INTRODUCTION
Colorectal cancer (CRC) is the third most prevalent cancer, accounting for 153020 new cases and 52550 deaths in the United States in 2023[1]. In addition, the incidence of CRC continues to increase in many nations, including China, which may be caused by alterations in the environment and lifestyle[2]. The prognosis of CRC varies, and the five-year survival rate of CRC patients ranges from 10%-90% for early- and late-stage CRC patients (TNM stages I and IV). However, most patients are initially diagnosed with stage II or III disease, the prognosis of these patients differs, and the predictive performance of the TNM staging system is not satisfactory for these patients[3,4], which makes it difficult to make decisions regarding adjuvant therapy for stage II or III CRC patients[5]. Thus, developing biomarkers or models for predicting the clinical outcome of CRC patients is currently a considerable challenge.
In recent years, single gene-based biomarkers, including genomic mutations, expression (mRNA, lncRNA, miRNA, etc.), and epigenetic and protein biomarkers, have been widely reported. For example, KRAS mutations are detected in 40% of CRC samples and are sensitive to specific treatment regimens[6]. The lncRNA LINRIS was reported to bind to the K139-linked ubiquitin chain of IGF2BP2 to regulate sugar metabolism in CRC[7]. The circRNA circEZH2 was shown to stabilize CREB1 mRNA via RNA methylation[8]. In addition, epigenetic modifications have also been widely reported as candidate targets for diagnosis, prognosis evaluation, and therapeutic applications. However, these studies focused mainly on the mechanisms underlying cancer development, and the clinical implications are still vague. One of the most important reasons is the robustness of the biomarker. Cancers, including CRC, are highly heterogeneous diseases with complex genomic, epigenomic, transcriptomic, metabolic, and proteomic features and tumor microenvironment interactions, which indicates that single genes are unreliable markers for prognosis prediction. Recently, multiple gene-based prognostic models have been emphasized for their robustness and reproducibility. Early in 2002, a prognostic tool named 'mammaprint' was developed according to the expression of 70 genes[9]. Subsequently, a large number of studies have retrospectively and prospectively validated the reproducibility of the model for guiding radiotherapy of early-stage breast cancer[10-12]. In CRC, Okuno et al[13] developed a signature using miRNAs to predict the response of CRC patients with stage II-III disease to FOLFOX adjuvant therapy, and the model had an area under the curve (AUC) of 0.89. Similarly, using 26 stem cell-related genes, a signature was constructed to predict the clinical outcome of CRC patients and may aid in predicting the response to immunotherapy[14]. Nevertheless, studies on these CRC models either lack validation experiments, which makes the models less robust, or lack investigation of signatures from multiple biological aspects, which makes the models less informative.
Epigenetic regulation is a complex system involving 229 genes that modulate the extent of gene expression by affecting the modification of DNA and histones. It is reportedly involved in multiple processes, including the development and progression of CRC[15], and investigations regarding its utilization for diagnosis and prognosis evaluation have been reported[16]. However, studies aimed at predicting the survival of CRC patients using epigenetic genes are rare. Thus, a prognostic model was developed using genes involved in epigenetic regulation in the TCGA dataset; the genes were evaluated by RNA-seq, and the results validated across cohorts from other platforms. Furthermore, associations between the constructed signature and genomic, clinical, transcriptomic, microenvironmental features and pharmaceutical signatures were identified. Single-cell sequencing revealed that the signature reflected different cell-cell interaction patterns.
MATERIALS AND METHODS
Data collection and patient enrollment
Data on gene expression, clinical features, mutations, gene copy number and DNA methylation for the TCGA-CRC cohort were retrieved from the cBioPortal[17] database. The GEO dataset was retrieved from the GEO database by searching the keywords 'colorectal cancer' and manually selecting datasets with sample sizes greater than 80 and sufficient survival information. The raw data of the GSE17526, GSE17537, GSE33113, GSE37892, GSE39048 and GSE39582 datasets were downloaded from the GEO database. The sample type was reviewed, and only primary tumor samples were retained. After background correction and normalization, the probes were annotated with gene symbols. If a probe corresponded to several different genes, the probe was excluded; if multiple probes corresponded to one gene, the probe with the highest signaling intensity was retained, and the other probes were excluded. All transcriptome data were log2 transformed.
Candidate gene selection, signature development and validation
Genes involved in epigenetic regulation were downloaded from the MSigDB[18] database, and Cox univariate regression was used to correlate the expression of these genes with overall survival. Genes that were significantly correlated with overall survival (P < 0.05) were retained as candidate genes. The combinations of these candidate genes were enumerated (with a gene number ≤ 10). For each combination, Cox multivariate regression was implemented for the TCGA cohort to develop a linear model. The signature was calculated as follows: Where ci is the coefficient of gene i and ei is the log2-transformed RSEM count. For validation, ei is the log2-transformed signaling intensity. For each combination, a log-rank sum test between groups was performed, and the combination with the most significant P value was used as the optimized panel. After selecting the panel, the coefficients were also determined. In each dataset, samples were classified into low- and high-risk groups according to the mean risk score in the dataset. Survival differences between groups were assessed in both the training and validation datasets.
Clinical, genomic, and transcriptomic feature identification
In the TCGA dataset, the signature values were compared between clinical groups using ANOVA. Cox multivariate regression analysis of the signature and other clinical indicators was performed, and the results were visualized using the R package 'survminer'. The nomogram/receiver operating characteristic (ROC) curve was visualized using the R packages 'rms' and 'pROC'. For transcriptome analyses, differentially expressed genes (DEGs) of high-/low-risk patients were identified with the following thresholds: |log2-fold change| > 1 and false discovery rate < 0.01. The enriched genes were evaluated with the R package 'clusterProfiler'[19]. Gene set enrichment analysis (GSEA) was implemented and visualized with the R packages 'GSEABase' and 'clusterProfiler'. For genome analyses, differentially mutated genes were identified with Fisher's exact test and visualized with 'GenVisR'[20]. Differential copy number analysis was performed with the Wilcoxon rank sum test, and the results were visualized with the 'ComplexHeatmap' R package.
Immune infiltration and drug response prediction
The TCGA transcriptome data were used for immune cell infiltration estimation. The TIMER2.0[21] website was used with default parameters, and algorithms including XCELL[22], EPIC[23], CIBERSORT[24], and QUANTISEQ[25] are available for use on this website. The differential abundance of cell types was assessed via student's t test and visualized with the R packages 'pheatmap', 'vioplot', and an in-house script. The response of each sample to different drugs was predicted with the algorithm 'oncopredict'[26] and compared with the level of immune infiltration.
