Published online Jul 15, 2024. doi: 10.4251/wjgo.v16.i7.2902
Revised: April 4, 2024
Accepted: May 21, 2024
Published online: July 15, 2024
Processing time: 176 Days and 6.6 Hours
Pancreatic ductal adenocarcinoma (PDAC) presents significant challenges in patient management due to a dismal prognosis, increasing incidence, and limited treatment options. In this regard, precision medicine, which personalizes treatments based on tumour molecular characteristics, has gained great interest. However, its widespread implementation is not fully endorsed in current recommendations. This review explores key molecular alterations in PDAC, while emphasizing differences between KRAS-mutated and KRAS-wild-type tumours. It assesses the practical application of precision medicine in clinical settings and outlines potential future directions with respect to PDAC. Actionable molecular targets are examined with the aim of enhancing our understanding of PDAC molecular biology. Insights from this analysis may contribute to a more refined and personalized approach to pancreatic cancer treatment, ultimately improving patient outcomes.
Core Tip: Management of patients with pancreatic cancer (PDAC) is a real challenge due to a poor prognosis, a rising incidence and few therapeutic options. Precision medicine is an approach that seeks to personalize treatments to the specific molecular characteristics of individual tumors. Despite a growing interest in applying molecular precision medicine for PDAC management, large scale precision medicine is not endorsed so far by the last recommendations. We review the main actionable molecular alterations found in PDAC, highlighting the differences between KRAS-mutated and KRAS-wild-type tumors, as well as precision medicine in clinical practice and future directions for precision medicine in PDAC.
- Citation: Boileve A, Smolenschi C, Lambert A, Boige V, Tarabay A, Valery M, Fuerea A, Pudlarz T, Conroy T, Hollebecque A, Ducreux M. Role of molecular biology in the management of pancreatic cancer. World J Gastrointest Oncol 2024; 16(7): 2902-2914
- URL: https://www.wjgnet.com/1948-5204/full/v16/i7/2902.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i7.2902
Pancreatic ductal adenocarcinoma (PDAC) management is challenging due to its very aggressive nature that, combined with limited therapeutic options, leads to poor overall survival (OS)[1,2]. For patients with PDAC, there is clearly an unmet need for the development of innovative strategies.
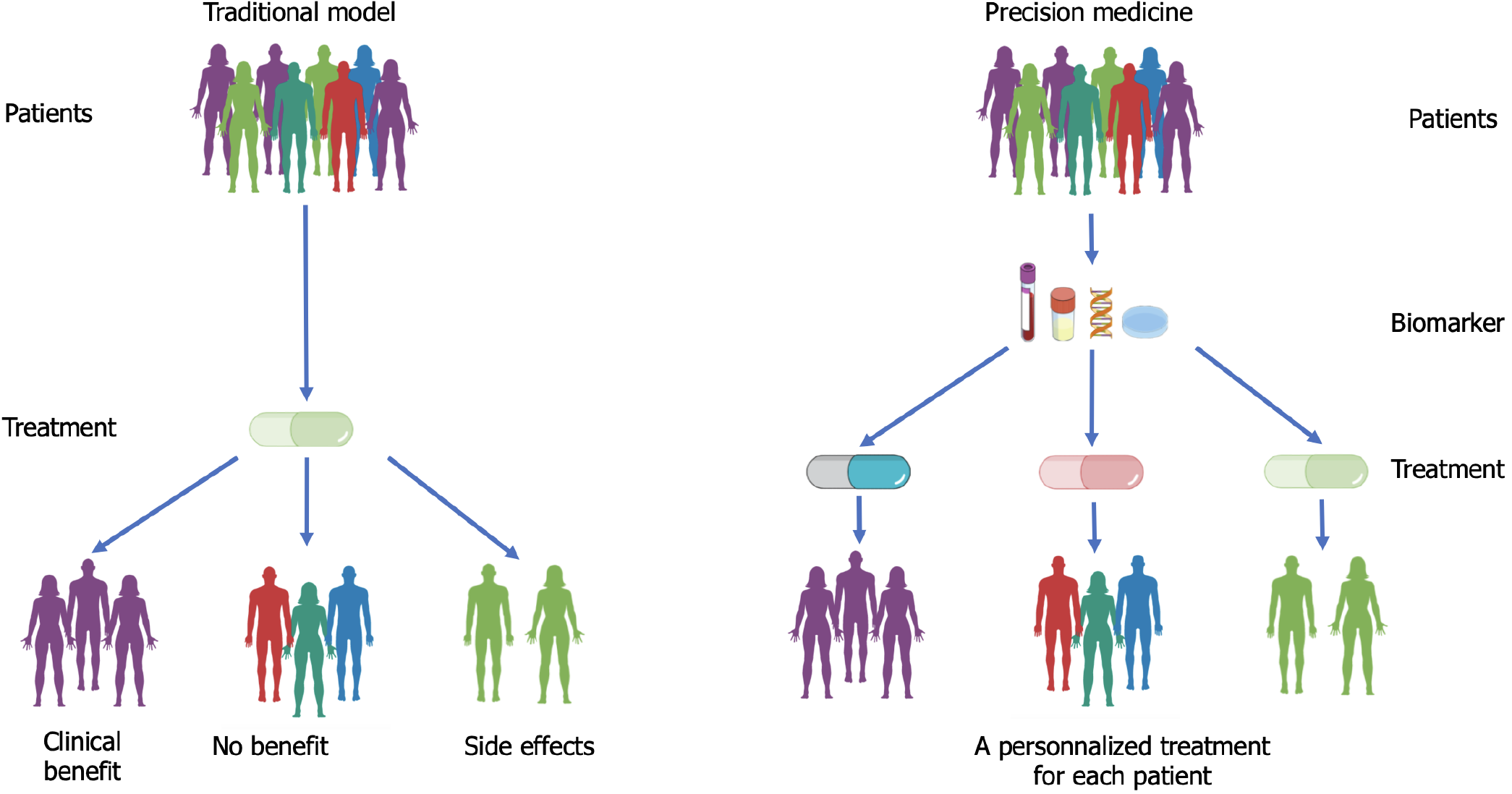
Over the years, the cancer treatment landscape has been evolving, with the emergence of a promising approach based on molecular precision medicine[3] (Figure 1). This approach seeks to personalize treatments to the specific genetic and/or molecular characteristics of individual tumours, offering the potential to improve treatment outcomes and enhance patient survival[4]. Recently, several prospective studies have been conducted to assess the feasibility and effectiveness of precision medicine through next-generation sequencing (NGS) in patients with various solid cancers[5-10]. It is worth noting that only 10% to 25% of patients in these studies received specific therapy based on molecular profiling, and few have as yet truly benefited clinically from this approach.
