbladder indices, imaging data, treatment history, treatment outcomes, adverse reactions, survival status, etc. We conducted long-term observation of patients through medical system inquiries and phone follow-up tracking. We divided the patients we collected into different groups, including patients treated with Kombo knife (Group A), patients treated with Kombo knife plus Sorafenib (Group B), and patients treated with chemoembolization plus Sorafenib (Group C).
The inclusion criteria for patients were as follows: (1) ≥ 18 years of age; (2) Met the diagnostic criteria in the 2020 edition of China's Chinese Society of Clinical Oncology Primary HCC Diagnosis and Treatment Guidelines; (3) Had no serious underlying disease (such as heart, brain, lung, or kidney dysfunction); and (4) Were in good compliance and were able to undergo regular follow-up and cooperate with the treatment.
The exclusion criteria for patients were as follows: (1) Had uncorrectable coagulation dysfunction; (2) Had concomitant systemic dysfunction or malignant tumors; (3) Were allergic or intolerant to targeted and immunomodulatory drugs; and (4) Could not be followed up or had incomplete data.
Patients were regularly followed up with regular assessments, including detailed medical history and physical examination, weekly routine blood and coagulation tests, biweekly liver and kidney function biochemical tests, and monthly liver computed tomography scans. Adverse reactions were assessed according to the Adverse Event Reporting Common Terminology Criteria for Adverse Events (CTCAE) version 4.0.
This study included 96 patients, and their clinical information is presented in Supplementary Table 1. The patients were divided into the following groups:
Group A included 32 patients who received treatment with the Kombo knife.
Group B included 32 patients who received combination therapy comprising a novel composite cold and hot ablation technique (Kombo knife) and sorafenib. Sorafenib was administered orally at a dosage of 400 mg twice daily.
Group C included 32 patients who received standard treatment comprising transarterial chemoembolization (TACE) combined with sorafenib. Sorafenib was administered orally at a dosage of 400 mg twice daily.
Observation indicators: The main observation indicators of the study were the objective response rate (ORR) and disease control rate (DCR). The calculation of ORR included complete remission (CR) and partial remission (PR); the calculation of DCR included CR, PR, and disease stabilization. The secondary outcome measures of this study included OS and progression-free survival (PFS). Approximately one month after diagnosis, the first follow-up and efficacy evaluation were conducted for each group of patients. Follow-up was subsequently conducted every two months for a total duration of one year. If the patient died, disease progression (PD) was considered. The 2010 version of the Modified Response Evaluation Criteria in Solid Tumors (mRECIST) evaluation criteria (Supplementary Table 2) was used here to assess treatment efficacy, while the CTCAE 5.0 version, published in 2017, was also used (Supplementary Table 3). The severity of each adverse event was evaluated.
After being diagnosed with HCC in Group A and Group B, blood samples were taken one day before blood collection and stored in anticoagulant tubes for the extraction of peripheral blood mononuclear cells (PBMCs); one month after treatment, blood samples were taken again and stored in anticoagulant tubes for the extraction of PBMCs. To serve as a control group, 32 healthy individuals, including 16 males and 16 females aged between 25 and 60 years (with an average age of 36.94 ± 10.65 years), were selected from the population undergoing physical examinations at this institution. The exclusion criteria were as follows: Had the diseases above, had abnormal liver and kidney function, or had chronic or autoimmune diseases.
Three milliliters of nonanticoagulated peripheral blood was collected under aseptic conditions and centrifuged for 10 min, after which the supernatant was collected after centrifugation. The serum was packed into cryovials and stored in a -70 °C ultralow temperature freezer for future use.
The anticoagulated peripheral blood, which was isolated under sterile conditions, was transferred to a 50 mL cen
The recovered cryopreserved PBMCs were stored in complete culture medium, after which the cell concentration was adjusted to 5 × 105/mL. Then, 2 mL of cell suspension was transferred to a fluorescence-activated cell sorting (FACS) tube. Three microlitres of anti-human CD3-BV 450 (652356, BD Bioscience, United States), anti-human CD4-antigen Presenting Cell-Cyanine 7 (CD4-APC-Cy7) (341115, BD Bioscience, United States), anti-human CD8-V 500-C (647458, BD Bioscience, United States), anti-human CD16-FITC (335035, BD Bioscience, United States), anti-human CD56-PE-Cy™7 (335791, BD Bioscience, United States), anti-human CD25-APC-R700 (659117, BD Bioscience, United States), anti-human Forkhead Box P3-Rhodamine B 780 (FoxP3-RB780) (568683, BD Bioscience, United States), anti-human CD45 Rat IgG2a-Phycoerythrin (PE)-Cyanine 5 (RO-PeCy5) (15-0457-42, Novusbio, Beijing, China), and anti-human CD45 Retinoic Acid-Fluorescein Isothiocyanate (RA-FITC) (ab18240, Abcam, United Kingdom) were mixed well and incubated in the dark at 4 °C for 30 min. Then, 2 mL of PBS (P4417, Sigma-Aldrich, Shanghai, China) was added, and the supernatant was discarded after centrifugation at 4 °C and 1500 × g for 10 min. Afterward, 2% paraformaldehyde (Sigma-Aldrich, Shanghai, China, catalog number 8187150100) was added to the PBS solution for fixation. Store at 4 °C in the dark and use the FACS Aria II flow cytometer [instrument parameters: Sorting speed of 70000 cells/s, sorting purity > 98%, fluorescence sensitivity: FITC < 125 Molecules of Equivalent Soluble Fluorochrome (MESF), PE < 125 MESF; BD Bioscience, United States] for detection within the following 24 h.
The levels of cytokines in peripheral blood were detected via enzyme-linked immunosorbent assay (ELISA). The specific steps were as follows: PBS was used to dilute the anti-human interleukin (IL)-2 antibody (ab270883, Abcam, United Kingdom), interferon (IFN)-γ antibody (ab224197, Abcam, United Kingdom), tumor necrosis factor (TNF)-α antibody (ab183218, Abcam, United Kingdom), IL-4 antibody (ab215089, Abcam, United Kingdom), IL-5 antibody (ab215536, Abcam, United States), IL-6 antibody (ab178013, Abcam, United Kingdom), IL-10 antibody (ab185986, Abcam, United Kingdom), and heat shock protein 70 (HSP70) antibody (ab133060, Abcam, United Kingdom) to the working concentration. The plates were immediately coated (coating solution, CB07100, Thermo Fisher, United States) with the diluted capturing antibodies added to each well in a volume of 100 μL. The mixture was incubated overnight at room temperature. The captured antibodies were removed, and 400 μL of wash solution (28360, Thermo Fisher, United States) was added to each well. The samples were washed thoroughly, after which the wash solution was removed. This washing procedure was repeated three times. After the final wash, absorbent paper was used to fully dry the detergent inside the tablet. Then, 300 μL of sample dilution solution (00-4202-56; Thermo Fisher, United States) was added to each well, which was sealed and incubated at room temperature for 1 h. One hundred microlitres of the tested sample or standard solution was added to each well. The samples were waterproofed and incubated at room temperature for 2 h. Then, 100 μL of detection antibody was added to each well. The samples were covered with new waterproof adhesive tape and incubated at room temperature for 2 h. Then, 100 μL of horseradish peroxidase (HRP) conjugated with streptavidin (35105ES60, Yek Shan Bio, Shanghai, China) was added to each well. The mixture was covered with a new waterproof adhesive strip and incubated at room temperature for 20 min, avoiding direct sunlight. Then, 100 μL of substrate solution was added to each well, and the plates were incubated at room temperature for 20 min in the dark. Then, 50 μL of termination solution (N600, Thermo Fisher, United States) was added to each well, and the samples were gently tapped to thoroughly mix without introducing bubbles. An M680 automatic microplate reader (Bio-Rad, United States) was used to read the absorbance at 450 nm. A standard curve was created based on the standard sample absorbance to calculate the test sample concentration.
Total RNA was extracted from cells using a TRIzol reagent kit (10296010, Invitrogen, Thermo Fisher, United States), and the quality and concentration of the RNA were determined using an ultraviolet-visible spectrophotometer (ND-1000, Nanodrop, United States). To detect mRNA expression, a PrimeScript™ real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) kit (RR086A, TaKaRa, Mountain View, CA, United States) was used for reverse transcription, and SYBR Premix Ex TaqTM (DRR820A, TaKaRa, United States) was used for RT-qPCR on a LightCycler 480 system (Roche Diagnostics, Pleasanton, CA, United States). Using GAPDH as the internal control for mRNA, Shanghai Universal Biotechnology Co., Ltd., designed and provided amplification primers, and the primer sequences are shown in Supplementary Table 4. In this study, the 2-ΔΔCt method was used, where ΔΔCT = ΔCt experimental group -ΔCt control group and ΔCt = target gene Ct - reference gene Ct, to quantify the fold difference in gene expression between the experimental and control groups.
Total proteins were extracted from human peripheral blood and mouse tumor tissues. The cultured cells were digested and collected using trypsin (T4799-5G; Sigma-Aldrich, Shanghai, China). Then, cell lysis was performed using enhanced RIPA lysis buffer containing a proteinase inhibitor (AR0108, Wuhan Boshide Biological Engineering Co., Ltd., Wuhan, China). The protein concentration was measured using a bicinchoninic acid protein quantitation kit (AR1189, Wuhan Boshido Company, Wuhan, China). Next, the proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride membranes, and blocked with 5% bovine serum albumin (BSA) (V900933, Merck, United States) at room temperature for 1 h. Then, diluted primary antibodies against PPIA (SAB2101855, 1/1000; Sigma-Aldrich, Shanghai, China), SLC29A3 (PA5-38039, 1/1000; Thermo Fisher, United States), and GAPDH (G9545, 1/2000; Sigma-Aldrich, Shanghai, China) were added separately and incubated overnight. The membrane was washed three times with phosphate-buffered saline with Tween (PBST) (3 × 5 min) before being incubated with an anti-mouse-HRP secondary antibody (Cat 7076, 1/5000; CST, United States) or an anti-rabbit-HRP secondary antibody (Cat 7074, 1/5000; CST, United States) at room temperature for 1 h. The membrane was washed three times (3 × 5 min) with PBST. The cells were washed with PBST, and an appropriate amount of enhanced chemiluminescence (ECL) working solution was added (Omt-01, Beijing Aomijia Medical Technology Co., Ltd., Beijing, China). membrane was incubated at room temperature for 1 min, the excess ECL reagent was removed, the membrane was sealed with plastic wrap, and the membrane was placed in a dark box. The membrane was developed and fixed after being exposed to X-ray film for 5-10 min. Finally, ImageJ analysis software was used to perform grayscale quantification of the bands in the Western blot images, with GAPDH serving as the reference.