Single-cell sequencing data processing
Processed single-cell sequencing data were downloaded from GSE178341[27]. Only tumor samples were retained, and samples retrieved from the same patients were regarded as different samples. After standard processing using Seurat as described in a previous study[28], the cell types were annotated. The R package 'ggalluvial' was used to create a Sankey diagram. The 'CellChat' package[29] was used for cell-cell interaction visualization, and the publicly available CellChatDB@Human list was used as a reference for ligand-receptor pairs.
RESULTS
Candidate signature genes
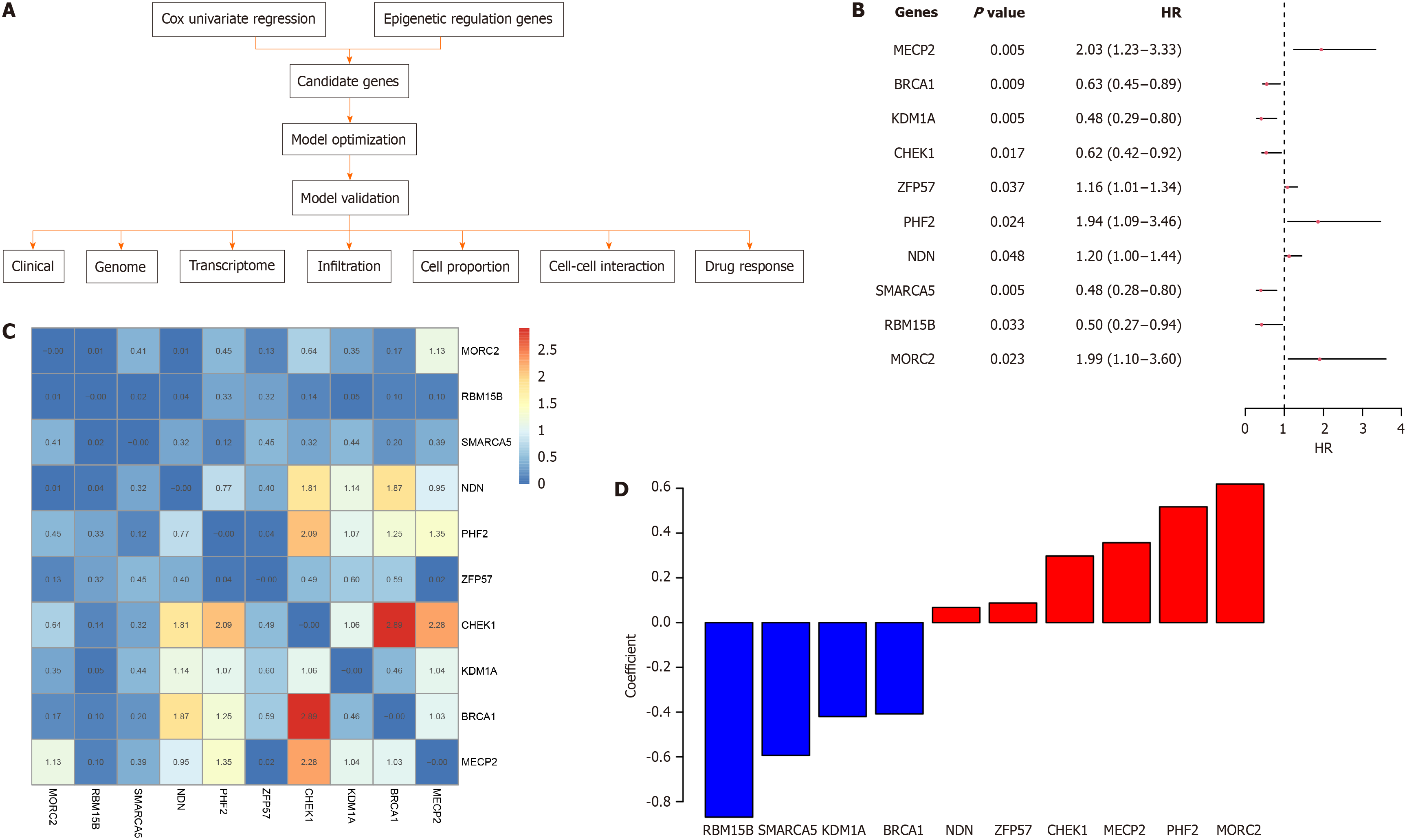
The schema of this study is depicted in Figure 1A. The 229 epigenetic regulation-related genes were retrieved from the MSigDB database, and Cox univariate regression was used to identify the survival-related genes in the TCGA-CRC cohort. A total of 21 survival-related genes were identified. To determine the optimal gene combination for signature construction, we considered all gene combinations with fewer than 11 genes. For each combination, a Cox multivariate regression model was developed, and samples were classified into low-/high-risk groups. Survival differences among the groups were compared, and the combination with the smallest P value and highest hazard ratio (HR) was identified as the optimized gene panel. This gene panel was comprised of 10 genes (MORC2, RBM15B, SMARCA5, NDN, PHF2, ZFP57, CHEK1, KDM1A, BRCA1, and MECP2). The HR and P values of these genes according to univariate regression are shown in Figure 1B. Most of the genes were not significantly correlated with one another (Figure 1C). The coefficients of the candidate genes are shown in Figure 1D.
Figure 1 Candidate genes for the signature.
A: Schema of this study; B: Hazard ratio, confidence interval, and P-value of each candidate gene; C: Cox univariate Correlation of the candidate genes, the number and color indicate the correlation P values of gene pairs; D: Coefficient of each gene in the model.