Today, precision medicine is now fully integrated into the management strategy for several types of tumours, including lung cancer[11]. For PDAC however, results with molecular precision medicine are controversial and despite the growing interest, its large scale application is discouraged in the latest recommendations[12,13]. Moreover, implementation of molecular precision medicine for PDAC remains challenging, due to the vast variety of KRAS alterations and the rapidly progressive nature of the disease[14]. In this review, we will explore the evolving landscape of molecular precision medicine in PDAC, highlighting the main molecular alterations found in PDAC, along with the molecularly-matched therapies and their potential benefit, and finally the remaining challenges and future directions.
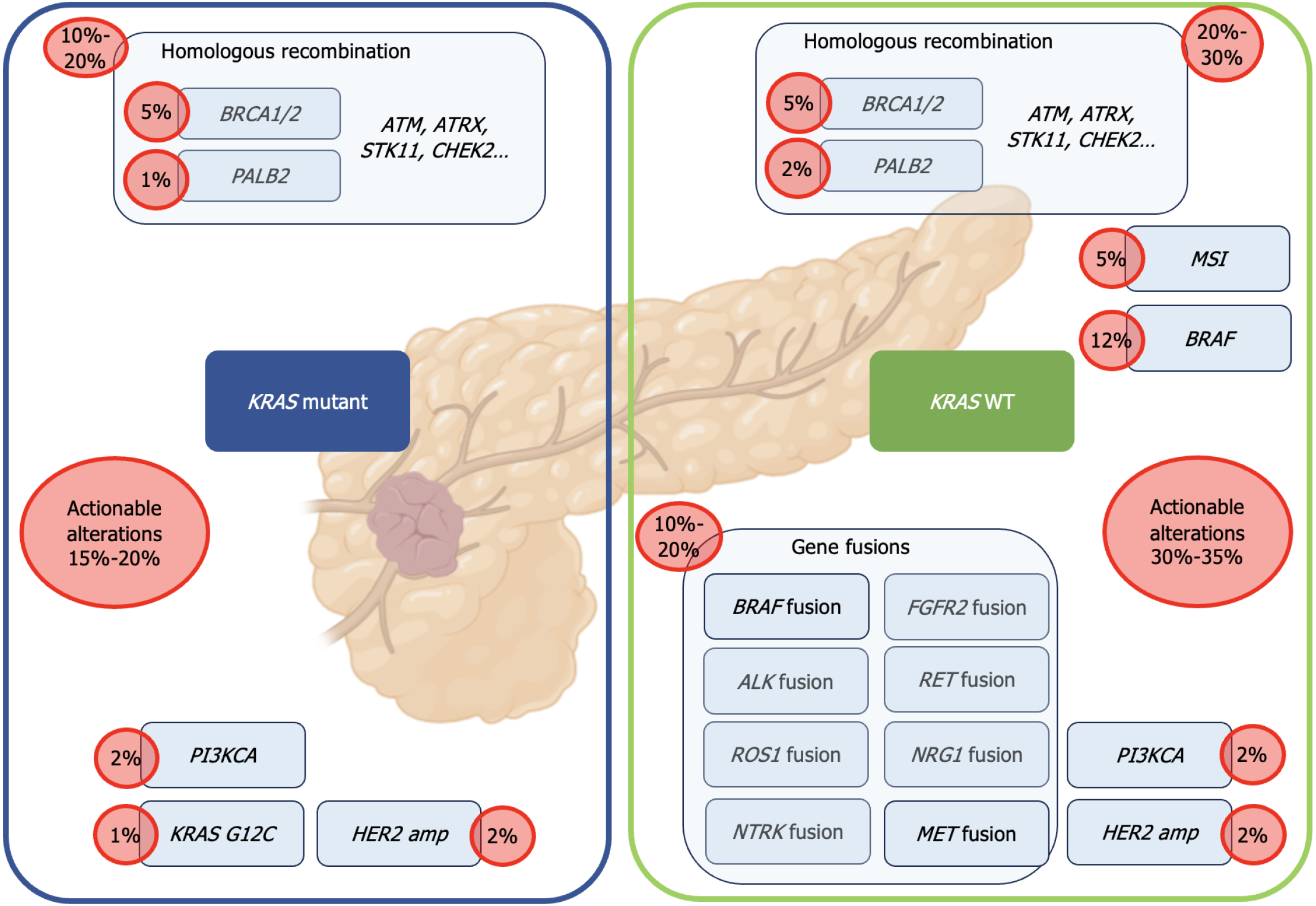
Molecular profiling in PDAC reveals the presence of several key genetic alterations involved in carcinogenesis, primarily in four genes: KRAS, TP53, SMAD4, and CDKN2A[14]. With the exception of the KRAS G12C mutation, which is present in less than 1%-2% of PDAC cases[15], none of these genetic alterations as yet have validated targeted therapies. However, several molecular profiling studies have demonstrated that up to 25% (ranging from 12% to 25%) of PDACs harbour actionable molecular alterations[14,16-21] (Figure 2). "Actionability" of a particular molecular alteration is defined as strong clinical or preclinical evidence of the predictive benefit from a specific therapy (in any type of cancer), as measured by the European Society for Medical Oncology Scale for Clinical Actionability of molecular Targets (ESCAT) classification[22] with ESCAT I to III being considered as actionable.