The human HCC cell lines JHH7 (CBP60204, Nanjing Kaibai Biotechnology Co., Ltd., Jiangsu, China) and Huh7 (CBP60202, Nanjing Kaibai Biotechnology Co., Ltd., Jiangsu, China) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 1% penicillin-streptomycin (Invitrogen, United States) and 10% fetal bovine serum (FBS; Gibco, Invitrogen, United States) at 37 °C in a 5% CO2 cell culture incubator. The growth medium was replaced every 3 d. When the cell culture reached 80% confluence, the cells were passaged using 0.25% trypsin/EDTA (Thermo Fisher, United States).
Add PBMCs to the culture bottle of JHH7 or Huh7 cells at a cell ratio of 10:1. The mixture was incubated in a constant temperature incubator.
The cells were divided into the following groups: JHH7 group, JHH7 + Before Kombo knife (pregnant PBMCs) from HCC patients before treatment with JHH7, JHH7 + After Kombo knife (cocultured PBMCs from HCC patients after treatment with JHH7), JHH7 + Before Kombo knife-Sorafenib (cocultured PBMCs from HCC patients before treatment with JHH7 and Sorafenib), and JHH7 + After Kombo knife-Sorafenib (cocultured PBMCs from HCC patients after treatment with JHH7 and Sorafenib). The exclusion criteria for the Huh7 group were as follows: Huh7 + Before Kombo knife (coculture of PBMCs from HCC patients before treatment with Huh7 cells); Huh7 + After Kombo knife (coculture of PBMCs from HCC patients after treatment with Huh7 cells); Huh7 + Before Kombo knife-Sorafenib (coculture of PBMCs from HCC patients before treatment with Huh7 cells and sorafenib); and Huh7 + After Kombo knife-Sorafenib (coculture of PBMCs from HCC patients after treatment with Huh7 cells and sorafenib).
A total of 50 μL of diluted Matrigel matrix gel (E6909, Merck, United States) was applied to the upper membrane surface of a Transwell insert. The solution was diluted at a ratio of 1:6. The mixture was incubated at 37 °C for 30 min to allow gel polymerization. JHH7 or Huh7 cells were cultured in RPMI 1640 + 10% FBS (Gibco, Carlsbad, CA, United States) medium until the logarithmic growth phase. Then, the cells were harvested and washed 1-2 times with PBS. The cells were resuspended in 5% serum-free RPMI 1640 medium (11875119, Gibco, Carlsbad, United States), and the concentration was adjusted to 5 × 105/mL. One hundred microlitres of cell suspension was added to the upper chamber of the Transwell plate, and 600 μL of RPMI 1640 medium containing 20% FBS was added to the lower chamber to avoid the formation of bubbles between the medium and the chamber. The Transwell plate was placed in a cell culture incubator and cultured for 24 h at 37 °C and 5% CO2. The Transwell chamber was removed, the culture medium in the wells was discarded, and the cells were washed twice with PBS. The cells were fixed with formaldehyde for 30 min, after which the chamber was air-dried appropriately. The cells were stained with 0.1% crystal violet (M031-1; Gibco, Shanghai, China) for 20 min, after which the nonmigrated cells were gently removed using a cotton swab. After washing three times with PBS, five fields were randomly observed under a 400 × microscope for cell counting.
Two hundred microliters of the supernatant was centrifuged for 5 min at 400 × g. After centrifugation, 120 μL of the supernatant was added to a 96-well plate. After the supernatants of the JHH7 and Huh7 cell cultures were treated with Triton X100 (100 ×, Sigma-Aldrich, Shanghai, China), the resulting culture was used as a positive control. Add 60 μL of lactate dehydrogenase detection working solution (BA1800, Shang Bao Biotechnology, Shanghai, China) to each well, mix well, and incubate at room temperature in the dark for 30 min (wrap the plate with aluminum foil and place it on a level shaker with slow agitation). The absorbance was measured at 490 nm using a reference wavelength of 600 nm for dual-wavelength detection.
PMBCs and HCC cells were cocultivated. Ficoll® PM 400 Lymphocyte separation solution (F4375-10G, Sigma-Aldrich, Shanghai) was added to the tube and centrifuged at 800 × g for 20 min. The monolayer of single cells was carefully aspirated and transferred to a new centrifuge tube. Then, a single-cell suspension of HCC cells was prepared, the suspension was added to the bottom of a centrifuge tube, and cell culture medium was added to resuspend the cells in the medium. The cells were collected, washed with precooled PBS and stained with Annexin V-FITC and PI from an apoptosis assay kit (APOAF-20TST, Sigma-Aldrich, Shanghai) for 10 min. The cell death rate (105 cells per sample) was detected with a CytoFlex flow cytometer (Beckman Coulter, Inc., United States). Cells in the upper right quadrant are Annexin V + PI-positive cells, which represent late apoptotic cells. Cells in the lower right quadrant, which represent early apoptotic cells, exhibited the Annexin V + PI phenotype. The cells in the lower left quadrant are Annexin V-PI-positive cells, which represent dead cells. The cells in the upper left quadrant are Annexin V-PI- cells, which are considered live cells[18]. FlowJo software (version 7.0; FlowJo LLC) was used to analyze the data, and the following specific analysis steps were used: FSC and SSC were set to logarithmic axis mode for reference; Annexin and PI channels were set as the X-axis and Y-axis, respectively; and the quadrant gating tool was used to define live cells and apoptotic cells. Usually, when calculating the cell death rate, the upper right quadrant and the lower right quadrant are used, representing the late apoptosis plus early apoptosis group (i.e., all Annexin V-positive groups).
Four- to five-week-old female BALB/c-nu/nu nude mice (purchased from Cyagen Biosciences, Inc., in Suzhou, China) were selected following the Regulations on the Administration of Experimental Animals in Medical Research and the Animal Research: Reporting of in vivo Experiments guidelines for animal management and studies and approval from the Institutional Animal Ethics Committee.
We collected JHH7 and Huh7 human HCC cells during the logarithmic growth phase and observed and counted the proportion of surviving cells that were greater than 95% under a microscope. The cell concentration was adjusted to 1.0 × 106/mL in PBS. Nude mice were injected with 0.1-0.2 mL of cell suspension, and the injection was administered quickly and gently subcutaneously on the right dorsal side. The skin inoculation site of nude mice was disinfected before and after injection to strictly control infection. We carefully observed the changes in the vaccination site of the naked mole-rats daily.
At 5 wk after subcutaneous tumor formation in nude mice, we subcutaneously injected patient-derived PBMCs at a concentration of 5.0 × 106/mL into the tumor-bearing mice. Every 7 d, we measured the size of the tumors and the body weight of the mice. The long axis (a) and short axis (b) of the tumor were measured using calipers, and the data were recorded to calculate the volume of the engrafted tumor. The nude mice were euthanized using a dose of 40 mg/kg body weight pentobarbital sodium administered via intraperitoneal injection. Following euthanasia, the transplanted tumor tissue on the raised skin of the back was completely dissected from the mice. Gross observation was also conducted to assess the color, texture, presence of capsules, and invasion of the surrounding tissues.
The mice were divided into the following groups: The Huh7 group, Huh7 + Before Kombo knife group (PBMCs from HCC patients before treatment with Kombo knife were injected into Huh7 tumors); the Huh7 + After Kombo knife group (PBMCs from HCC patients after treatment with Kombo knife were injected into Huh7 tumors); the Huh7 + Before Kombo knife-Sorafenib group (PBMCs from HCC patients before treatment with Kombo knife and sorafenib were injected into Huh7 tumors); and the Huh7 + After Kombo knife-Sorafenib group (PBMCs from HCC patients after treatment with Kombo knife and sorafenib were injected into Huh7 tumors).
After fully soaking the fresh transplanted tumor tissue in 4% paraformaldehyde solution (V900894, Merck, United States) for at least one day, the required parts of the tissue were cut flat under sterile conditions with a surgical knife, and the processed tissues were grouped, labeled and dehydrated in a dehydration box. The tissues were soaked in wax and embedded in an embedding machine. The corresponding tissue blocks were labeled, and the wax blocks were sub
Sections were dewaxed in water from paraffin and stained with hematoxylin (H8070, Beijing Solabao Technology Co., Ltd., Beijing, China) for cell nuclei staining and eosin (G1100, Beijing Solabao Technology Co., Ltd., Beijing, China) for cytoplasm staining. Then, dehydration and coverslipping were performed, and images were collected under a microscope for data analysis.
The paraffin sections were deparaffinized with water. The tissue sections were placed in a repair box containing EDTA antigen retrieval buffer (pH 9.0) (C9999, Merck, United States), and antigen retrieval was performed in a microwave oven. After cooling, the slides were placed in PBS (pH 7.4) and washed three times on a destaining shaker for 15 min each. Endogenous peroxidase activity was blocked with a 3% hydrogen peroxide solution (H1009, Merck, United States), and the sections were blocked with serum BSA. Primary antibodies against Ki-67 (1:100; human, 34330; CST, United States), CD34 (1:100; human, ab81289; Abcam, United Kingdom), programmed cell death 1 (PD-1) (1:100; human, 86163; CST, United States), programmed cell death ligand 1 (PD-L1) (1:100; human, 13684; CST, United States), and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) (1:100; human, 53560; CST, United States) were applied to the sections, after which the slides were incubated with the cells in a humid box at 4 °C overnight. After the sections were slightly dried, the corresponding goat anti-mouse or anti-rabbit secondary antibodies (conjugated with HRP) were applied from the imm
The paraffin was removed from the sections by soaking them in water and performing repair. After the sections were slightly air-dried, the membrane-breaking working solution (IH0018, Raygene Bio, Beijing, China) was added to cover the tissues in the circle. The appropriate amounts of Reagent 1 (TdT) and Reagent 2 (dUTP) were obtained from the TUNEL Kit (40308ES20; Yisheng Bio, Shanghai, China) according to the number of sections and the size of the tissue blocks, after which the reagents were mixed at a ratio of 2:29. The mixture was added to the surface of the tissue in the circle, the slices were placed flat in a wet box and incubated at 37 °C for 2 h under constant temperature conditions. A 3% hydrogen peroxide solution was prepared with methanol (hydrogen peroxide:methanol = 1:9) to block endogenous peroxidase activity. Each slice was covered with a suitable amount of reagent 3 (converter-POD) on the tissue, after which color development and counterstaining were performed with DAB (34002, Thermo Fisher, United States) and hematoxylin. Then, the slides were dehydrated and mounted, images were observed, the samples were collected and analyzed under a microscope, and the number of apoptotic cells was calculated.
When the "survival" package in R was used to construct KM survival curves, a two-sided log-rank test was used to analyze the difference in survival between the high-risk and low-risk groups. Additionally, univariate and multivariate Cox proportional hazards models were established to estimate the relative hazards of the prognostic factors.