Signature performance in training datasets
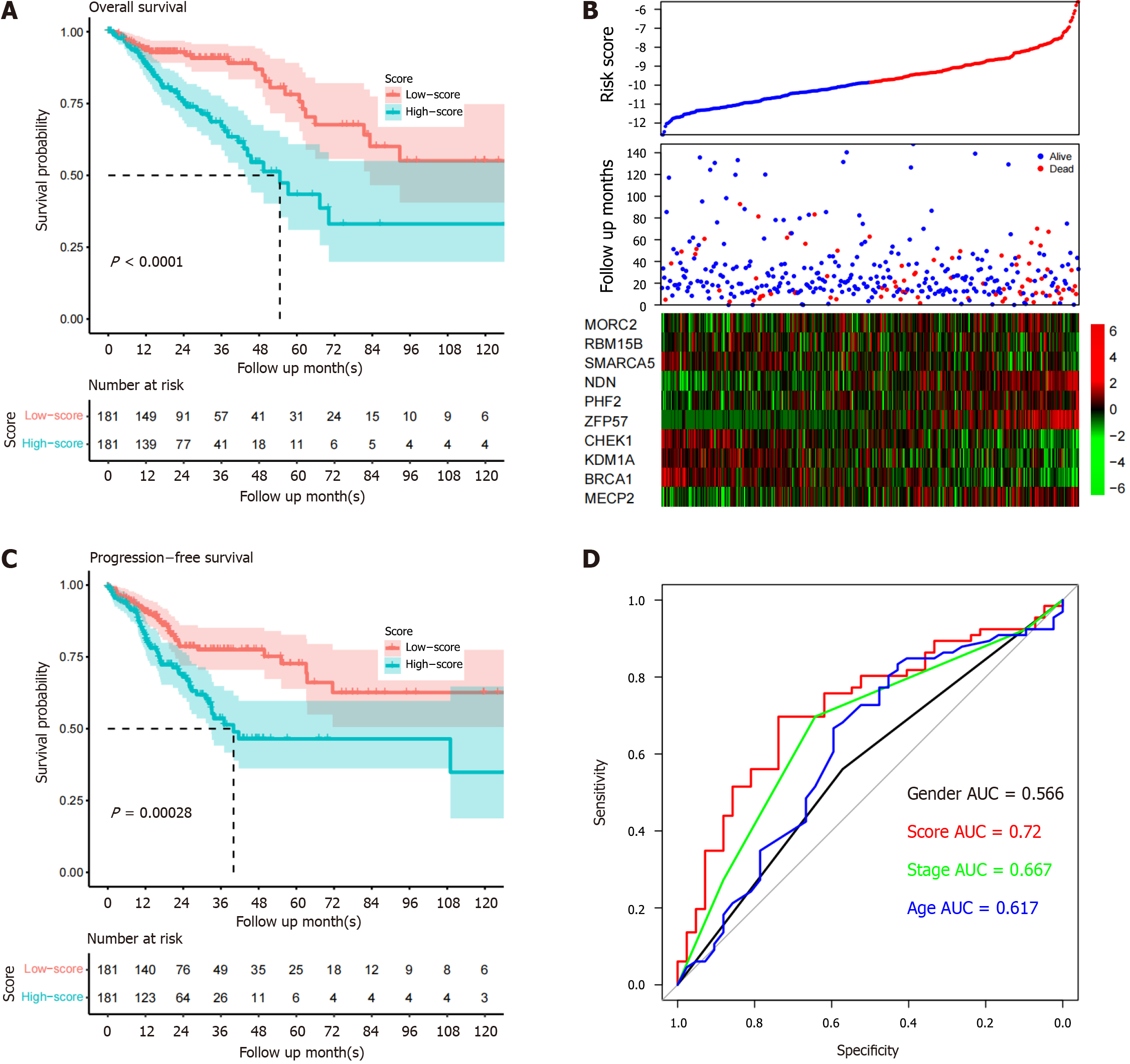
The performance of the signature was evaluated in the TCGA dataset. After separating the samples into high- and low-risk groups, the survival differences were assessed. The survival time of high-risk patients (median survival months: 43.2) was significantly shorter than that of low-risk patients (median survival months: 83.2, P < 0.0001; Figure 2A). The high-risk patients exhibited MECP2, ZFP57, PHF2, NDN, and MORC2 overexpression and low RBM15B, SMARCA5, CHEK1, KDM1A, and BRCA1 expression (Figure 2B). The difference in progression-free survival between the groups was also analyzed, and the trend resembled that of overall survival (Figure 2C). To evaluate the predictive power of the model, the area under the ROC curve (AUC) of the model for predicting five-year overall survival was compared with that of other clinical indicators. The AUCs of age, sex, TNM stage, and the signature were 0.617, 0.566, 0.667, and 0.720, respectively (Figure 2D). In conclusion, the signature is a powerful biomarker for predicting CRC patient survival in the training dataset.
Figure 2 Signature performance in the TCGA dataset.
A: Overall survival period of high-risk samples is significantly shorter compared to low-risk samples; B: Heatmap of candidate genes suggests that high-risk samples have high expression of certain genes; C: Progression-free survival of group samples is similar to the overall survival pattern; D: Five-year survival receiver operating characteristic curve of the model and other clinical indicators. AUC: Area under the curve.
Validation of signature performance across datasets
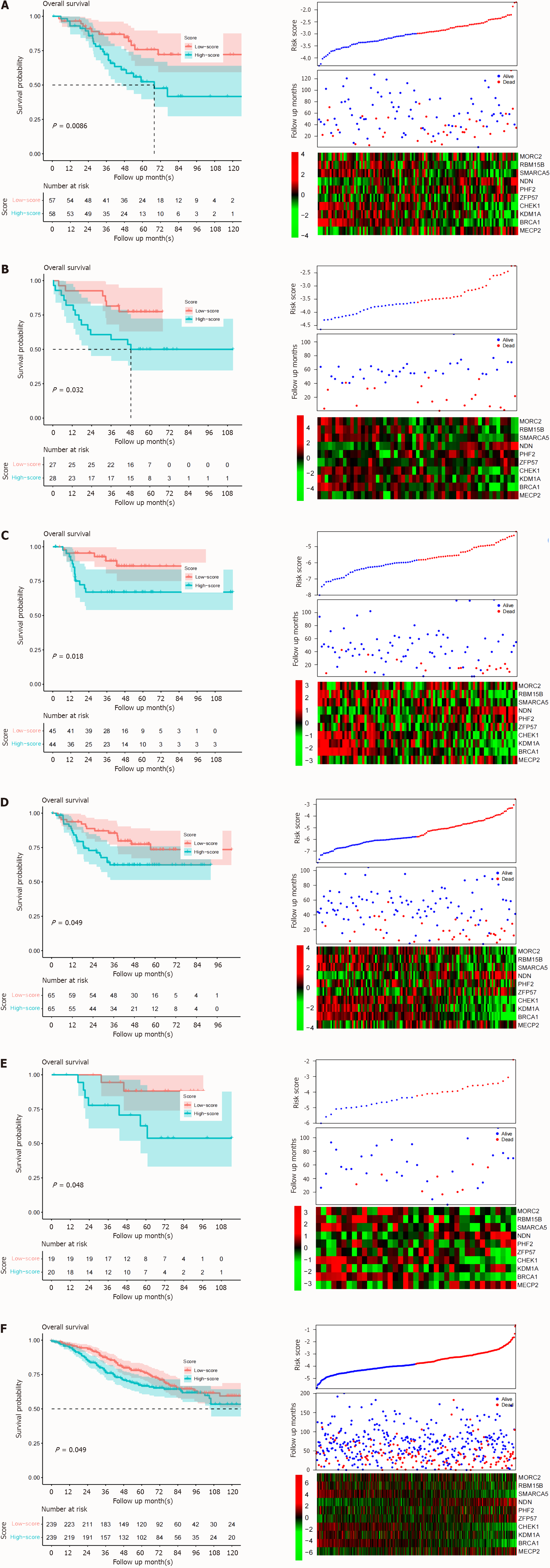
The high predictive value of the signature in the training dataset may have resulted from overfitting since the candidate genes and signatures were developed based on the training dataset, the TCGA-CRC dataset. Thus, the reproducibility of the signature was evaluated in six independent datasets-GSE17526, GSE17537, GSE33113, GSE37892, GSE39048, and GSE39582 from the GEO database. The risk scores of all the samples in all the datasets were calculated. The samples in each dataset were further divided into low- and high-risk groups with the same sample size. The differences in survival among the groups in each dataset were compared. As expected, in all these validation datasets, worse survival was observed in the high-risk group, while a prolonged survival period was detected in the low-risk group (Figure 3, top panel). In accordance with the TCGA dataset, the gene expression pattern of the validation datasets also showed a similar pattern (Figure 3, bottom panel), suggesting the reproducibility of the signature across datasets and platforms.