The main actionable alterations primarily involve the DNA damage response and repair pathways (BRCA1, BRCA2, PALB2, ATM), targeted by platinum agents or poly (ADP-ribose) polymerase (PARP) inhibitors. The randomized phase III Polo study demonstrated a significant improvement in progression-free survival on platinum-based chemotherapy for patients with germline BRCA1/2-mutation and metastatic PDAC receiving the PARP inhibitor Olaparib[12,23]. Consequently, guidelines now recommend the testing of PDAC patients for pathogenic germline alterations in BRCA1 and BRCA2. To date, olaparib remains the only approved molecularly-targeted therapy in PDAC.
Clinical benefits from and significant responses to biomarker-based tumour agnostic therapies have also been reported for PDAC. These include immune checkpoint inhibitors for high microsatellite instability tumours[24,25] or neurotrophic tropomyosin receptor kinase (NTRK) inhibitors for NTRK1, NTRK2, NTRK3, or ROS1 fusions[26,27]. Additionally, patients with PDAC carrying BRAFV600E mutations have been shown to benefit from treatment with a RAF-MEK targeted therapy[28] or, in the case of FGFR2 fusion, from FGFR2 inhibitors[29]. Durable responses to zenocutuzumab were also observed in cases of PDAC with NRG1 fusion[30]. However, while the anti-HER2 antibody-drug conjugate trastuzumab-deruxtecan is effective in a very large range of tumours with HER2 amplification, results are disappointing in PDAC with only 4% objective responses and 36% of tumours showing controlled progression at 12 wk[31]. The major actionable molecular alterations found in PDAC are summarized in Table 1 along with their current ESCAT classification.
| Biomarker | Targeted therapy | ESCAT classification | Ref. |
| BRCA1, BRCA2 germinal | Olaparib | IA | [87] |
| BRCA1, BRCA2 germinal/somatic | Rucaparib | IB | [86,88] |
| PALB2 germinal/somatic | Rucaparib | IB | [88] |
| NTRK fusions | Larotrectinib, entrectinib | IC | [27,89] |
| MSI high | Pembrolizumab | IC | [25] |
| RET fusions | Pralseltinib, selpercatinib | IC | [90,91] |
| NRG1 fusions | Zenocutuzumab | IIB | [30] |
| KRASG12C | Sotorasib, adagrasib | IIB | [55,56] |
| FGFR2 Fusions | Pemigatinib, infigratinib, futibatinib | IIIA | [92,93] |
| PIK3CA mutations | Alpelisib, buparlisib | IIIA | [94,95] |
| BRAFV600Emutation | Dabrafenib + trametinib, Binimetinib + encorafenib | IIIA | [96,97] |
| ALK/ROS1 fusions | Crizotinib, ceritinib, alectinib | IIIA | [98-100] |
| ERBB2 amplification | Trastuzumab + pertuzumab, T-DM1 | IIIA | [101,102] |
Improved overall response rates, progression-free survival, and median OS have been observed in metastatic PDAC patients with actionable molecular alterations treated with corresponding molecularly-matched treatment, compared (though not randomized in most cases) to those not receiving targeted therapy[20,21,32,33]. However, it remains to be demonstrated whether molecular profiling could improve therapeutic decision-making in PDAC patients.
Several prospective studies have attempted to assess the feasibility and clinical benefits of molecular precision medicine in patients with various advanced solid tumours[5-8]. A wide range of actionable molecular alterations has been described for most tumour types, and are found in 25% to 90% of patients[5-8]. However, only 10% to 25% of patients in these studies received molecularly-matched treatment based on precision medicine[5-10]. In PDAC, while around 25% of tumours were found with actionable targets[20,21,34], the number of patients actually receiving a molecularly-matched treatment could be as low as 5%[21]. Additionally, two clinical trials designed to evaluate precision medicine strategies (across all solid tumour types) showed controversial results[5,6]. Nevertheless, one prospective randomized precision medicine trial conducted in patients with breast cancer did report improved progression-free survival obtained from administering targeted therapies according to genomic data but only when molecular alterations were classified as level I/II according to the ESCAT scale [(Hazard Ratio (HR) 0.41, P < 0.001][8]. This has been confirmed in the data from the precision medicine program at Gustave Roussy[35].
In their retrospective study, Pishvaian et al[21] reported the OS results of the Know Your Tumour program for pancreatic cancers and found a positive impact on OS of molecularly-matched treatments, when compared to patients with an actionable molecular alteration who did not receive the corresponding targeted therapy (2.58 years vs 1.51 years). Similarly, the median OS was better when compared to that of patients without actionable molecular alterations who received standard-of-care treatment (2.58 years vs 1.32 years, HR = 0.34, P < 0.0001). There was no significant difference in OS between patients with actionable molecular alterations who did not receive the corresponding targeted therapy and those without actionable molecular alterations who received standard-of-care treatment (HR = 0.82, P = 0.10).