All experiments were conducted independently at least three times, and the data are expressed as the mean ± SD. A paired sample t-test or an independent sample t-test was used to analyze the differences between groups. Tumor size was compared between groups of nude mice using 2-way ANOVA. All the statistical analyses were performed using GraphPad Prism 8.0[19]. A P value less than 0.05 was considered to indicate statistical significance.
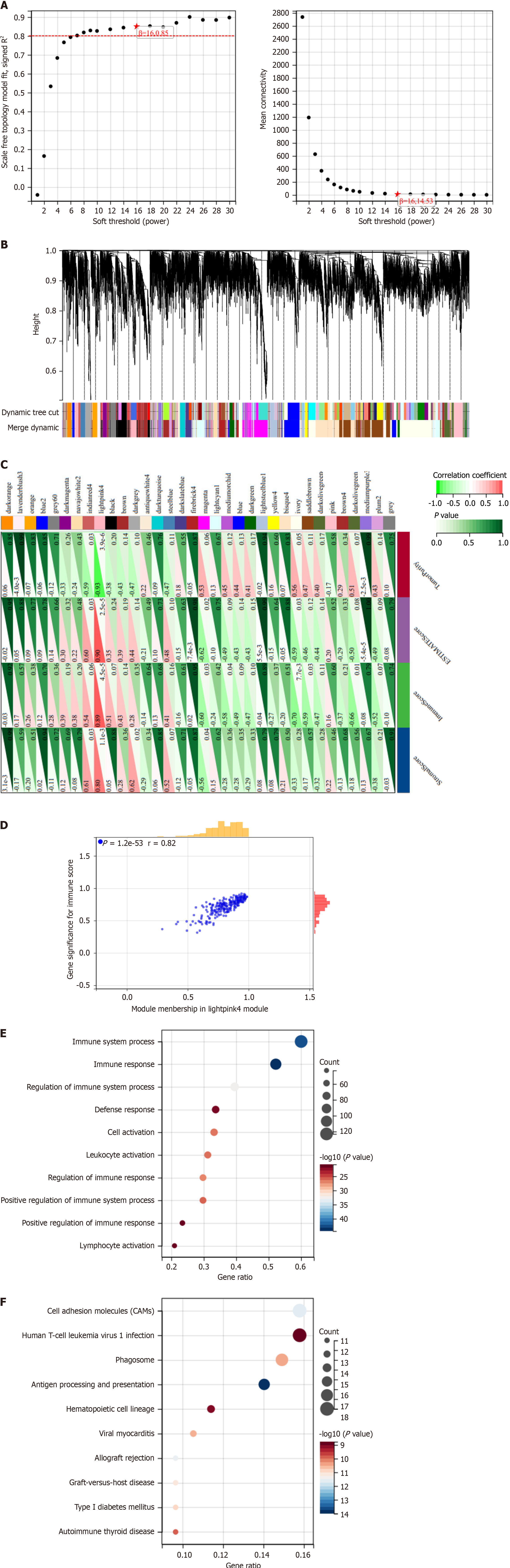
We downloaded RNA-seq and clinical information on HCC from TCGA database and selected RNA-seq data from 13 HCC patient samples from patients who received ablative therapy. A coexpression network of these patients was constructed using the WGCNA algorithm and included 9938 genes, with β set to 16 to meet the scale-free network criterion. When the minimum module size was set to 40 and the module merging threshold was set to 0.25, 34 modules were identified (Figure 1A), as shown in the clustered dendrogram (Figure 1B).
To further analyze these data, we employed the ESTIMATE analysis technique to calculate algorithm scores, immune scores, matrix scores, and tumor purity and then combined them with the WGCNA data above as clinical features to obtain correlation results. The module correlation analysis results suggested that the light pink module was significantly correlated with four clinical feature parameters. There were significant correlations between these modules and the matrix score [correlation coefficient (cor) = 0.8, P = 1.1e-3], immune score (cor = 0.89, P = 4.5e-5), ESTIMATE score (cor = 0.90, P = 2.5e-5), and tumor purity (cor = -0.92, P = 3.6e-6; Figure 1C and D). The bright pink 4-module contains 220 genes, which we preliminarily define as IRGs.
We performed GO and KEGG pathway analyses of 220 IRGs in the 4 modules highlighted in bright pink. GO analysis revealed that the modular IRGs were mainly associated with immune system processes, the immune response, leukocyte activation, and lymphocyte activation (Figure 1E). Moreover, KEGG pathway analysis revealed significant correlations between modular IRGs and cell adhesion molecules, human T-cell leukemia virus 1 infection, antigen processing, and presentation (Figure 1F). Functional enrichment analysis further demonstrated the association between genes in the bright pink module and immune-related pathways. For the first time, through the combination of WGCNA and immune infiltration analysis, we discovered 220 IRGs associated with ablative therapy for HCC.
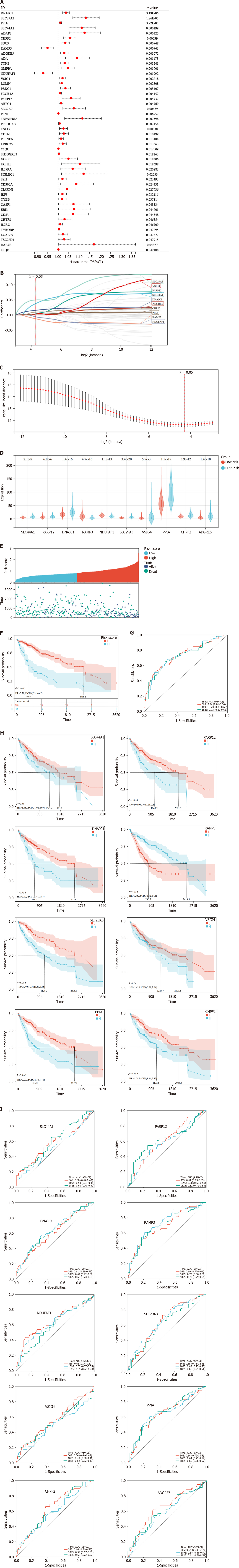
Through collaborative analysis of WGCNA and immune infiltration, we identified 220 IRGs. We first performed univariate Cox regression analysis of these 220 IRGs in 371 patients with TCGA HCC to identify key genes significantly associated with prognosis. We identified 48 IRGs that were significantly associated with OS (P < 0.05; Table 1 and Figure 2A). Next, we employed LASSO Cox analysis with a lambda value of 0.05 to further integrate survival time, survival status, and gene expression data. This analysis resulted in the reduction of the independent prognostic gene set to ten genes: Solute carrier family 44 member 1 (SLC44A1), poly(ADP-ribose) polymerase family member 12 (PARP12), DnaJ heat shock protein family (Hsp40) member C1 (DNAJC1), receptor activity modifying protein 3 (RAMP3), NADH: Ubiquinone oxidoreductase complex assembly Factor 1 (NDUFAF1), SLC29A3, V-Set and immunoglobulin domain containing 4 (VSIG4), PPIA, chondroitin polymerizing factor 2 (CHPF2), and adhesion G protein-coupled receptor E5 (ADGRE5; Table 1 and Figure 2B and C).
| Gene | Univariate Cox regression analysis | LASSO Cox, Coefficient | |||
| HR | HR 0.95L | HR 0.95H | P value | ||
| ADGRE5 | 1.029 | 1.012 | 1.048 | 1.07E-03 | 3.15E-05 |
| CHPF2 | 1.057 | 1.025 | 1.089 | 3.90E-04 | 9.24E-03 |
| DNAJC1 | 1.037 | 1.021 | 1.053 | 3.19E-06 | 0.015 |
| NDUFAF1 | 0.929 | 0.887 | 0.974 | 1.99E-03 | -0.006 |
| PARP12 | 1.061 | 1.018 | 1.105 | 4.74E-03 | 3.51E-03 |
| PPIA | 1.011 | 1.006 | 1.016 | 3.92E-05 | 2.94E-03 |
| RAMP3 | 0.949 | 0.921 | 0.979 | 7.63E-04 | -0.013 |
| SLC29A3 | 1.132 | 1.070 | 1.198 | 1.86E-05 | 0.049 |
| SLC44A1 | 1.113 | 1.052 | 1.177 | 1.99E-04 | 0.038 |
| VSIG4 | 1.021 | 1.007 | 1.034 | 2.22E-03 | 3.10E-03 |
Based on the linear combination of the expression levels of these 10 genes multiplied by their respective LASSO coefficients, we calculated the patient's RS. The formula for calculating the RS was as follows: RS = 0.0376SLC44A1 + 0.0035PARP12 + 0.0154DNAJC1 - 0.0127RAMP3 - 0.0061NDUFAF1 + 0.0495SLC29A3 + 0.0031VSIG4 + 0.0029PPIA + 0.0092CHPF2 + 3.15e-05ADGRE5.
According to the above formula, we obtained the RS of 365 HCC patients in the TCGA cohort (of 371 patients in the TCGA dataset, 6 patients with a survival time of 0 were excluded, leaving 365 patients). The optimal cutoff value for calculating the RS was determined simultaneously by setting the minimum sample size for groups to be greater than 25% and the maximum sample size for groups to be smaller than 75%; this optimal value was determined to be 1.08. All patients were divided into high-risk groups (high risk) or low-risk groups (low risk) based on the optimal cutoff value for the RS. The high-risk group comprised 268 patients, while the low-risk group comprised 97 patients.
Moreover, we analyzed the relationships between the changes in the expression of 10 IRGs and different RS. RAMP3 and NDUFAF1 are protective factors, and their expression levels were significantly lower in the high-risk group than in the low-risk group. The remaining eight genes were harmful factors, with significantly greater expression in the high-risk group than in the low-risk group (Figure 2D). Furthermore, we observed a significant decrease in patient survival rates as the RS increased (Figure 2E).
By comparing the KM curves, we observed a significant difference in prognosis between the two groups (P = 2.4e-12), with a significantly lower survival rate in the high-risk group than in the low-risk group (Figure 2F). We also conducted ROC analysis at 1, 3, and 5 years, and the results showed that the area under the curve (AUC) values for 1-year, 3-year, and 5-year survival prediction were 0.74, 0.73, and 0.73, respectively (Figure 2G). The research results indicate that the greater the risk factor is, the worse the patient prognosis and the lower the survival rate.