Figure 3 Signature verification.
The performance of the signature was assayed from GEO database. A: GSE17526; B: GSE17537; C: GSE33113; D: GSE37892; E: GSE39048; F: GSE39582. Similar to the training dataset, the upper panel is the survival curve, the middle is the follow-up information and sorted risk scores. The bottom panel is the candidate gene expression heatmap.
Clinical features associated with the signature
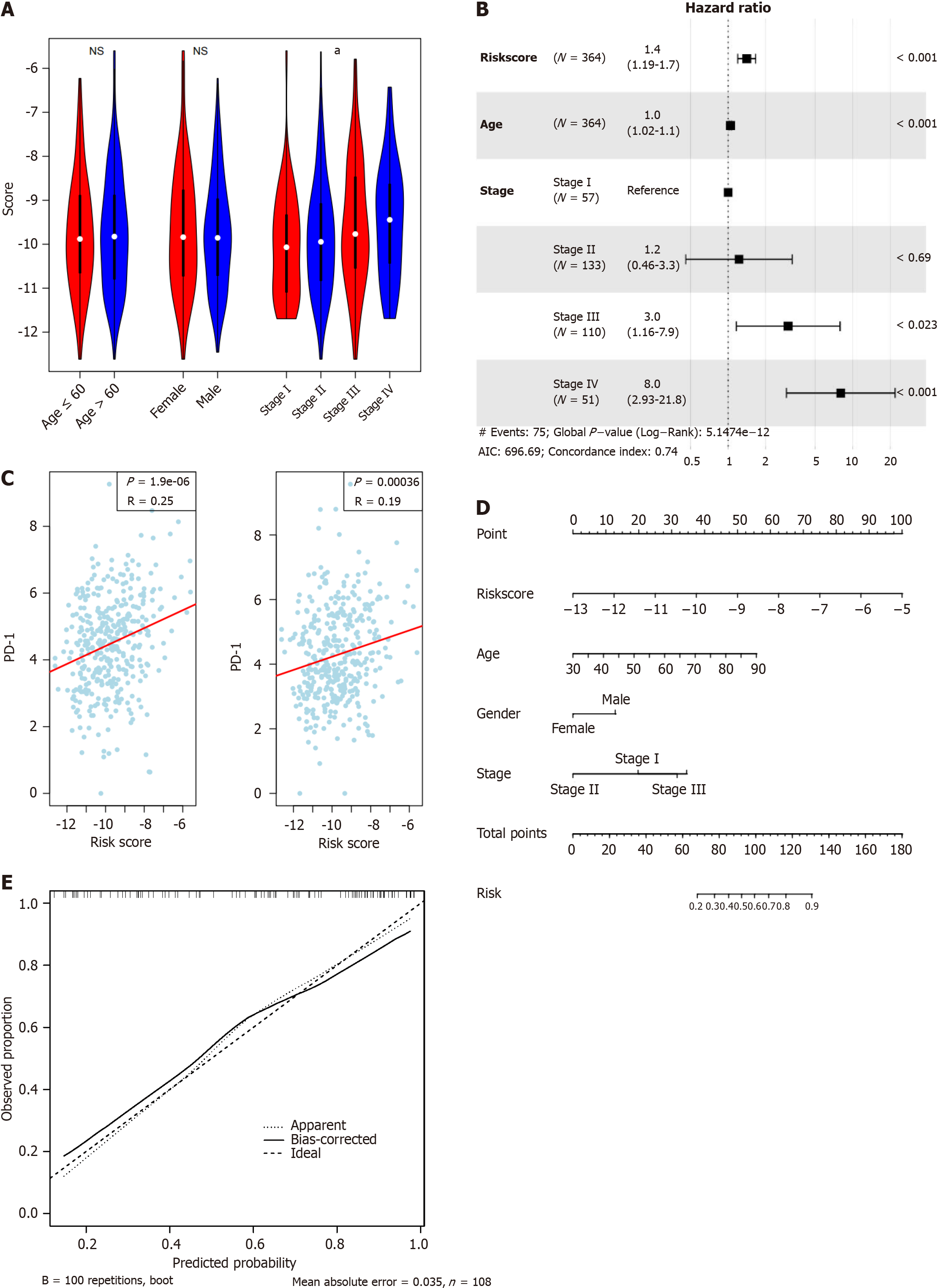
After investigating the performance and robustness of the signature, we also analyzed the relationships between the signature and clinical indicators in the TCGA dataset. As shown in Figure 4A, the signature was not correlated with age or sex but was significantly associated with TNM stage (ANOVA), with higher stages accompanied by higher risk values. Cox multivariate regression analysis demonstrated that the model was an independent predictor of overall survival in patients with CRC (Figure 4B). As an important clinical indicator, PD-1/PD-L1 expression is a biomarker for the microenvironment and the response to immunotherapy. As shown in Figure 4C, the signature was positively and significantly associated with PD-1 and PD-L1 gene expression (P = 1.9e-6 and 3.6e-4, respectively). To simplify the clinical use of the signature, a nomogram for predicting five-year survival was generated (Figure 4D) with a calibration curve (Figure 4E). The signature contribution ranged from 0 to 100 points, which spanned a larger range than other clinical indicators. The results above indicate that the model is a valuable biomarker and serves as an independent indicator for CRC.
Figure 4 Clinical association of the signature.