A similar study conducted in a reference tertiary centre involved 28% of patients with an actionable molecular alteration, 41% of whom (only 10% of the whole cohort) received a molecularly-matched treatment[36]. While no significant improvement in survival was noted, the number of treated patients was limited. In specific subgroups (Microsatellite instability[24] or FGFR2[29]), a significant improvement in survival was however noted. Thus, while PDAC may not as yet have benefited greatly from the advent of precision medicine, efforts are ongoing to improve the prognosis of patients with actionable molecular alterations. Remaining challenges are the rapid progression of the disease along with general state degradation, the vast proportion of tumours having powerfully oncogenic and largely undruggable KRAS mutations, and the large range of different actionable molecular alterations for which no Food and Drug Administration or European Medicines Agency-approved drug yet exists.
For an optimal implementation of precision medicine in routine healthcare, several points need to be considered.
First, the increasing number of molecular alterations identified in tumours, including PDAC, amplifies the challenge of their clinical interpretation. The actionability of any given molecular alteration evolves with clinical and preclinical evidence. To facilitate the interpretation of these data, a Molecular Tumour Board (MTB) has now been organized in some hospitals[37]. This MTB brings together clinicians, geneticists, molecular pathologists, genetic counsellors, bioinformaticians, and even researchers[38]. It reviews the patient's clinical, pathological, and molecular results and makes recommendations for targeted treatment options. It also classifies alterations according to ESMO recommendations using the ESCAT classification[22] and may guide the patient toward inclusion in precision medicine clinical trials.
Second, tumour-based molecular profiling can reveal incidental germline variants that are clinically significant in up to 12% of patients[39,40]. This can raise significant challenges to physicians who may not be prepared to handle these incidental findings, which can have significant implications for the patient's relatives, especially in PDAC with the involvement of BRCA1/2. Therefore, it is essential that NGS-based tumour tests are associated with appropriate pre-test counselling to inform patients about the possibility of identifying a hereditary cancer predisposition. Several studies have shown that, even in the context of advanced cancer, the majority of patients wish to be informed about such incidental findings[39,40]. For such patients, a post-test genetic counselling should be organized to plan for confirmatory germline testing with informed consent, counsel the patient on the associated risks for themselves and their relatives, and even invite relatives for result confirmation consultations. MTB could help guide patients towards oncogenetics testing to ensure that no potential germline mutation is left unexplored.
The third point for consideration in the implementation of precision medicine is NGS of circulating tumour DNA (e.g. liquid biopsy) shown to offer several advantages over tissue-based NGS in cancer molecular profiling. Indeed, tissue-based tumour sequencing, the current gold standard for detecting actionable molecular abnormalities in patients with solid cancers[41], has several limitations including screening failures due to limited tissue availability (e.g., after a biopsy) and an inability to capture spatial and temporal tumour heterogeneity, which can compromise accurate treatment selection. Furthermore, obtaining a tumour biopsy is challenging and can lead to adverse events not encountered with blood sampling[42,43]. Liquid biopsy on the other hand is non-invasive, easily feasible and reproducible, and is representative of the patient's entire tumour molecular landscape[44,45]. It allows the detection of genomic alterations with high accuracy compared to tissue analysis[44,46], and importantly captures spatial and temporal heterogeneity, a well-established characteristic of malignant tumours. The molecular profile obtained from a tumour biopsy is limited to a single site (space) and is frozen in time. Liquid biopsy has the advantage of providing access to a "genomic pool" from multiple metastatic sites in the patient[44], allowing for more precise genotyping in metastatic patients. Indeed, tumour progression and systemic treatment regimens can promote clonal evolution and increased tumour heterogeneity, leading to discordance between archived tissue and liquid biopsy results. However, one limitation of liquid biopsy is the possibility of false negatives (i.e., not identifying an alteration of interest actually present in the tumour). Indeed, Sugimori et al[47] showed fluctuations in KRAS detection in advanced PDAC patients under treatment. More data from prospective studies on the use of liquid biopsy for molecular profiling are now needed to validate its use for non-invasive serial sampling of tumour material, which could become part of PDAC management. One promising liquid biopsy test developed by Foundation Medicine® (Massachusetts, United States) can detect targetable alterations using NGS to study 324 cancer-associated genes, including short pathogenic variants, copy number alterations, and some rearrangements, as well as mutational burden. The development of such liquid biopsy tests will likely facilitate precision medicine in PDAC.
The fourth point for consideration is access to molecular biology technologies. A recent study assessed access to molecular biology technologies in Europe and showed profound disparities between countries, some of which thus had limited access for patients to targeted drugs[48]. While immunohistochemistry and basic genomic techniques such as fluorescence in situ hybridization, polymerase chain reaction, and microsatellite instability analysis were widely available in routine clinical practice, access to more novel and advanced technologies, including large-scale NGS panels, was highly heterogeneous in Europe and within each country. Even in high-income countries, large high-throughput sequencing panels remain largely unavailable in routine clinical practice and are limited to clinical trials, research or tertiary centres. Thus, despite the existence of targeted therapies, the search for additional biomarkers in every patient is currently not permitted in routine medical practice. With the advent of new targeted therapies requiring associated biomarkers, the diffusion of molecular profiling is essential to ensure equal access to healthcare throughout Europe.
The final point for consideration is drug access issues, which at least in part explain the low percentage of patients treated with target therapies. In France, despite European marketing authorizations and studies proving their clinical effectiveness, certain innovative drugs such as larotrectinib[48] have been issued unfavourable opinions by French authorities with regards their reimbursement. Precision medicine goes hand in hand with associated medications, so advocating for NGS for all PDAC patients would only make sense should patients be assured access to treatments. While some clinical trials undertaken in France are allowing patients to receive such treatments, the restrictive inclusion criteria of these trials effectively exclude a certain number of patients.