We simultaneously predicted the prognostic ability of 10 IRGs and identified seven harmful genes significantly associated with HCC prognosis, namely, SLC44A1, PARP12, DNAJC1, SLC29A3, PPIA, CHPF2, and ADGRE5, as well as two protective genes, RAMP3 and NDUFAF1. Among these genes, low expression of RAMP3 and NDUFAF1 is associated with poor prognosis, while high expression of the other seven genes is associated with poor prognosis (Figure 2H). Next, we performed ROC analysis on these 10 IRGs. Using the ROC curve function in the pROC, we conducted ROC analysis at the 1-, 3-, and 5-year time points and obtained the final AUC results. Specifically, the AUC values for SLC44A1 at 1, 3, and 5 years were 0.58, 0.53, and 0.60, respectively. The AUC values for PARP12 at 1, 3, and 5 years were 0.61, 0.58, and 0.62, respectively. The AUC values for DNAJC1 at 1, 3, and 5 years were 0.61, 0.64, and 0.63, respectively. The AUC values for RAMP3 at 1, 3, and 5 years were 0.69, 0.73, and 0.70, respectively. The AUC values for NDUFAF1 at 1, 3, and 5 years were 0.65, 0.62, and 0.59, respectively. The AUC values for SLC29A3 at 1, 3, and 5 years were 0.65, 0.66, and 0.61, respectively. The AUC values for VSIG4 at 1, 3, and 5 years were 0.56, 0.49, and 0.52, respectively. The AUC values for 1, 3, and 5 years after PPIA were 0.64, 0.64, and 0.66, respectively. The AUC values for CHPF2 at 1, 3, and 5 years were 0.64, 0.59, and 0.62, respectively. The AUC values for ADGRE5 at 1, 3, and 5 years were 0.65, 0.58, and 0.61, respectively (Figure 2I).
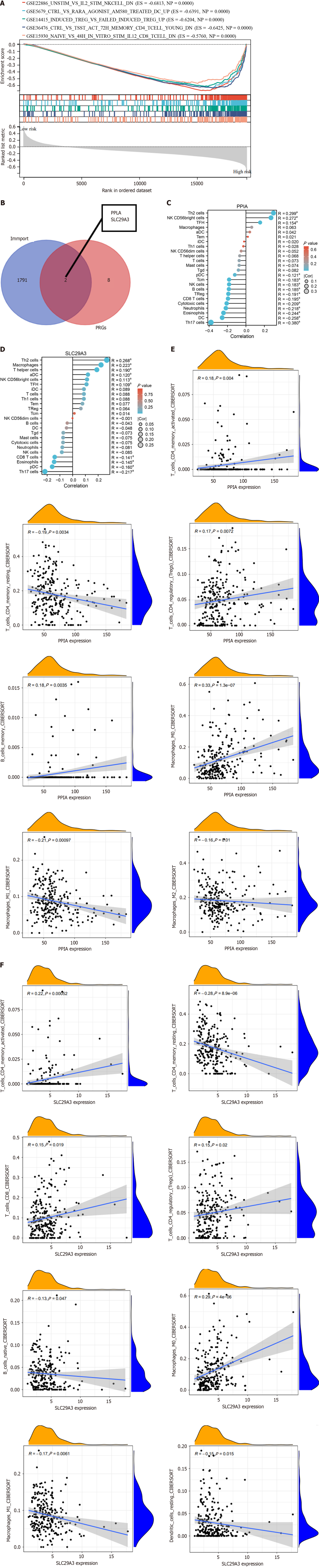
To further investigate the correlation between these genes and immune status, we first conducted GSEA on the high-risk and low-risk groups using the ImmuneSigDB and analyzed the relationship between cell types and states within the immune system. The results revealed that more than 400 immune markers were involved, with the five most common immune markers being natural killer (NK) cells, Dendritic Cells (DCs), regulatory T cells (Tregs), CD4 memory T cells, and CD8+ T cells (Figure 3A).
Moreover, we downloaded 1793 IRGs from the ImmPort website and performed an intersection analysis of 9 genes significantly associated with prognosis. The results revealed 2 overlapping genes, PPIA and SLC29A3 (Figure 3B). Using a dataset of 371 HCC patients from the TCGA, we analyzed the correlation between PPIA and SLC29A3 expression and 24 immune cell types. The results revealed that these two genes were strongly correlated with the Th2 subset of CD4+ T cells (Figure 3C and D). The Th2 subset can generate a series of cytokines, such as IL-4, IL-5, and IL-13, which promote the proliferation and invasion of tumor cells and inhibit immune cell resistance to tumor cells[20]. These findings are consistent with the prognostic curve results we obtained for PPIA and SLC29A3.
In addition, we performed immune infiltration analysis using CIBERSORT on the mRNA of 13 patients who underwent ablation and 235 patients who did not receive ablation treatment in the TCGA dataset. Then, we separately examined the relationships between PPIA or SLC29A3 and 22 types of immune cells in the dataset. The results showed that PPIA was associated with seven immune cell types, including memory CD4+ T cells, Tregs, memory B cells, and M0-M2 macrophages (Figure 3E), while SLC29A3 was significantly associated with eight immune cell types, including memory CD4+ T cells, CD8+ T cells, Tregs, memory B cells, M0-M1 macrophages, and DCs (Figure 3F).
In summary, we identified two genes, PPIA and SLC29A3, that are significantly associated with immune cell infiltration.
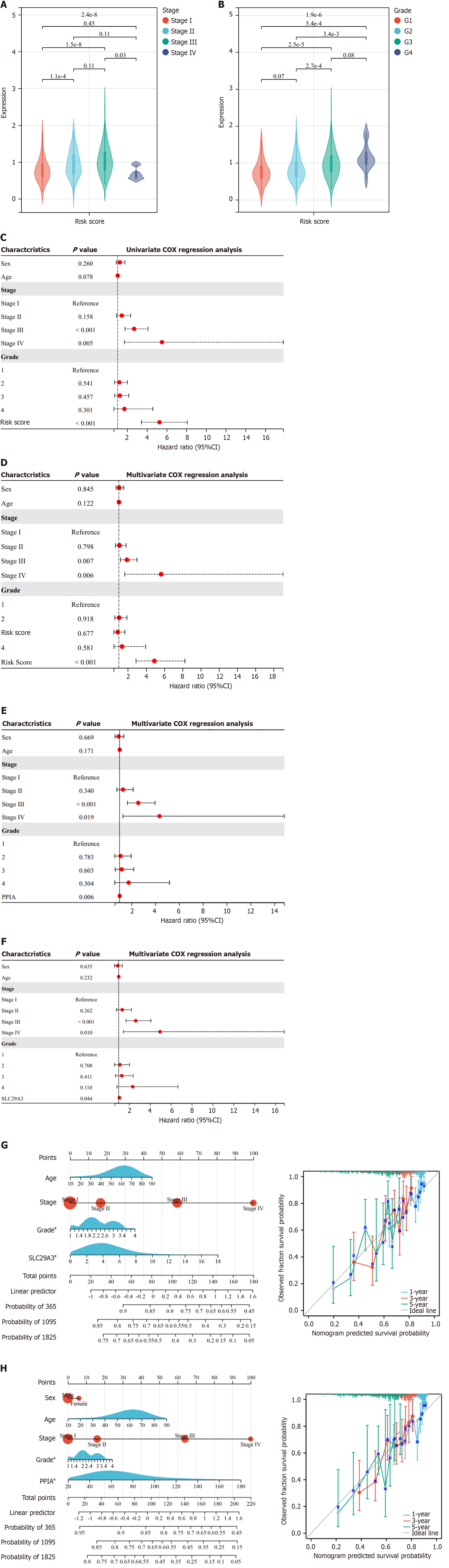
After further exploring the independent prognostic genes, we found significant correlations between RS, pathological grade, and tumor stage (Figure 4A and B). Using univariate and multivariate Cox regression analyses, we combined the RS with various clinical features, such as sex, age, stage, and grade. Through this approach, we determine the prognostic performance of the RS. The results of univariate Cox regression analysis showed that RS [hazard ratio (HR) = 5.250, P < 0.001] and stage (stage III, HR = 2.676, P < 0.001; stage IV, HR = 5.496, P = 0.005) were risk factors for the deterioration of HCC (Figure 4C). The results of the multivariate analysis indicated that the RS (HR = 4.877, P < 0.001) is an independent factor for assessing PD and is not influenced by other clinical characteristics (Figure 4D). Moreover, we performed multivariate Cox regression analyses of PPIA and SLC29A3 separately, and the results showed that neither was influenced by clinical characteristics (P < 0.05; Figure 4E and F).
We included PPIA and SLC29A3 as candidate markers according to the constructed nomogram. The overall C-index of the PPIA model was 0.673, and the C-index of SLC29A3 was 0.667. The results indicate that both genes have good predictive potential. The prediction of OS at 1, 3, and 5 years showed that the 1-year and 3-year survival rates predicted by the nomogram were consistent with the best predictive performance (Figure 4G and H). Therefore, PPIA and SLC29A3 have excellent prognostic ability and can be referred to as prognostic-related genes.
In summary, we first identified gene modules that were significantly correlated with immune infiltration by analyzing data from HCC patients treated with ablation therapy. Then, using machine learning, we identified 9 genes that can significantly predict patient prognosis. Ultimately, we found that the key genes most closely related to immunity were PPIA and SLC29A3. The expression level of PPIA is significantly elevated in HCC tissues[21]. Moreover, PPAAs may be involved in the immune evasion of HCC cells, thereby inhibiting the immune system's attack of HCC cells[21]. HCC cells may regulate the expression of SLC29A3, inhibiting the immune system's attack on HCC cells and thus promoting the development and metastasis of HCC[22]. In addition, according to the immune infiltration analysis, PPIA was significantly associated with CD4+ T cells, T Tregs, and macrophages, while SLC29A3 was significantly associated with CD4+ T cells, CD8+ T cells, T Tregs, B cells, macrophages, and DCs.
The tumor ablation procedure is a minimally invasive method commonly used to treat liver tumors[23]. The Kombo knife compound thermal ablation technique is an innovative research and development technique that is internationally original and domestically innovative. Sorafenib is an oral, small-molecule, targeted therapy that inhibits the formation and proliferation of tumor blood vessels by targeting the vascular endothelial growth factor receptor on tumor endothelial cells. It blocks the blood supply and tumor growth, thus exhibiting antitumor effects[24]. To improve patient survival, we administered treatment with the Kombo knife in combination with the oral administration of sorafenib.