A: The signature is statistically correlated with TNM stage, but not age and gender; B: Cox multivariate regression of the signature and clinical indicators suggests that the signature is an independent indicator for survival; C: The signature is positively associated with both PD-1 and PD-L1 gene expression; D: Nomogram for five-year survival; E: Calibrate curve revealed the signature is a valuable marker for predicting survival of colorectal cancer. aP < 0.05; cP < 0.001.
Genomic and transcriptomic features of the signature
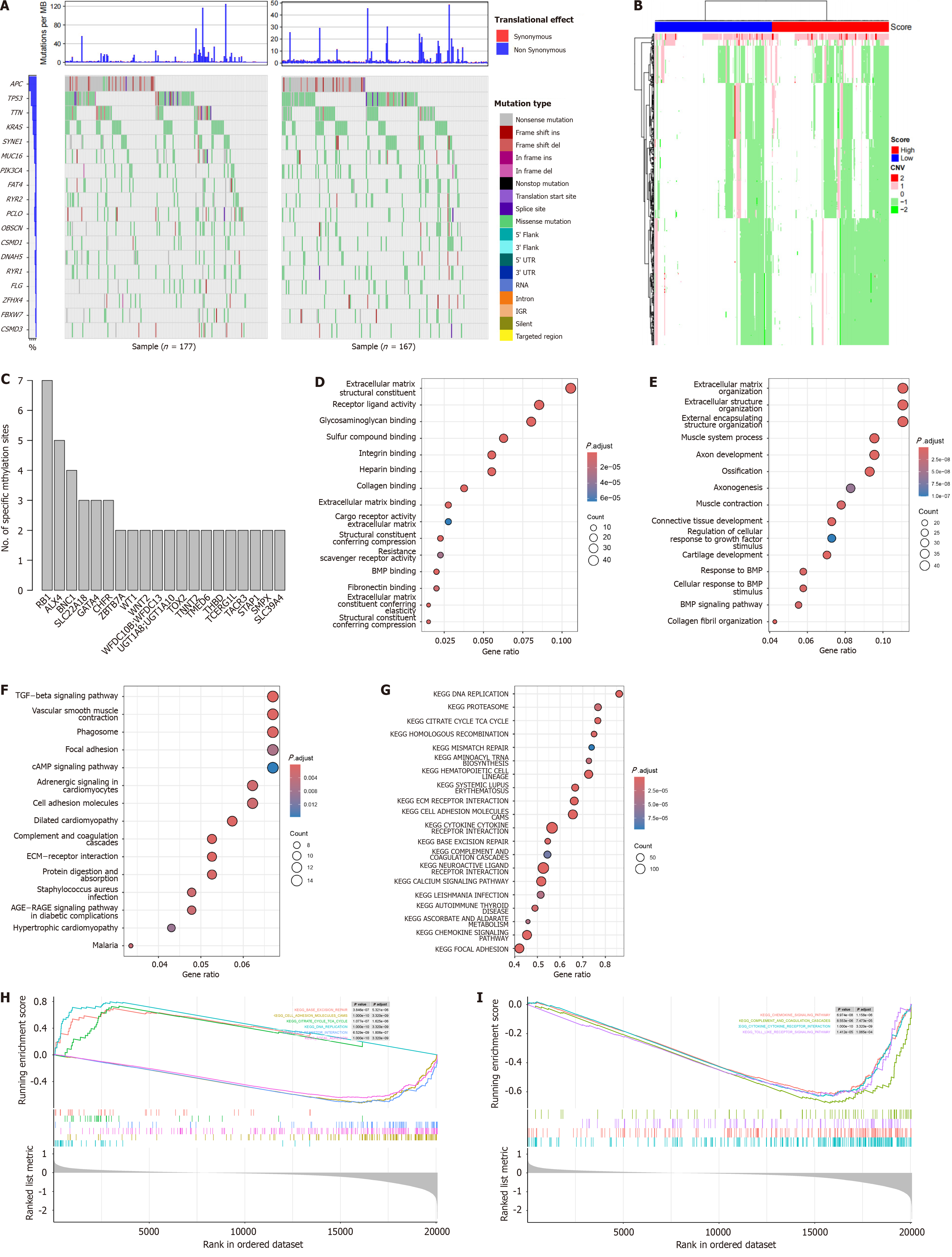
Next, the relationships between the signature and genomic/epigenetic/transcriptomic features were evaluated. Despite the fact that the mutation patterns at the gene level of the groups were similar (Figure 5A), the differential copy number analyses revealed that the high-risk group had a significantly greater proportion of samples with genomic amplification and deletion than did the low-risk group (Figure 5B), suggesting genome instability in the high-risk group. Differential DNA methylation CpG sites were also identified, and these sites were enriched in RB1, ALX4, BNC1, SLC22A18, GATA4, and CHFR (Figure 5C). DEGs between groups were identified, and enriched molecular functions, biological processes, and Kyoto Encyclopedia of Genes and Genomes pathways were involved in multiple processes that are correlated with carcinogenesis and CRC development (Figure 5D-F). GSEA was also performed (Figure 5G). In addition to canonical cancer-related pathways (Figure 5H, DNA replication, the tricarboxylic acid cycle cycle, DNA repair, etc.), immune-related pathways including chemokine, complement and coagulation cascades, and cytokine-cytokine interaction pathways were also differentially enriched (Figure 5I). Overall, the signature revealed the genomic, epigenetic, and transcriptomic features of CRC.
Figure 5 Genome and transcriptome feature of the signature.
A: Mutational landscape of the high and low-risk samples; B: Differential copy number variation was shown; C: Enriched differentially methylated sites across genes were visualized; D: Gene Ontology analyses of differentially expressed genes in molecular function; E: Biological process; F: Kyoto Encyclopedia of Genes and Genomes pathways were shown; G: Gene set enrichment analysis revealed that the enriched pathways; H: Canonical cancer-related pathways; I: Immune-related pathways.