In 2020, it was estimated that nearly 20 million new cancer cases occurred worldwide[49,50]. Studies have revealed that approximately one in seven human cancers carries a KRAS gene mutation, making it one of the major human oncogenes[51-54]. Targeting KRAS has been a major focus of cancer research over the past four decades, particularly since the fundamental discovery by Ostrem et al[54] of a KRAS G12C inhibitor[54], which led to the clinical validation of two targeted therapies sotorasib and adagrasib[55,56]. There is a wide diversity of KRAS gene mutations[57]. While the most frequent alteration in PDAC affects codon G12, representing 93% of cases, the amino acid substitution differs among patients[58,59]. The most common substitution is G12D (41%), followed by G12V (34%) then G12R (16%) and only 1.1% for the G12C that is actionable[58,59]. The presence of the KRAS mutation is associated with a poor response to treatment and confers an unfavourable overall prognosis and shorter survival in PDAC patients[60-62]. Its targeting therefore represents a challenging yet crucial goal in the development of effective new therapies.
In 2018, sotorasib (AMG510) became the first KRAS G12C inhibitor to enter human clinical trials and demonstrated its safety and clinical effectiveness. Hong et al[55] reported the results of a phase 1 multicentre open-label trial[55] involving sotorasib in patients with advanced solid tumours (locally advanced or metastatic) harbouring the KRAS G12C mutation. A total of 129 patients (59 with lung cancer; 42 with colorectal cancer; and 28 with other tumour types) were included in dose escalation and expansion cohorts. Interesting results in terms of clinical efficacy were confirmed in phase II, and those specific for pancreatic cancer patients are presented in Table 2 (38 patients)[62]. The second inhibitor adagrasib (MRTX849) has also been tested in phase I and II trials[56,63]. Among the 18 evaluable patients with PDAC, 33% showed a confirmed objective response, with a disease control rate of 81% and a median progression free survival of 5.4 months. To enhance the activity of KRAS G12C inhibitors, vertical inhibition strategies (double or triple with anti-SHP2, anti-MEK, and/or anti-EGFR) could be an interesting strategy[64,65]. Reports of promising results of combining anti-KRAS G12C and anti-EGFR inhibitors have been made concerning colorectal cancer[66,67] though not as yet with regards pancreatic cancer. However, considering that KRAS G12C mutated tumours represent only a small proportion of all PDAC patients, the development of other strategies is critical in this patient group.
| Adagrasib (n = 21)[57] | Sotorasib (n = 38)[63] | |
| Objective response rate (95%CI) | 33 (15-57) | 21 (10-37) |
| Disease control rate | 81 | 84 |
| Median duration of response (months) | NA | 5.7 (1.6-non evaluable) |
| Progression-free survival (months) (95%CI) | 5.4 (3.9-8.2) | 4.0 (2.8-5.6) |
| Overall survival (months) (95%CI) | 8.0 (5.2-11.8) | 6.9 (5.0-9.1) |
| Grade 3-4 toxicities | 27 | 16 |
| Dose reduction | 40 (solid tumours) | 13 |
| Discontinuation for toxicity | 0 | 0 |
Alternative strategies have already been developed in this regard. The first involved the development of other selective inhibitors against more common codons, such as G12D. A selective and potent KRAS G12D inhibitor, MRTX1133[68], with non-covalent and picomolar binding affinity to the protein was developed by the Mirati® team. Pre-clinical efficacy testing gave good results in both cell lines and murine models[68,69]. The second strategy involved the development of direct pan-specific KRAS inhibitors and proteolysis targeting chimeras (PROTAC), capable of sparing NRAS and HRAS[15]. The PROTACs are an emerging class of drugs that specifically degrade proteins through the cellular protein degradation system[70]. They interact simultaneously with a protein of interest and an E3 ligase, forming a ternary complex that allows the E3 ligase to ubiquitinate and induce degradation of the target protein[71]. It remains to be established whether targeting all three RAS isoforms simultaneously would be compatible with regards to therapeutic window in patients (i.e., with manageable toxicities). A third indirect inhibition strategy has been developed to interfere with nucleotide exchange and KRAS activation through the inhibition of SHP2 or SOS1[72-76].
Patients with KRAS wild-type (KRASWT) tumours represent a specific subgroup of patients, accounting for approximately 10% to 15% of all PDAC cases[34,60,77,78]. In a recently published study[78], these patients showed a better prognosis than those for whom KRAS was mutated, with an OS of 24 months vs 17.5 months, despite a higher OS for the KRAS mutants in this study compared to that generally reported in this population. The KRASWT subtype is also characterized by a higher prevalence of actionable alterations, ranging from 30% to 40% in various studies[34,60,77]. An analysis of molecular alterations based on tumour tissue from around 2500 patients demonstrated that KRASWT pancreatic cancer more commonly harboured mutations in DNA damage repair genes (i.e., ATM, BAP1, BRCA2, FANCE, PALB2, and RAD50), chromatin remodeling genes (ARID1A, ARID2, KMT2C, KMT2D, PBRM1, SETD2, and SMARCA4), cell cycle control genes (CDKN2A, CCND1, and CCNE1), and BRAF[77]. Other important molecular phenotypes such as high microsatellite instability (4.7% vs 0.7%; P < 0.05) and high tumour mutation burden (4.5% vs 1.0%, P <0.05) were also more frequently observed in KRASWT pancreatic cancer in this study[77]. KRASWT PDAC harbour a distinct molecular landscape with the frequent detection of therapeutically targetable oncogenic fusions, including ALK, BRAF, FGFR2, MET, NRG1, and RET fusions, which have been reported in 19% to 67% of KRASWT pancreatic cancer compared with 0% to 1% of KRAS-mutated PDAC[34,77,79,80]. Patients with KRASWT pancreatic cancer with therapeutically targetable fusion proteins who received targeted therapy, including entrectinib or larotrectinib for NTRK fusion, afatinib for NRG1 fusion, crizotinib for MET fusion, selpercatinib for RET fusion and erdafitinib for FGFR2 fusion, have experienced durable responses[34,79-81].