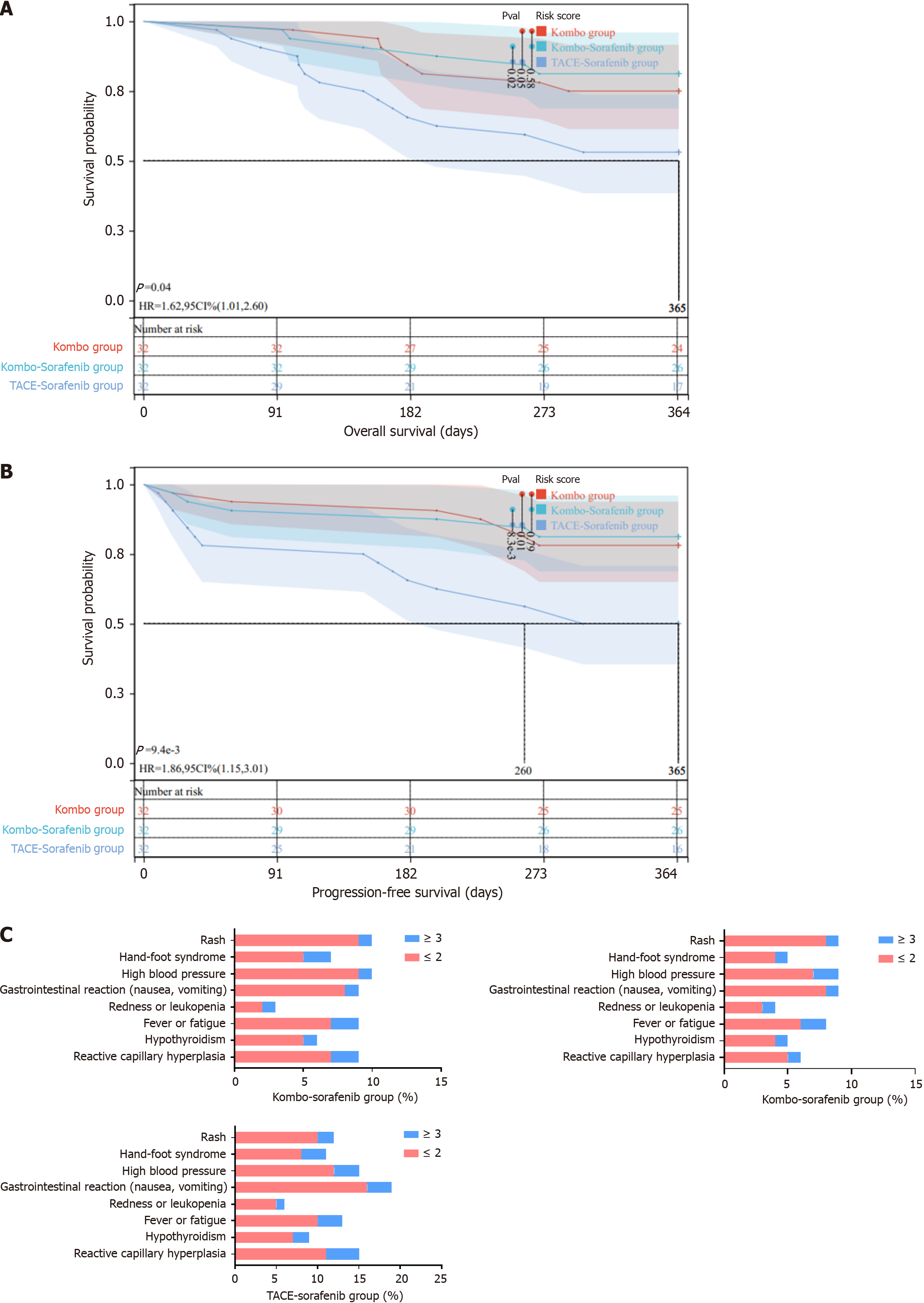
To evaluate the efficacy and safety of combination therapy, we followed up with 32 patients who received treatment with Kombo knife (Group B), 32 patients who received combination therapy with Kombo knife and sorafenib (Group C), and 32 patients with HCC who received traditional TACE combined with sorafenib therapy (Group C). Supplementary Table 1 shows that there were no significant differences in age, sex, alpha-fetoprotein levels, Child-Pugh classification, vascular invasion, or distant metastasis among the groups. The efficacy of the treatment was evaluated mainly on the basis of the ORR and DCR. According to the 2010 version of the mRECIST evaluation criteria for solid tumors, a 6-month follow-up was conducted from the start of treatment. In Group A, the ORR was 59.4%, and the DCR was 87.5%; in Group B, the ORR was 65.6%, and the DCR was 90.6%; and in Group C, the ORR was 46.9%, and the DCR was 78.8% (Table 2). After performing the chi-square test (χ2), significant differences were observed in the ORR between Groups A and C and between Groups B and C. Furthermore, there were significant differences in DCR among the three groups (Table 3), indicating that the combination of the Kombo knife and sorafenib has significant advantages and better treatment outcomes for HCC.
| Efficacy | Combo knife | Combo knife + sorafenib group | TACE + sorafenib |
| CR | 1 (3.1) | 1 (3.1) | 0 (0.0) |
| PR | 18 (56.3) | 20 (62.5) | 15 (46.9) |
| SD | 9 (28.1) | 8 (25.0) | 10 (31.3) |
| PD | 4 (12.5) | 3 (9.4) | 7 (21.8) |
| ORR | 59.4 | 65.6 | 46.9 |
| DCR | 87.5 | 90.6 | 78.7 |
Moreover, we calculated the OS for both groups. Survival began on the first day of combined therapy and ended at the 1-year follow-up. If the patient died, the follow-up was automatically terminated. The log-rank test was used to assess the significance of the differences in prognosis among the three groups (A, B, and C). In the end, we found that both Group A and Group B had better prognoses than Group C did, and Group B had a better prognosis than Group A did (Figure 5A).
Regarding PFS, Group B had a longer PFS than did the other two groups (Figure 5B), indicating a positive prognosis for patients with HCC treated with the Kombo Knife combined with sorafenib. Fifty-two out of 96 patients (54.1%) experienced adverse reactions during the one-year follow-up. Among them, there were 17 patients (53.1%) in Group A, 15 patients (46.9%) in Group B, and 20 patients (66.7%) in Group C. We statistically analyzed treatment-related adverse reactions, including reactive capillary proliferation, gastrointestinal reactions (nausea, vomiting), hypothyroidism, reduced red and white blood cell counts, fever, fatigue, hypertension, hand-foot reactions, and rash (Figure 5C). The rate of adverse reactions was relatively lower in Group A and Group B than in Group C, and no unexpected adverse reactions were observed. The above results indicate that combining the novel composite cold and hot ablation technique with sorafenib is safe for treating HCC patients.
Bioinformatics prediction revealed that genes significantly associated with HCC prognosis closely correlate with immune cells. Therefore, immunity is crucial in treating HCC, and T cells are its main component. The novel composite cold and hot ablation technique is a composite cold and heat ablation technique, and a large amount of HSP70 can be released after combined cold and heat treatment. It can induce the differentiation of myeloid-derived suppressor cells into mature DCs, increasing immune presentation and effector T-cell activation[25].
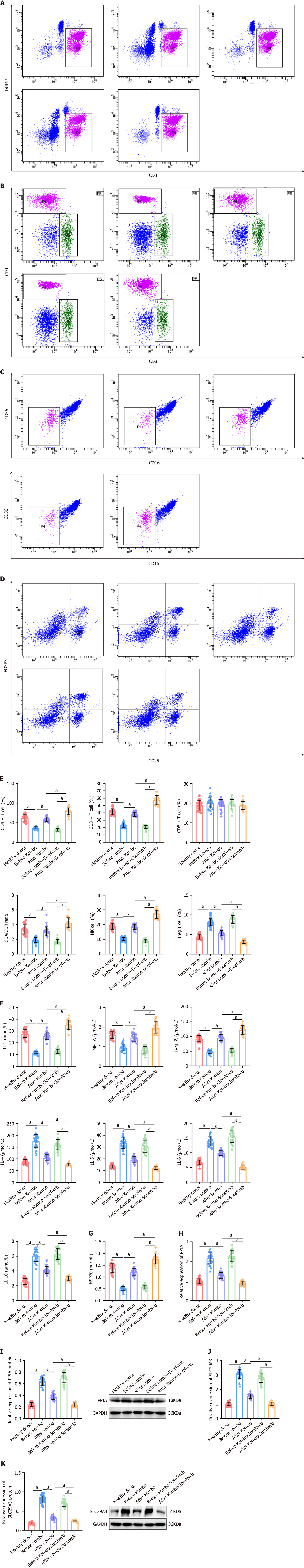
This study compared immune function before and after treatment in 32 patients with HCC who received combination treatment with Kombo knife and sorafenib and 32 who received treatment with only Kombo knife. A group of 32 healthy volunteers served as the control group. The numbers of CD8+ T cells, CD4+ T cells, CD3+ T cells, Tregs, and NK cells among PBMCs were quantified using flow cytometry. The results showed that, compared to those before treatment, the numbers of CD3+ T cells, CD4+ T cells, the CD4/CD8 ratio, and NK cells significantly decreased after treatment, while the number of Treg cells significantly increased. The change in the number of CD8+ T cells was not significant. However, compared with Kombo knife monotherapy, combination therapy significantly increased the number of CD3+ T cells and CD4+ T cells, the CD4/CD8 ratio, and the number of NK cells, while the number of Treg T cells decreased significantly. We also found that combination therapy improved the increase in the number of immune cells (Figure 6A-E).
We detected the levels of the Th1 cytokines IL-2 and IFN-γ, TNF-α and the Th2 cytokines IL-4, IL-5, IL-6, and IL-10 in the peripheral blood of patients before and after treatment using an ELISA. The results indicate that before treatment, the Th1 cytokine level is significantly lower in HCC patients, while the Th2 cytokine level is significantly greater. After treatment, the concentration of Th1 cytokines increased, while the concentration of Th2 cytokines decreased (Figure 6F). HSP70 is a protein that plays a role in immune coordination. We also measured the levels of HSP70 in the peripheral blood of patients and healthy volunteers. We observed a significant decrease in HSP70 Levels before treatment and a significant increase after treatment. In addition, the effect of combination therapy was more significant than that of monotherapy (Figure 6G).
Finally, via RT-qPCR analyses, we detected the expression of the immune genes PPIA and SLC29A3, which are significantly associated with the prognosis of HCC, in patients’ PBMCs. The results showed that the expression of PPIA and SLC29A3 significantly increased after treatment compared to that in the control group. After treatment, compared with those in the monotherapy group, the expression of PPIA and SLC29A3 in the combination therapy group was significantly lower. We also utilized Western blot analysis to assess the expression of the immune genes PPIA and SLC29A3, which are significantly associated with the prognosis of liver cancer, in PBMCs of the patients. Consistently, we observed a similar trend in their expression levels (Figure 6H-K).
Therefore, this study preliminarily demonstrated that combining novel composite cold and hot ablation techniques (Kombo knife) and sorafenib treatment could significantly improve the peripheral blood immune function of HCC patients.