Infiltrating immune cell and the signature
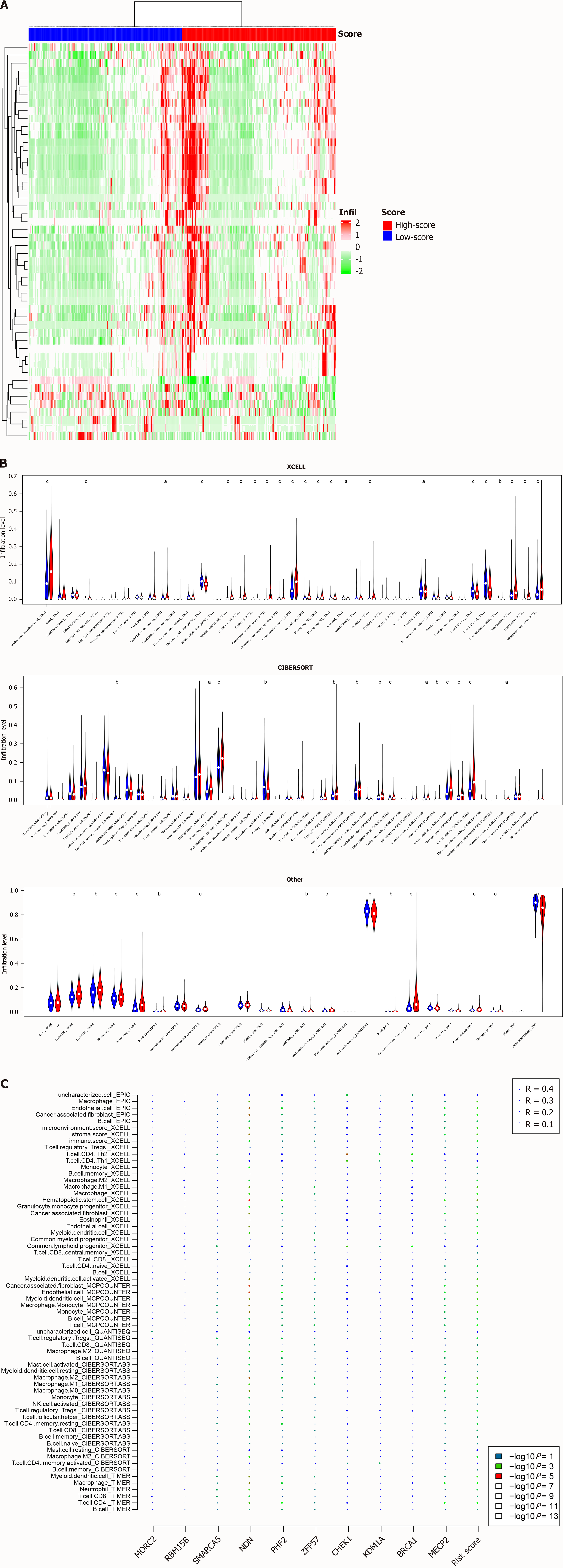
GSEA showed that the signature was correlated with immune pathways, which prompted us to investigate whether the signature reflected the immune cell infiltration status. In the TCGA dataset, the relative abundance of immune cells was estimated with several algorithms, including XCELL, CIBERSORT, EPIC, and TIMER. Significantly differentially infiltrated immune cell types were identified. As shown in Figure 6A, each group had a unique infiltration pattern; some were highly infiltrated in the high-risk group, while others were more abundant in the low-risk group. For example, macrophages (M0/M1/M2), naïve CD4+ T cells, and cancer-associated fibroblasts were highly infiltrated in samples from the high-risk group, while NK T cells and Th1 and Th2 cells were significantly more abundant in samples from the low-risk group (Figure 6B). Although different algorithms may have different prediction results, a high proportion of immune cells showed significant differences in infiltration levels between groups. The relationships among the candidate genes, signature score, and the infiltration of immune cell types are visualized in Figure 6C. Most candidate genes were not significantly associated with the infiltration levels of immune cells, while the signature score was, highlighting the advantage of a signature based on multiple genes.
Figure 6 Immune abundance and the signature.
A: Differentially infiltrated cell types in TCGA dataset were identified; B: A high proportion of cell types according to different algorithms were significantly infiltrated between high and low-risk groups; C: correlation analyses revealed that the correlation was contributed by several genes. aP < 0.05, bP < 0.01, cP < 0.001.
Immune landscape and the signature
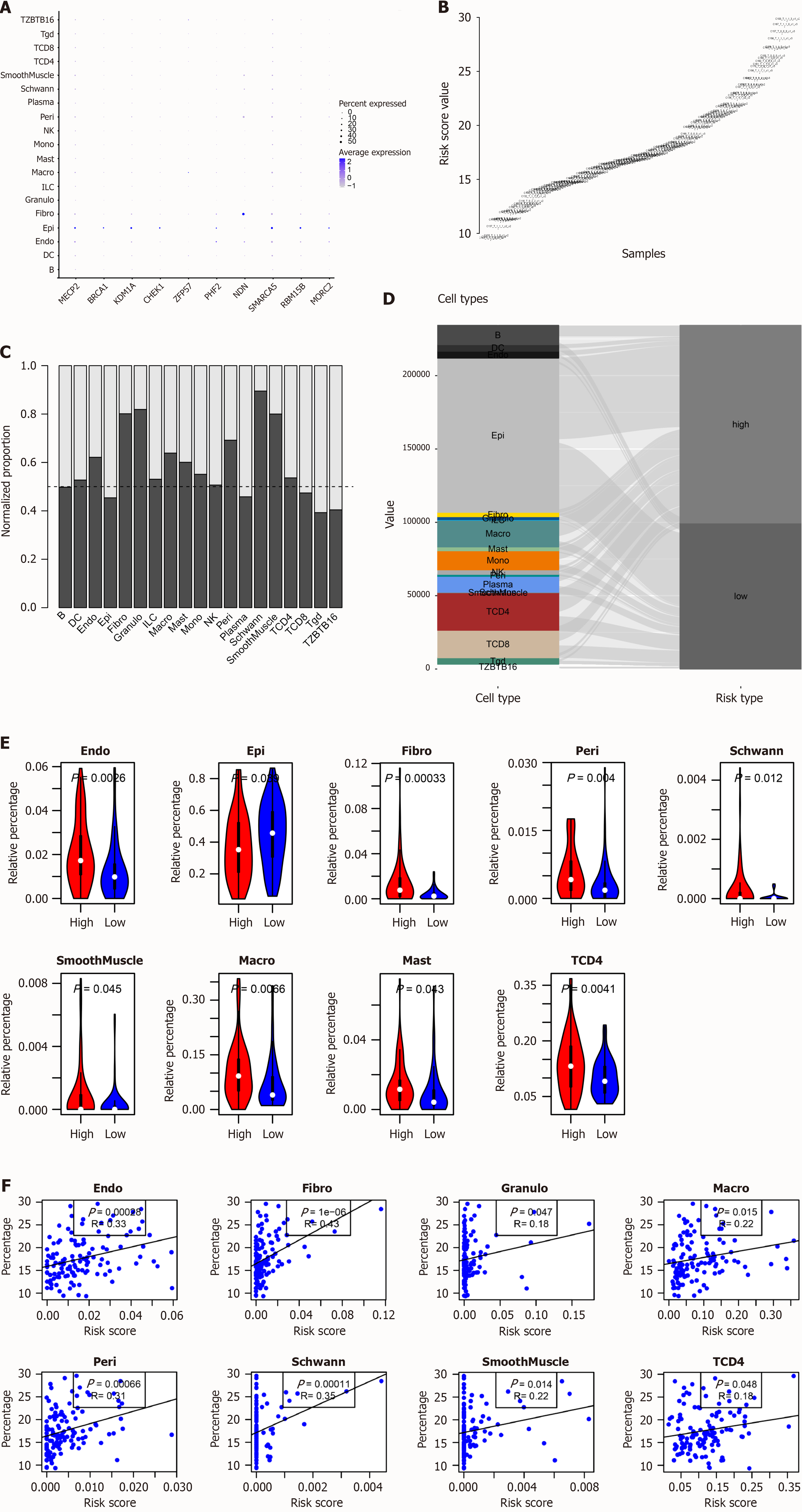
The abundance of infiltrating immune cells was estimated based on the specific biomarkers in each immune cell type, but single-cell sequencing comprehensively revealed the immune infiltration status. Thus, the single-cell sequencing dataset GSE178341, consisting of samples from 62 patients, was used for further analyses. The samples were processed using standard procedures as described in previous reports. The candidate genes were enriched in basal cells, including fibroblasts and epithelial and endothelial cells, but not immune cells (Figure 7A). To evaluate the risk score, pseudo-bulk RNA-seq analysis was implemented, and the data were transformed to the same format of the TCGA dataset. The risk scores of each sample were calculated (Figure 7B), and the samples were also equally divided into high- and low-risk groups. Despite both groups having all cell types, the proportion (normalized according to the cell number) was different between risk groups for some cell types (Figure 7C and D). According to consistent estimation algorithms used in the TCGA dataset, a significantly greater proportion of CD4+ T cells and