One proposed strategy accounting for the difficulty in rendering routine NGS for all PDAC patients, is to first determine KRAS status through targeted sequencing on the basis of which NGS would only be performed for KRASWT patients[77]. The impact of identifying KRASWT tumours appears to exceed the greater proportion of actionable molecular alterations. Indeed, nimotuzumab, a monoclonal antibody directed against EGFR, was effective when combined with gemcitabine for wild-type KRAS tumours with an acceptable tolerance profile in Asian patients[82]. Specific treatments should therefore be developed and/or validated for these tumour subtypes.
One team determined transcriptomic signatures, based on gene expression profiles in patient-derived organoids, primary cell cultures, and xenografts. They first correlated the transcriptomic profiles with concurrent gemcitabine sensitivity analyses, which then permitted the identification of tumours showing particular sensitivity to different treatments, including gemcitabine, oxaliplatin, 5-FU, irinotecan, and docetaxel[83-85]. A statistical approach was used to derive a multi-gene signature predictive of gemcitabine sensitivity from these preclinical models (GemPred signature)[83]. This signature was finally tested in a single-centre cohort and validated in a large multicentre cohort of resectable PDAC patients[83]. The same approach was used for metastatic PDAC and allowed good patient stratification[84]. The same team then developed a sensitivity signature for FOLFIRINOX (specifically for 5-FU, irinotecan, and oxaliplatin)[85]. A clinical trial is currently ongoing (PACsign, NCT05475366) to evaluate the strategy of guiding first-line treatment of patients based on these predictive signatures.
To conclude, the spectrum of actionable molecular alterations is evolving rapidly, with the development of KRAS inhibitors that could potentially provide actionability for almost all PDAC tumours. The higher frequency of actionable alterations harboured by KRASWT tumours provides numerous therapeutic opportunities. Considering the potential clinical benefit of molecularly-matched treatments, along with their improved tolerance rates compared to che
| 1. | Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, Adenis A, Raoul JL, Gourgou-Bourgade S, de la Fouchardière C, Bennouna J, Bachet JB, Khemissa-Akouz F, Péré-Vergé D, Delbaldo C, Assenat E, Chauffert B, Michel P, Montoto-Grillot C, Ducreux M; Groupe Tumeurs Digestives of Unicancer; PRODIGE Intergroup. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817-1825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4838] [Cited by in RCA: 5612] [Article Influence: 400.9] [Reference Citation Analysis (1)] |
| 2. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9819] [Article Influence: 4909.5] [Reference Citation Analysis (1)] |
| 3. | Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzrock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat Rev. 2020;86:102019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 409] [Article Influence: 81.8] [Reference Citation Analysis (0)] |
| 4. | Le Tourneau C, Borcoman E, Kamal M. Molecular profiling in precision medicine oncology. Nat Med. 2019;25:711-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 5. | Massard C, Michiels S, Ferté C, Le Deley MC, Lacroix L, Hollebecque A, Verlingue L, Ileana E, Rosellini S, Ammari S, Ngo-Camus M, Bahleda R, Gazzah A, Varga A, Postel-Vinay S, Loriot Y, Even C, Breuskin I, Auger N, Job B, De Baere T, Deschamps F, Vielh P, Scoazec JY, Lazar V, Richon C, Ribrag V, Deutsch E, Angevin E, Vassal G, Eggermont A, André F, Soria JC. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017;7:586-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 519] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 6. | Le Tourneau C, Delord JP, Gonçalves A, Gavoille C, Dubot C, Isambert N, Campone M, Trédan O, Massiani MA, Mauborgne C, Armanet S, Servant N, Bièche I, Bernard V, Gentien D, Jezequel P, Attignon V, Boyault S, Vincent-Salomon A, Servois V, Sablin MP, Kamal M, Paoletti X; SHIVA investigators. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 701] [Cited by in RCA: 802] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 7. | Trédan O, Wang Q, Pissaloux D, Cassier P, de la Fouchardière A, Fayette J, Desseigne F, Ray-Coquard I, de la Fouchardière C, Frappaz D, Heudel PE, Bonneville-Levard A, Fléchon A, Sarabi M, Guibert P, Bachelot T, Pérol M, You B, Bonnin N, Collard O, Leyronnas C, Attignon V, Baudet C, Sohier E, Villemin JP, Viari A, Boyault S, Lantuejoul S, Paindavoine S, Treillleux I, Rodriguez C, Agrapart V, Corset V, Garin G, Chabaud S, Pérol D, Blay JY; ProfiLER investigators. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30:757-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 8. | Andre F, Filleron T, Kamal M, Mosele F, Arnedos M, Dalenc F, Sablin MP, Campone M, Bonnefoi H, Lefeuvre-Plesse C, Jacot W, Coussy F, Ferrero JM, Emile G, Mouret-Reynier MA, Thery JC, Isambert N, Mege A, Barthelemy P, You B, Hajjaji N, Lacroix L, Rouleau E, Tran-Dien A, Boyault S, Attignon V, Gestraud P, Servant N, Le Tourneau C, Cherif LL, Soubeyran I, Montemurro F, Morel A, Lusque A, Jimenez M, Jacquet A, Gonçalves A, Bachelot T, Bieche I. Genomics to select treatment for patients with metastatic breast cancer. Nature. 2022;610:343-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 90] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 9. | Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1826] [Cited by in RCA: 2465] [Article Influence: 308.1] [Reference Citation Analysis (0)] |