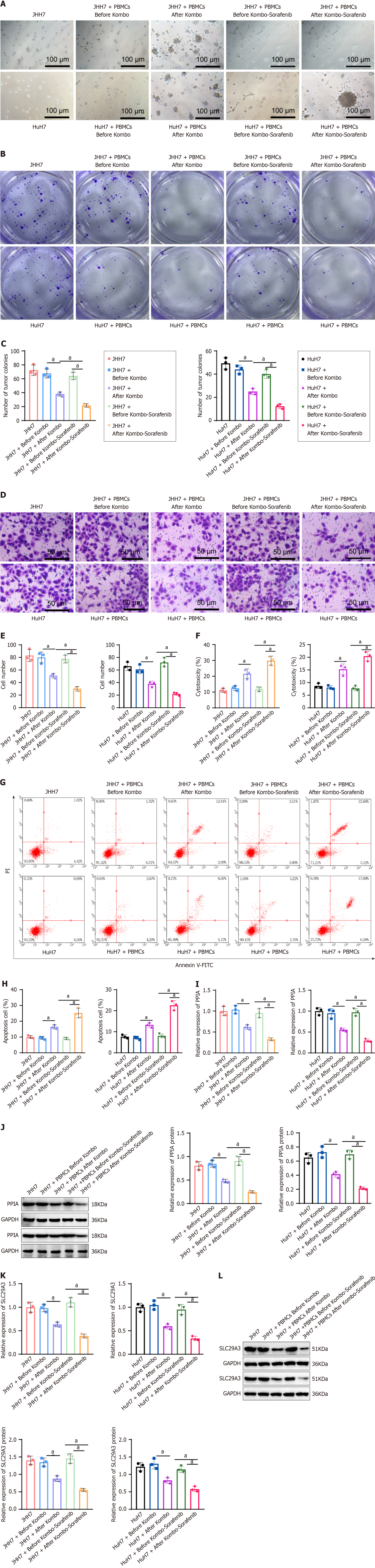
We found that combining Kombo knife and sorafenib increases the number of immune cells and the level of Th1 factors in the patient's peripheral blood. To further validate the influence of this combination treatment on HCC cells, we treated the HCC cell lines JHH7 and HuH7 with pre- and posttreatment with PBMCs (with a ratio of PBMCs:HCC = 10:1) separately or in combination therapy. The results showed that the combination therapy group had more immune cell aggregation than the single treatment group and inhibited tumor colony formation (Figure 7A-C).
Transwell assays were used to assess the effect of different PBMC combinations on the invasive ability of HCC cells. The results showed a decrease in the invasive ability of HCC cells in both treatment groups, with a more significant decrease in the group receiving combined treatment (Figure 7D and E). In addition, the cytotoxicity in the treatment group significantly increased, and the cytotoxicity in the combination treatment group became even more pronounced (Figure 7F). To detect the degree of apoptosis in HCC cells, we used flow cytometry to separate mononuclear cells and HCC cells and then used an Annexin V/PI staining kit to detect apoptosis. The results showed that the HCC cells in both treatment groups underwent significant apoptosis, with the combined treatment group showing more pronounced apoptosis (Figure 7G and H). We used RT-qPCR analyses to detect the expression of PPIA and SLC29A3. The results showed that the expression of PPIA and SLC29A3 significantly decreased after treatment compared to before treatment, and the decrease was more significant after combination therapy. Similarly, this expression trend was also observed in the Western blot experiments (Figure 7I-L).
Our results indicate that combination therapy comprising Kombo knife and sorafenib can be used to treat PBMCs and alleviate the proliferation and migration of HCC cells.
To further confirm the synergistic effect of the Kombo knife in combination with sorafenib on immune function among patients and to enhance the inhibition of HCC tumors, we simulated the impact of the patient's immune system on tumors in mice.
According to the above cell experiments, both types of HCC cells produced good results. To further confirm the tumor formation ability of both types of HCC cells in mice, logarithmic phase JHH7 or HuH7 cells were injected into the backs of BALB/c-nu/nu mice. As shown in Supplementary Figure 1, tumor formation was observed in mice, and both cell types exhibited good tumorigenic effects. Therefore, we selected the HuH7 cell line for subsequent experiments.
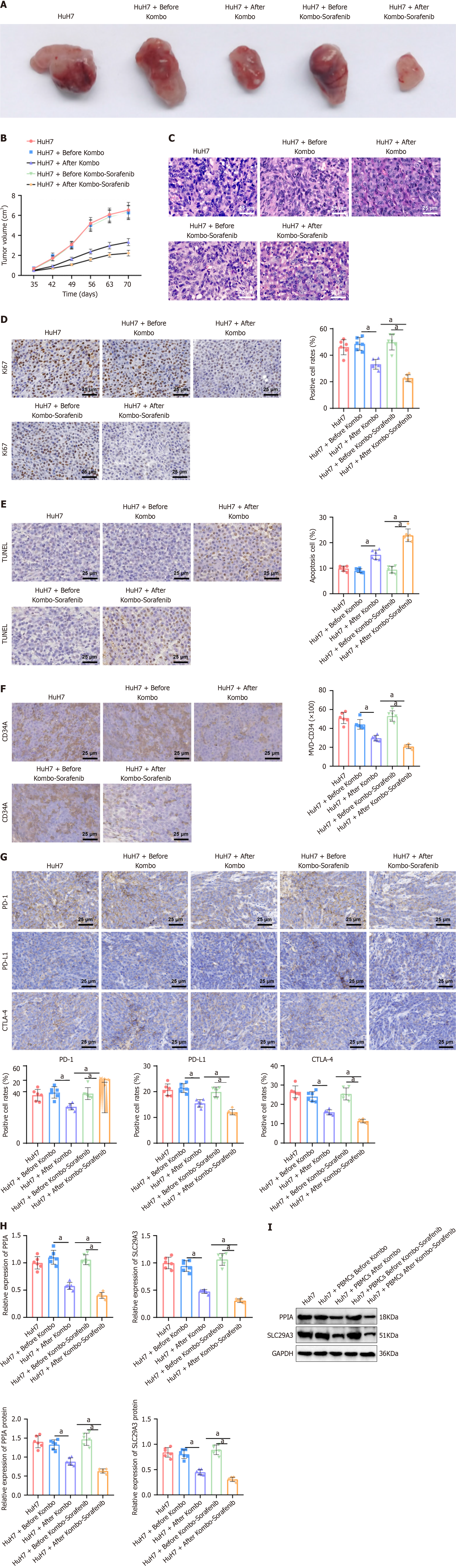
In the fifth week, PBMCs were collected from patients before and after treatment with Kombo knife alone or in combination with sorafenib and were injected into tumors. Starting from the 5th week, we observed the formation of tumors in the mice every week. The results showed that, compared to that in the group injected with HCC cells alone, the improvement in tumor formation was not significant when the mice were injected with PBMCs before treatment. However, when the mice were injected with PBMCs after treatment, the tumor volume significantly decreased, and the combination therapy group showed a more significant reduction in tumor volume than the monotherapy group did (Figure 8A and B). In the 10th week, the mice were euthanized. Histology with hematoxylin and eosin staining revealed typical tumor changes in the cells of the control and PBMC groups before treatment. The tumor cells were densely arranged and disorderly, with occasional trabecular structures. The cell nuclei appeared large, dark, and aberrant, and pathological nuclear division was observed. Focal necrosis could be observed. The number of tumor cells notably decreased in the treatment group, and the cells were loosely arranged (Figure 8C).
IHC staining analysis revealed that the expression of Ki-67, a protein associated with proliferative activity, was decreased in excised subcutaneous transplant tumor tissues after treatment. Furthermore, the combined treatment had an even more significant effect (Figure 8D). Furthermore, the TUNEL assay detected apoptotic cell nuclear fragmentation in the subcutaneous transplanted tumor tissue. Under the microscope, after DAB staining, both treatment groups showed apoptotic cells with dense brown-yellow staining in the nucleus, with the combination therapy group having more apoptotic cells than the single treatment group (Figure 8E).
In malignant tumors, there is a rapid increase in the formation of blood vessels, along with the occurrence of blood vessel curling and dilation. The phenomenon of concentrated neovascularization at the edge of the tumor was observed. Therefore, we tested whether the infiltration of these cells altered the tumor microvessel density (MVD). Compared to that in patients treated with monotherapy, immunostaining for CD34, a marker of endothelial cells, revealed a significant decrease in the MVD in tumor tissues injected with HCC cells and PBMCs after treatment (Figure 8F).
Immune checkpoints (ICs), as immunological regulators, can be "hijacked" and continuously activated by tumors, suppressing antitumor immunity and promoting tumor development[26]. Immunohistochemical analysis of PD-1, PD-L1, and CTLA-4 immune checkpoint expression in tumor tissue showed that treatment with PBMCs significantly reduced the levels of immune checkpoint genes, and combination therapy had more significant effects (Figure 8G).
Using RT-qPCR analyses, the expression of PPIA and SLC29A3 in tumor tissues was detected. The results showed that after treatment, the expression of PPIA and SLC29A3 decreased significantly, and the decrease was even more significant after combined treatment. The trend observed in the Western blot experiments aligns with that of the qRT-PCR results (Figure 8H and I).
Research indicates that after combination therapy with Kombo knife and sorafenib, PBMCs can significantly inhibit tumor formation in mice.
HCC is a prevalent malignant tumor, and there are ongoing challenges associated with its treatment[27]. The combination of state-of-the-art composite cold and heat ablation technology (Kombo Knife) with the drug sorafenib has demonstrated promising potential in the treatment of liver cancer, thus addressing this critical concern[22]. The Kombo Knife effectively induces cold and heat ablation to destroy tumor cells, whereas sorafenib can inhibit both tumor growth and blood vessel formation. Hence, the combination of these two treatment methods can offer liver cancer patients a more efficacious and all-encompassing treatment strategy[28].
Bioinformatics technology plays an important role in tumor research[29]. The TCGA database is a crucial resource that plays a pivotal role in immune infiltration research[30]. By analyzing the gene expression data from tumor samples, researchers have the ability to evaluate the infiltration of immune cells within tumor tissue. This analysis allows for a deeper investigation into the connection between immune regulation and tumor development[31]. These findings offer crucial insights for the advancement of immunotherapy and personalized treatment[32].