macrophages were observed in samples of high-risk patients (Figure 7E). In addition, mast cells, endothelial cells, fibroblasts, pericytes, and other cells were also significantly more abundant in the high-risk group, while epithelial cells were significantly more abundant in the low-risk group. The Pearson correlation between the risk score and the proportion of these cells showed a similar pattern (Figure 7F). In summary, the single-cell sequencing results suggested that the signature reflects the microenvironmental heterogeneity of CRC.
Figure 7 Single cell infiltration of the signature.
A: The candidate genes were mainly expressed in ‘basal’ cells; B: After calculating risk scores of each sample, the normalized proportion of high/low-risk groups was visualized; C: A bar plot; D: Sankey diagram; E: The signature is significantly associated with several cell types according to the single cell sequencing data by dividing high and low-risk groups; F: The signature is significantly associated with several cell types according to the single cell sequencing data by Pearson correlation.
Cell-cell interaction and the signature
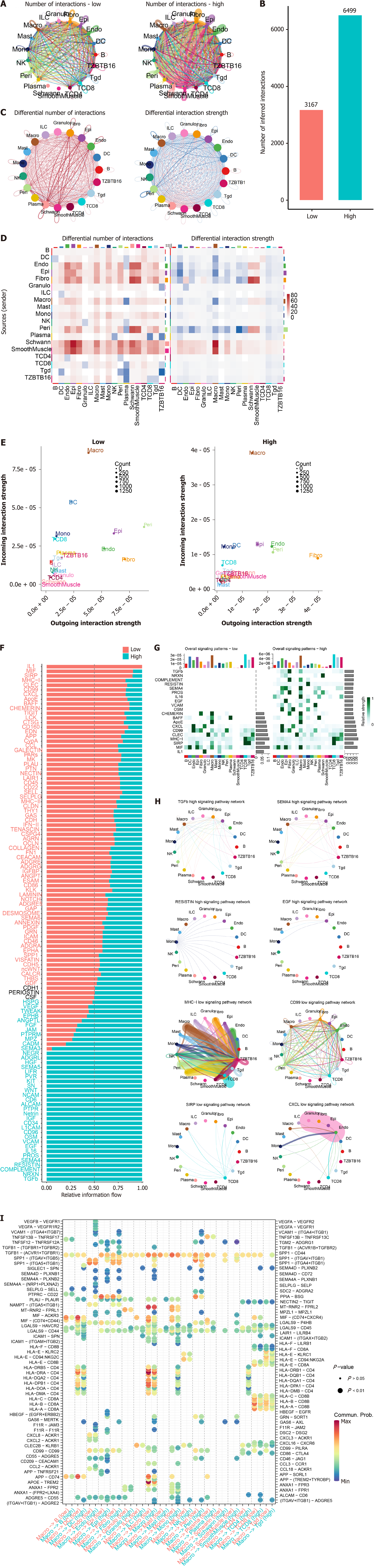
As mentioned above, the signature reflected immune cell abundance, which prompted us to analyze whether the signature reflected the heterogeneity of cell-cell interactions in CRC. Overall, the interaction patterns of the samples from the low- and high-risk groups were similar (Figure 8A), but the number of cell-cell interactions was greater in the high-risk group (Figure 8A and B). Differential interaction revealed that although the interaction number of high-risk group was greater (especially between basal cells such as fibroblasts, epithelial cells, and endothelial cells), the interaction strength decreased (Figure 8C and D). Notably, macrophage interactions were enriched in pathways that were specifically related to the high-risk group (Figure 8E-H). The interactions between macrophages and other cell types at the pathway level were compared (Figure 8I), and most pathways differed. In summary, the signature revealed heterogeneity in cell-cell interactions, especially interactions between basal cells and macrophages.
Figure 8 Cell-cell interaction and the signature.
A: Although most interactions exist in both low and high-risk samples; B: The interaction number is significantly different; C and D: Despite that the interaction number is higher in high-risk samples, the interaction strength is lower; E: The macrophage is noticed as the most significantly altered immune cell among the interactions; F: The pathway level interaction information flow was visualized for both groups, overall; G: Top 10 for each group; H: Among these interactions, it is noticed that macrophage is especially active in high-risk samples while interactions involved CD8+ T cell were increased in the low-risk group; I: Specific pathways and cell type interaction landscape also revealed that macrophage is activated in high-risk samples.