| 10. | Tuxen IV, Rohrberg KS, Oestrup O, Ahlborn LB, Schmidt AY, Spanggaard I, Hasselby JP, Santoni-Rugiu E, Yde CW, Mau-Sørensen M, Nielsen FC, Lassen U. Copenhagen Prospective Personalized Oncology (CoPPO)-Clinical Utility of Using Molecular Profiling to Select Patients to Phase I Trials. Clin Cancer Res. 2019;25:1239-1247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 11. | Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, Peters S, Planchard D, Smit EF, Solomon BJ, Veronesi G, Reck M; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:339-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 370] [Article Influence: 185.0] [Reference Citation Analysis (0)] |
| 12. | Conroy T, Pfeiffer P, Vilgrain V, Lamarca A, Seufferlein T, O'Reilly EM, Hackert T, Golan T, Prager G, Haustermans K, Vogel A, Ducreux M; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Pancreatic cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:987-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 189] [Article Influence: 94.5] [Reference Citation Analysis (0)] |
| 13. | Neuzillet C, Gaujoux S, Williet N, Bachet JB, Bauguion L, Colson Durand L, Conroy T, Dahan L, Gilabert M, Huguet F, Marthey L, Meilleroux J, de Mestier L, Napoléon B, Portales F, Sa Cunha A, Schwarz L, Taieb J, Chibaudel B, Bouché O, Hammel P; Thésaurus National de Cancérologie Digestive (TNCD); Société Nationale Française de Gastroentérologie (SNFGE); Fédération Francophone de Cancérologie Digestive (FFCD); Groupe Coopérateur multidisciplinaire en Oncologie (GERCOR); Fédération Nationale des Centres de Lutte Contre le Cancer (UNICANCER); Société Française de Chirurgie Digestive (SFCD); Société Française d’Endoscopie Digestive (SFED); Société Française de Radiothérapie Oncologique (SFRO); Association de Chirurgie Hépato-Bilio-Pancréatique et Transplantation (ACHBT); Association Française de Chirurgie (AFC). Pancreatic cancer: French clinical practice guidelines for diagnosis, treatment and follow-up (SNFGE, FFCD, GERCOR, UNICANCER, SFCD, SFED, SFRO, ACHBT, AFC). Dig Liver Dis. 2018;50:1257-1271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 14. | Lowery MA, Jordan EJ, Basturk O, Ptashkin RN, Zehir A, Berger MF, Leach T, Herbst B, Askan G, Maynard H, Glassman D, Covington C, Schultz N, Abou-Alfa GK, Harding JJ, Klimstra DS, Hechtman JF, Hyman DM, Allen PJ, Jarnagin WR, Balachandran VP, Varghese AM, Schattner MA, Yu KH, Saltz LB, Solit DB, Iacobuzio-Donahue CA, Leach SD, O'Reilly EM. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res. 2017;23:6094-6100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 145] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | Hofmann MH, Gerlach D, Misale S, Petronczki M, Kraut N. Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants. Cancer Discov. 2022;12:924-937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 179] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 16. | Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM; Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2848] [Cited by in RCA: 2534] [Article Influence: 281.6] [Reference Citation Analysis (0)] |
| 17. | Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, Feiler HS, Ko AH, Olshen AB, Danenberg KL, Tempero MA, Spellman PT, Hanahan D, Gray JW. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500-503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1536] [Cited by in RCA: 1350] [Article Influence: 96.4] [Reference Citation Analysis (0)] |
| 18. | Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168-1178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1041] [Cited by in RCA: 1462] [Article Influence: 146.2] [Reference Citation Analysis (0)] |
| 19. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA; Australian Pancreatic Cancer Genome Initiative, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1982] [Article Influence: 198.2] [Reference Citation Analysis (1)] |
| 20. | Tarabay A, Hollebecque A, Smolenschi C, Antoun L, Klotz C, Fuerea A, Perret A, Gouriou C, Burtin P, Belkhodja H, Boige V, Malka D, Ducreux M. Molecular precision medicine in pancreatic cancer: A single-center experience. J Clin Oncol. 2022;40:605-605. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 21. | Pishvaian MJ, Blais EM, Brody JR, Lyons E, DeArbeloa P, Hendifar A, Mikhail S, Chung V, Sahai V, Sohal DPS, Bellakbira S, Thach D, Rahib L, Madhavan S, Matrisian LM, Petricoin EF 3rd. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020;21:508-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 362] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 22. | Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, Ng CKY, Bedard PL, Tortora G, Douillard JY, Van Allen EM, Schultz N, Swanton C, André F, Pusztai L. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol. 2018;29:1895-1902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 507] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 23. | Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park JO, Hochhauser D, Arnold D, Oh DY, Reinacher-Schick A, Tortora G, Algül H, O'Reilly EM, McGuinness D, Cui KY, Schlienger K, Locker GY, Kindler HL. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med. 2019;381:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1137] [Cited by in RCA: 1614] [Article Influence: 269.0] [Reference Citation Analysis (0)] |