However, predicting the crucial role of immune genes in tumor treatment is imperative[33]. Numerous studies have shown a close correlation between the overexpression of PPIA and the growth, proliferation, invasion, and metastasis capabilities of HCC[21,34,35]. Additionally, PPIA is involved in the regulation of apoptosis in HCC cells, as its high expression levels can suppress tumor cell apoptosis, thereby enhancing the survival capacity of HCC cells[36]. Importantly, PPIA also interacts with other molecules in the tumor microenvironment, such as inflammatory factors and extracellular matrix components, to modulate the invasion and metastasis capabilities of HCC cells[37,38]. SLC29A3 is a transporter protein that plays a significant role in nucleotide transport and metabolic regulation. The role of SLC29A3 has been found to be closely associated with the development and prognosis of various tumors, such as colorectal cancer and lung cancer[39,40]. One study confirmed that the upregulation of SLC29A3 promotes the proliferation, invasion, and migration of pancreatic cancer cells and is associated with poor prognosis[41]. Another study revealed that SLC29A3 affects cytokine secretion and the inflammatory response by regulating the TLR-MAPK signaling pathway[42]. These studies provide important experimental and clinical evidence, thus confirming the critical role of PPIA and SLC29A3 in immune regulation and offering insights for a deeper understanding of the underlying mechanisms by which PPIA and SLC29A3 function in HCC. Research studies have indicated that the genes PPIA and SLC29A3 play vital roles in the process of immune infiltration[43]. PPIA regulates the function and activation of immune cells, whereas SLC29A3 controls the migration and antimicrobial activity of monocytes[44]. The expression levels of these genes may be correlated with treatment efficacy in combination therapy with Kombo and sorafenib[45]. Thus, the prediction of immune gene exp
Recent research has indicated the crucial role of predicting immune genes in understanding immune infiltration and the significance of disease treatment[47]. The immune genes PPIA and SLC29A3 have been shown to have crucial impacts on immune infiltration[43]. Studying these genes allows us to improve our understanding of the immune system's res
Research on the clinical efficacy of combination therapy has made significant progress in improving treatment safety, PFS, and the ORR[49]. However, in previous studies, both cabozantinib and sorafenib were used as monotherapies. Thus, this study aimed to investigate the enhanced therapeutic effects of combining cabozantinib and sorafenib in liver cancer treatment[50]. Our research findings demonstrated that combination therapy comprising cabozantinib and sorafenib has significant advantages, as it improved treatment outcomes and notably prolonged OS and PFS in liver cancer patients. Additionally, the ORR was significantly increased, demonstrating the safety of this combined treatment. Through experimental validation, we observed that combination therapy significantly enhances peripheral blood immune function in liver cancer patients, leading to a more active immune system that aids in resisting further tumor growth and metastasis. Furthermore, the combination treatment also markedly inhibited the proliferation and migration ability of liver cancer cells. The experimental results indicated a significant reduction in the proliferation and migration ability of liver cancer cells in the combination therapy group, demonstrating its superior efficacy compared to that of cabozantinib or sorafenib alone in suppressing the malignant behavior of tumors. Importantly, our study also revealed that the combination of cabozantinib and sorafenib significantly inhibited tumor formation in a mouse model. Following the combined treatment, the mouse tumor growth rate noticeably decreased, and the tumor volume significantly decreased, further confirming the effectiveness of the combination therapy. Moreover, combination therapy has been found to significantly enhance immune infiltration and immune cell generation, thereby augmenting the responsiveness of the immune system[51]. These findings suggest that combination therapy has the potential to significantly improve treatment efficacy and patient survival rates[52].
Coculture of PBMCs with liver cancer cells from patients has shown promise as a therapeutic strategy for liver cancer[53]. Our research has validated the effectiveness of this treatment method through both in vitro experiments and mouse models[54]. In particular, PBMCs significantly inhibit the proliferation, invasion, and migration of liver cancer cells. Additionally, PBMCs play a crucial role in modulating ICs[55]. These findings offer valuable insights and references for the development of immune therapy strategies targeting PBMCs[56].
Although this study yielded significant findings (Figure 9), it is crucial to acknowledge the potential biases and limi
This study explores an advanced approach to managing hepatocellular carcinoma (HCC), integrating data analytics from The Cancer Genome Atlas (TCGA) and innovative treatments, notably the Kombo Knife and sorafenib regimen. It emphasizes the need for holistic management strategies in HCC treatment, leveraging genomic insights.
The motivation behind this research is to address key challenges in HCC treatment by exploring the therapeutic potential of combined Kombo Knife and sorafenib regimen. The study aims to solve significant issues in HCC management by understanding the immunological genes and mechanisms through TCGA data analysis, contributing to future research advancements in this domain.
The primary objective is to identify immunological genes and the underlying mechanisms of the Kombo Knife and sorafenib regimen in HCC treatment. By using TCGA data and machine learning, the study seeks to enhance the understanding and effectiveness of HCC treatments, marking a significant stride in oncological research.
The study employs a combination of weighted gene coexpression network analysis, machine learning, and RNA sequencing data from TCGA to evaluate immune attributes in postablation HCC. Additionally, it critically examines the therapeutic landscape and safety metrics of the combined treatment across cellular and animal models.
Significant findings include the identification of immune genes, specifically peptidylprolyl isomerase A and solute carrier family 29 member 3, as key prognostic markers. The combined treatment of Kombo Knife with sorafenib showed enhanced outcomes like prolonged progression-free survival and increased overall response rate in HCC.
The study concludes that the innovative combination of Kombo Knife with sorafenib, rooted in immune-centric strategies, is a promising modality in HCC treatment. It underscores the effectiveness of integrating genomic data analysis with progressive therapeutic approaches.
Future research should focus on further elucidating the immunological mechanisms behind the combined treatment, exploring its broader applicability, and refining the integration of genomic data for personalized HCC management. This direction promises to unlock new therapeutic frontiers in oncology.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Arslan M, Turkey; Kishida Y, Japan S-Editor: Li L L-Editor: A P-Editor: Zhao YQ
| 1. | Verma N, Yumeen S, Raggio BS. Ablative Laser Resurfacing. 2023 Apr 23. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. [PubMed] |
| 2. | Tan LL, Loganathan N, Agarwalla S, Yang C, Yuan W, Zeng J, Wu R, Wang W, Duraiswamy S. Current commercial dPCR platforms: technology and market review. Crit Rev Biotechnol. 2023;43:433-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 3. | Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021;134:783-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1624] [Cited by in RCA: 1783] [Article Influence: 445.8] [Reference Citation Analysis (1)] |
| 4. | Lu L, Mullins CS, Schafmayer C, Zeißig S, Linnebacher M. A global assessment of recent trends in gastrointestinal cancer and lifestyle-associated risk factors. Cancer Commun (Lond). 2021;41:1137-1151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 183] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 5. | Shi X, Singh S, Lin E, Li H. Chimeric RNAs in cancer. Adv Clin Chem. 2021;100:1-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Mutebi M, Anderson BO, Duggan C, Adebamowo C, Agarwal G, Ali Z, Bird P, Bourque JM, DeBoer R, Gebrim LH, Masetti R, Masood S, Menon M, Nakigudde G, Ng'ang'a A, Niyonzima N, Rositch AF, Unger-Saldaña K, Villarreal-Garza C, Dvaladze A, El Saghir NS, Gralow JR, Eniu A. Breast cancer treatment: A phased approach to implementation. Cancer. 2020;126:2365-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 81] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 7. | Zhou Y, Wu C, Wang B, Xu Y, Zhao H, Guo Y, Wu X, Yu J, Rao L, Wang X, Yu F. Characterization difference of typical KL1, KL2 and ST11-KL64 hypervirulent and carbapenem-resistant Klebsiella pneumoniae. Drug Resist Updat. 2023;67:100918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 36] [Reference Citation Analysis (0)] |
| 8. | Irfan M, Delgado RZR, Frias-Lopez J. The Oral Microbiome and Cancer. Front Immunol. 2020;11:591088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 179] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 9. | Singh S, Numan A, Somaily HH, Gorain B, Ranjan S, Rilla K, Siddique HR, Kesharwani P. Nano-enabled strategies to combat methicillin-resistant Staphylococcus aureus. Mater Sci Eng C Mater Biol Appl. 2021;129:112384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Yin K, Patten D, Gough S, de Barros Gonçalves S, Chan A, Olan I, Cassidy L, Poblocka M, Zhu H, Lun A, Schuijs M, Young A, Martinez-Jimenez C, Halim TYF, Shetty S, Narita M, Hoare M. Senescence-induced endothelial phenotypes underpin immune-mediated senescence surveillance. Genes Dev. 2022;36:533-549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 11. | Won JE, Byeon Y, Wi TI, Lee CM, Lee JH, Kang TH, Lee JW, Lee Y, Park YM, Han HD. Immune checkpoint silencing using RNAi-incorporated nanoparticles enhances antitumor immunity and therapeutic efficacy compared with antibody-based approaches. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, Galvez-Reyes J, Kalekar RL, Mulugeta N, Kapner KS, Raghavan MS, Borah AA, Liu N, Väyrynen SA, Costa AD, Ng RWS, Wang J, Hill EK, Ragon DY, Brais LK, Jaeger AM, Spurr LF, Li YY, Cherniack AD, Booker MA, Cohen EF, Tolstorukov MY, Wakiro I, Rotem A, Johnson BE, McFarland JM, Sicinska ET, Jacks TE, Sullivan RJ, Shapiro GI, Clancy TE, Perez K, Rubinson DA, Ng K, Cleary JM, Crawford L, Manalis SR, Nowak JA, Wolpin BM, Hahn WC, Aguirre AJ, Shalek AK. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184:6119-6137.e26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 311] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 13. | Yang H, Zhao L, Zhang Y, Li FF. A comprehensive analysis of immune infiltration in the tumor microenvironment of osteosarcoma. Cancer Med. 2021;10:5696-5711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 14. | Seki T, Yang Y, Sun X, Lim S, Xie S, Guo Z, Xiong W, Kuroda M, Sakaue H, Hosaka K, Jing X, Yoshihara M, Qu L, Li X, Chen Y, Cao Y. Brown-fat-mediated tumour suppression by cold-altered global metabolism. Nature. 2022;608:421-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 143] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 15. | Bapat SP, Whitty C, Mowery CT, Liang Y, Yoo A, Jiang Z, Peters MC, Zhang LJ, Vogel I, Zhou C, Nguyen VQ, Li Z, Chang C, Zhu WS, Hastie AT, He H, Ren X, Qiu W, Gayer SG, Liu C, Choi EJ, Fassett M, Cohen JN, Sturgill JL, Crotty Alexander LE, Suh JM, Liddle C, Atkins AR, Yu RT, Downes M, Liu S, Nikolajczyk BS, Lee IK, Guttman-Yassky E, Ansel KM, Woodruff PG, Fahy JV, Sheppard D, Gallo RL, Ye CJ, Evans RM, Zheng Y, Marson A. Obesity alters pathology and treatment response in inflammatory disease. Nature. 