Predicted drug response and the signature
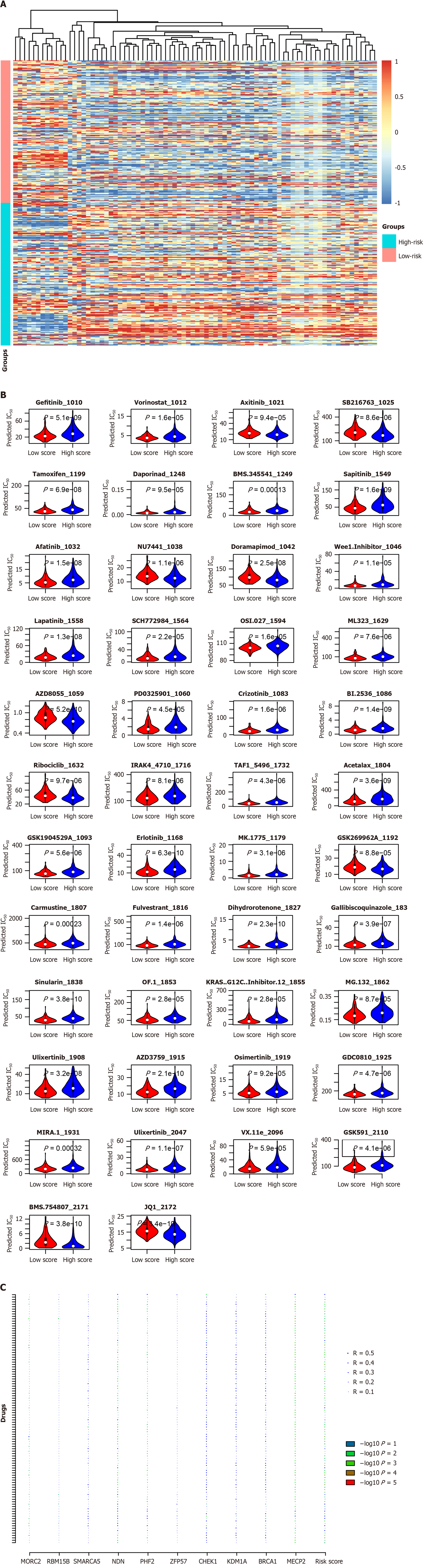
As described above, the signature reflects the multiple features of CRC, and these statuses are correlated with drug response. Therefore, the correlation between the signature and drug sensitivity was evaluated. In the TCGA dataset, the predicted IC50 values of drugs for each sample were calculated, and the correlation between the signature score and drug sensitivity was assessed. As shown in Figure 9A, low-risk patients were significantly more sensitive to most drugs, while high-risk patients were more sensitive to some other drugs. For example, the KRAS G12C inhibitor and afatinib was predicted to benefit low-risk patients, while AZD8055 and ribociclib were predicted to be more effective for high-risk patients (Figure 9B). The correlations among candidate genes, signatures, and IC50 values were analyzed. The results revealed that NDN and MECP2 contribute to drug sensitivity (Figure 9C). Collectively, these results indicate that the signature may be used as a potential biomarker for predicting the treatment response of CRC.
Figure 9 Drug response and the signature.
A: Drugs with different predicted IC50 values were identified and visualized with a heatmap; B: Detailed information was showed with vioplot; C: The drug response prediction was mostly contributed by NDN, PHF2, and MECP2 according to the correlation analyses.
DISCUSSION
Among the candidate genes, MORC2 was reported to bind -446 to -213 bp of the NDRG1 promoter and inhibit its transcription to promote the development of CRC[30]. The gene expression level of NDN, which is regulated by DNA methylation, was reported to activate the WNT signaling pathway and thus facilitate the cell cycle in CRC[31]. PHF2 was shown to facilitate p53-mediated cell death in cancers, including CRC and gastric cancer[32]. As a member of the zinc finger family of transcription factors, ZNF57 was reported to promote distant metastasis in CRC but not lymphatic metastasis in CRC[33]. Antona et al[34] reported that disrupting the gene expression of KDM1A alters the phenotypes of CRC, including carcinogenesis, stemness, and apoptosis. Similarly, MECP2 depletion also changes the progression phenotypes of CRC by binding to SPI1 and facilitating metastasis and stemness[35]. The other genes, including SMARCA5[36], RBM15B[37], BRCA1[38] and CHEK1[39], have also been reported to be important genes for the development of cancers, including CRC. These results suggest that the identified candidate genes all have experimentally validated functions in CRC, which explains why the model is reproducible and effective for predicting the clinical outcomes of CRC patients.
According to the single-cell sequencing dataset, the candidate genes were mainly expressed in fibroblasts, epithelial cells, and endothelial cells, which are basal cells, not immune cells (T cells or B cells), suggesting that the model is a reflection of CRC cells rather than other cell types. On the other hand, NDN, PHF2, and MECP2 contributed to immune infiltration. The prediction of the drug response was mainly influenced by NDN, PHF2, and MECP2, suggesting the important roles of these genes in the model. According to the gene expression correlation analyses, most genes were not significantly differentially expressed, suggesting that the model integrated complementary information from these genes. Therefore, it is suspected that other genes may reflect the biological status of CRC from other aspects.
In infiltration analyses based on bulk RNA-seq data from the TCGA dataset, most algorithms indicated that the high-risk group had a significantly greater percentage of macrophages, including macrophages, M1 macrophages and M2 macrophages, than did the low-risk group. However, according to QUANTISEQ, the levels of M1 macrophages were not significantly different between the groups, which may be due to the different biomarkers used for estimation. Single-cell sequencing data also revealed that samples from high-risk patients had a greater proportion of macrophages than samples from low-risk patients. M1 macrophages are engaged in proinflammatory processes, but M2 macrophages promote anti-inflammatory processes[40]. A high level of anti-inflammatory M2 macrophages explains the poor prognosis of patients in the high-risk group. Consistently, the most cell-cell interaction network differed between the low/high-risk groups. However, it is unclear why M1 macrophage levels also increase. We suspect that the M1/M2 ratio may be the key factor.
The main limitation of this study is that the sample data were retrospectively collected, and clinical and treatment information was missing, which made comprehensive analysis of the model unfeasible. Another limitation is that the drug response values (IC50 values) were predicted according to an algorithm, which makes the data less persuasive. Despite this, this study provides a novel signature and insight into the prognosis of CRC. Despite this limitation, the results were validated in seven different datasets, indicating that the signature is robust across centers and platforms rather than being a false-positive. Prospectively studies mostly have rigorous restrictions in terms of enrollment. We believe that the model can be further validated in prospective studies. Although the model was robust across datasets, the prognostic value of the signature differed. One of the points for improving performance is to use a standardized platform, in which case a training set would be used to determine the optimized cutoff value. In this study, different cohorts had different cutoff values (the median risk score value for each cohort) due to the difference in data types (signal intensity in microarrays or normalized count values in RNA-seq). If drug response data are available, we can also evaluate the value of the signature in predicting the response to therapeutic regimens. Thus, in our future work, we will prospectively collect more samples to further evaluate the potential clinical utility of the signature.