2022;604:337-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 128] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 16. | Su W, Wright CM, Lee DY, Kim M, Anstadt EJ, Teo BK, Carlson DJ, Lukens JN, Eisbruch A, Lin A. Stricter Postoperative Oropharyngeal Cancer Radiation Therapy Normal Tissue Dose Constraints Are Feasible. Pract Radiat Oncol. 2022;12:e282-e285. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Lv M, Chen M, Zhang R, Zhang W, Wang C, Zhang Y, Wei X, Guan Y, Liu J, Feng K, Jing M, Wang X, Liu YC, Mei Q, Han W, Jiang Z. Manganese is critical for antitumor immune responses via cGAS-STING and improves the efficacy of clinical immunotherapy. Cell Res. 2020;30:966-979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 530] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 18. | Zhou KX, Huang S, Hu LP, Zhang XL, Qin WT, Zhang YL, Yao LL, Yu Y, Zhou YQ, Zhu L, Ji J, Zhang ZG. Increased Nuclear Transporter KPNA2 Contributes to Tumor Immune Evasion by Enhancing PD-L1 Expression in PDAC. J Immunol Res. 2021;2021:6694392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 19. | Zhang QF, Li J, Jiang K, Wang R, Ge JL, Yang H, Liu SJ, Jia LT, Wang L, Chen BL. CDK4/6 inhibition promotes immune infiltration in ovarian cancer and synergizes with PD-1 blockade in a B cell-dependent manner. Theranostics. 2020;10:10619-10633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 20. | Zhu X, Zhu J. CD4 T Helper Cell Subsets and Related Human Immunological Disorders. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 217] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 21. | Mou L, Jia C, Wu Z, Xin B, Liang Zhen CA, Wang B, Ni Y, Pu Z. Clinical and Prognostic Value of PPIA, SQSTM1, and CCL20 in Hepatocellular Carcinoma Patients by Single-Cell Transcriptome Analysis. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3844] [Article Influence: 961.0] [Reference Citation Analysis (3)] |
| 23. | Izzo F, Palaia R, Albino V, Amore A, di Giacomo R, Piccirillo M, Leongito M, Nasto A, Granata V, Petrillo A, Lastoria S. Hepatocellular carcinoma and liver metastases: clinical data on a new dual-lumen catheter kit for surgical sealant infusion to prevent perihepatic bleeding and dissemination of cancer cells following biopsy and loco-regional treatments. Infect Agent Cancer. 2015;10:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Musk SR, Downes CS, Johnson RT. Caffeine induces uncoordinated expression of cell cycle functions after ultraviolet irradiation. Accelerated cycle transit, sister chromatid exchanges and premature chromosome condensation in a transformed Indian muntjac cell line. J Cell Sci. 1988;90:591-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Theivanthiran B, Yarla N, Haykal T, Nguyen YV, Cao L, Ferreira M, Holtzhausen A, Al-Rohil R, Salama AKS, Beasley GM, Plebanek MP, DeVito NC, Hanks BA. Tumor-intrinsic NLRP3-HSP70-TLR4 axis drives premetastatic niche development and hyperprogression during anti-PD-1 immunotherapy. Sci Transl Med. 2022;14:eabq7019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 26. | Li B, Chan HL, Chen P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr Med Chem. 2019;26:3009-3025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 312] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 27. | Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1284] [Cited by in RCA: 1206] [Article Influence: 402.0] [Reference Citation Analysis (41)] |
| 28. | Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 848] [Article Influence: 212.0] [Reference Citation Analysis (0)] |
| 29. | Lu Z, Fei L, Hou G. A pan-cancer analysis of the oncogenic role of ERCC6L. BMC Cancer. 2022;22:1347. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 30. | Liu X, Zhan Y, Xu W, Liu X, Geng Y, Liu L, Da J, Wang J, Zhang X, Jin H, Liu Z, Guo S, Zhang B, Li Y. Prognostic and immunological role of Fam20C in pan-cancer. Biosci Rep. 2021;41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 31. | Zhang ZJ, Huang YP, Liu ZT, Wang YX, Zhou H, Hou KX, Tang JW, Xiong L, Wen Y, Huang SF. Identification of immune related gene signature for predicting prognosis of cholangiocarcinoma patients. Front Immunol. 2023;14:1028404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Scheiner B, Pinter M. Reply to: "The CRAFITY score: A new guidepost for prognosis prediction in patients with hepatocellular carcinoma undergoing immunotherapy". J Hepatol. 2022;76:1233-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 33. | Jiang H, Zhang X, Wu Y, Zhang B, Wei J, Li J, Huang Y, Chen L, He X. Bioinformatics identification and validation of biomarkers and infiltrating immune cells in endometriosis. Front Immunol. 2022;13:944683. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 34. | Li L, Yu S, Chen J, Quan M, Gao Y, Li Y. miR-15a and miR-20b sensitize hepatocellular carcinoma cells to sorafenib through repressing CDC37L1 and consequent PPIA downregulation. Cell Death Discov. 2022;8:297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 35. | Wang S, Li M, Xing L, Yu J. High expression level of peptidylprolyl isomerase A is correlated with poor prognosis of liver hepatocellular carcinoma. Oncol Lett. 2019;18:4691-4702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Cheng S, Luo M, Ding C, Peng C, Lv Z, Tong R, Xiao H, Xie H, Zhou L, Wu J, Zheng S. Downregulation of Peptidylprolyl isomerase A promotes cell death and enhances doxorubicin-induced apoptosis in hepatocellular carcinoma. Gene. 2016;591:236-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Gu Y, Wang C, Chen S, Tang J, Guo X, Hu W, Cui A, Zhang D, Yu K, Chen M. A Critical Role of Peptidylprolyl Isomerase A Pseudogene 22/microRNA-197-3p/Peptidylprolyl Isomerase A Axis in Hepatocellular Carcinoma. Front Genet. 2021;12:604461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Li X, Gu M, Hu Q, Weng Y, Cai X. Development and validation of metabolic models for predicting survival and immune status of hepatocellular carcinoma patients. Adv Clin Exp Med. 2023;32:1423-1439. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 39. | Song NS, Pei ZD, Fu G. MiR-1224-5p acts as a tumor suppressor via inhibiting the malignancy of rectal cancer through targeting SLC29A3. IUBMB Life. 2020;72:2204-2213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 40. | Chen X, Zhang L, Ren S, Li X, Zhou F, Li W, Gao G, He Y, Zhou C. Genomic polymorphisms of SLC29A3 associated with overall survival in advanced non-small-cell lung cancer treated with gemcitabine. Med Oncol. 2014;31:865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Mohelnikova-Duchonova B, Brynychova V, Hlavac V, Kocik M, Oliverius M, Hlavsa J, Honsova E, Mazanec J, Kala Z, Melichar B, Soucek P. The association between the expression of solute carrier transporters and the prognosis of pancreatic cancer. Cancer Chemother Pharmacol. 2013;72:669-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Shiloh R, Lubin R, David O, Geron I, Okon E, Hazan I, Zaliova M, Amarilyo G, Birger Y, Borovitz Y, Brik D, Broides A, Cohen-Kedar S, Harel L, Kristal E, Kozlova D, Ling G, Shapira Rootman M, Shefer Averbuch N, Spielman S, Trka J, Izraeli S, Yona S, Elitzur S. Loss of function of ENT3 drives histiocytosis and inflammation through TLR-MAPK signaling. Blood. 2023;142:1740-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Cohen YC, Zada M, Wang SY, Bornstein C, David E, Moshe A, Li B, Shlomi-Loubaton S, Gatt ME, Gur C, Lavi N, Ganzel C, Luttwak E, Chubar E, Rouvio O, Vaxman I, Pasvolsky O, Ballan M, Tadmor T, Nemets A, Jarchowcky-Dolberg O, Shvetz O, Laiba M, Shpilberg O, Dally N, Avivi I, Weiner A, Amit I. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat Med. 2021;27:491-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 156] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 44. | Kumar S, Singh SK, Viswakarma N, Sondarva G, Nair RS, Sethupathi P, Sinha SC, Emmadi R, Hoskins K, Danciu O, Thatcher GRJ, Rana B, Rana A. Mixed lineage kinase 3 inhibition induces T cell activation and cytotoxicity. Proc Natl Acad Sci USA. 2020;117:7961-7970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 45. | Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 820] [Article Influence: 205.0] [Reference Citation Analysis (0)] |
| 46. | Ma B, Wang K, Liang Y, Meng Q, Li Y. Molecular Characteristics, Oncogenic Roles, and Relevant Immune and Pharmacogenomic Features of EVA1B in Colorectal Cancer. Front Immunol. 2022;13:809837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 47. | Song Q, Zhou R, Shu F, Fu W. Cuproptosis scoring system to predict the clinical outcome and immune response in bladder cancer. Front Immunol. 2022;13:958368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 109] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 48. | Yanjun Z, Jin S, Qiuyue L, Tao L. Risk factors and the establishment of nomogram model for incisional-wound infection in patients with bladder cancer after radical resection. Asian J Surg. 2023;46:1418-1419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 49. | Bosma NA, Warkentin MT, Gan CL, Karim S, Heng DYC, Brenner DR, Lee-Ying RM. Efficacy and Safety of First-line Systemic Therapy for Metastatic Renal Cell Carcinoma: A Systematic Review and Network Meta-analysis. Eur Urol Open Sci. 2022;37:14-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 50. | Shanks GD. Historical 8-Aminoquinoline Combinations: Not All Antimalarial Drugs Work Well Together. Am J Trop Med Hyg. 2022;107:964-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 51. | Chen Z, Zeng L, Liu G, Ou Y, Lu C, Yang B, Zuo L. Construction of Autophagy-Related Gene Classifier for Early Diagnosis, Prognosis and Predicting Immune Microenvironment Features in Sepsis by Machine Learning Algorithms. J Inflamm Res. 2022;15:6165-6186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 52. | Wolchok JD, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, Ferrucci PF, Smylie M, Butler MO, Hill A, Márquez-Rodas I, Haanen JBAG, Guidoboni M, Maio M, Schöffski P, Carlino MS, Lebbé C, McArthur G, Ascierto PA, Daniels GA, Long GV, Bas T, Ritchings C, Larkin J, Hodi FS. Long-Term Outcomes With Nivolumab Plus Ipilimumab or Nivolumab Alone Versus Ipilimumab in Patients With Advanced Melanoma. J Clin Oncol. 2022;40:127-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 752] [Article Influence: 250.7] [Reference Citation Analysis (0)] |
| 53. | Liu Y, Zhang Q, Xing B, Luo N, Gao R, Yu K, Hu X, Bu Z, Peng J, Ren X, Zhang Z. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell. 2022;40:424-437.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 237] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 54. | Xu X, Gao W, Li L, Hao J, Yang B, Wang T, Bai X, Li F, Ren H, Zhang M, Zhang L, Wang J, Wang D, Zhang J, Jiao L. Annexin A1 protects against cerebral ischemia-reperfusion injury by modulating microglia/macrophage polarization via FPR2/ALX-dependent AMPK-mTOR pathway. J Neuroinflammation. 2021;18:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 155] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 55. | Zhang H, Fu L. The role of ALDH2 in tumorigenesis and tumor progression: Targeting ALDH2 as a potential cancer treatment. Acta Pharm Sin B. 2021;11:1400-1411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 99] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 56. | Sathe A, Grimes SM, Lau BT, Chen J, Suarez C, Huang RJ, Poultsides G, Ji HP. Single-Cell Genomic Characterization Reveals the Cellular Reprogramming of the Gastric Tumor Microenvironment. Clin Cancer Res. 2020;26:2640-2653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 239] [Article Influence: 47.8] [Reference Citation Analysis (0)] |