| 24. | Taïeb J, Sayah L, Heinrich K, Kunzmann V, Boileve A, Cirkel G, Lonardi S, Chibaudel B, Turpin A, Beller T, Hautefeuille V, Vivaldi C, Mazard T, Bauguion L, Niger M, Prager GW, Coutzac C, Benedikt Westphalen C, Auclin E, Pilla L. Efficacy of immune checkpoint inhibitors in microsatellite unstable/mismatch repair-deficient advanced pancreatic adenocarcinoma: an AGEO European Cohort. Eur J Cancer. 2023;188:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 39] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 25. | Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, Geva R, Gottfried M, Penel N, Hansen AR, Piha-Paul SA, Doi T, Gao B, Chung HC, Lopez-Martin J, Bang YJ, Frommer RS, Shah M, Ghori R, Joe AK, Pruitt SK, Diaz LA Jr. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol. 2020;38:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2120] [Cited by in RCA: 1995] [Article Influence: 399.0] [Reference Citation Analysis (0)] |
| 26. | Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, van Tilburg CM, Nagasubramanian R, Berlin JD, Federman N, Mascarenhas L, Geoerger B, Dowlati A, Pappo AS, Bielack S, Doz F, McDermott R, Patel JD, Schilder RJ, Tahara M, Pfister SM, Witt O, Ladanyi M, Rudzinski ER, Nanda S, Childs BH, Laetsch TW, Hyman DM, Drilon A. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 695] [Article Influence: 139.0] [Reference Citation Analysis (0)] |
| 27. | Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, Blakely CM, Seto T, Cho BC, Tosi D, Besse B, Chawla SP, Bazhenova L, Krauss JC, Chae YK, Barve M, Garrido-Laguna I, Liu SV, Conkling P, John T, Fakih M, Sigal D, Loong HH, Buchschacher GL Jr, Garrido P, Nieva J, Steuer C, Overbeck TR, Bowles DW, Fox E, Riehl T, Chow-Maneval E, Simmons B, Cui N, Johnson A, Eng S, Wilson TR, Demetri GD; trial investigators. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21:271-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 1128] [Article Influence: 188.0] [Reference Citation Analysis (0)] |
| 28. | Li HS, Yang K, Wang Y. Remarkable response of BRAF (V600E)-mutated metastatic pancreatic cancer to BRAF/MEK inhibition: a case report. Gastroenterol Rep (Oxf). 2022;10:goab031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 29. | Helal C, Valéry M, Ducreux M, Hollebecque A, Smolenschi C. FGFR2 fusion in metastatic pancreatic ductal adenocarcinoma: Is there hope? Eur J Cancer. 2022;176:168-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 30. | Schram AM, Odintsov I, Espinosa-Cotton M, Khodos I, Sisso WJ, Mattar MS, Lui AJW, Vojnic M, Shameem SH, Chauhan T, Torrisi J, Ford J, O'Connor MN, Geuijen CAW, Schackmann RCJ, Lammerts van Bueren JJ, Wasserman E, de Stanchina E, O'Reilly EM, Ladanyi M, Drilon A, Somwar R. Zenocutuzumab, a HER2xHER3 Bispecific Antibody, Is Effective Therapy for Tumors Driven by NRG1 Gene Rearrangements. Cancer Discov. 2022;12:1233-1247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 108] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 31. | Meric-Bernstam F, Makker V, Oaknin A, Oh D-Y, Banerjee SN, Gonzalez Martin A, Jung KH, Lugowska IA, Manso L, Manzano A, Melichar B, Siena S, Stroyakovskiy D, Anoka C, Ma Y, Puvvada SD, Lee J-Y. Efficacy and safety of trastuzumab deruxtecan (T-DXd) in patients (pts) with HER2-expressing solid tumors: DESTINY-PanTumor02 (DP-02) interim results. J Clin Oncol. 2023;41:LBA3000-LBA3000. [DOI] [Full Text] |
| 32. | Tsimberidou AM, Hong DS, Ye Y, Cartwright C, Wheler JJ, Falchook GS, Naing A, Fu S, Piha-Paul S, Janku F, Meric-Bernstam F, Hwu P, Kee B, Kies MS, Broaddus R, Mendelsohn J, Hess KR, Kurzrock R. Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson Precision Medicine Study. JCO Precis Oncol. 2017;2017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 89] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 33. | Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33:3817-3825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 348] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 34. | Ben-Ammar I, Rousseau A, Nicolle R, Tarabay A, Boige V, Valery M, Pudlarz T, Malka D, Gelli M, Fernandez-De-Sevilla E, Fuerea A, Tanguy ML, Rouleau E, Barbe R, Mathieu JRR, Jaulin F, Smolenschi C, Hollebecque A, Ducreux M, Boileve A. Precision medicine for KRAS wild-type pancreatic adenocarcinomas. Eur J Cancer. 2024;197:113497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 35. | Martin-Romano P, Mezquita L, Hollebecque A, Lacroix L, Rouleau E, Gazzah A, Bahleda R, Planchard D, Varga A, Baldini C, Postel-Vinay S, Friboulet L, Loriot Y, Verlingue L, Geraud A, Camus MN, Nicotra C, Soria JC, André F, Besse B, Massard C, Italiano A. Implementing the European Society for Medical Oncology Scale for Clinical Actionability of Molecular Targets in a Comprehensive Profiling Program: Impact on Precision Medicine Oncology. JCO Precis Oncol. 2022;6:e2100484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 36. | Tarabay A, Boileve A, Smolenschi C, Antoun L, Valery M, Fuerea A, Perret A, Burtin P, Cosconea S, Belkhodja H, Malka D, Boige V, Hollebecque A, Ducreux M. Precision Medicine in Pancreatic Ductal Adenocarcinoma: The Impact of Targeted Therapies on Survival of Patients Harboring Actionable Mutations. Biomedicines. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 37. | Vasseur D, Sassi H, Bayle A, Tagliamento M, Besse B, Marzac C, Arbab A, Auger N, Cotteret S, Aldea M, Blanc-Durand F, Géraud A, Gazzah A, Loriot Y, Hollebecque A, Martín-Romano P, Ngo-Camus M, Nicotra C, Ponce S, Sakkal M, Caron O, Smolenschi C, Micol JB, Italiano A, Rouleau E, Lacroix L. Next-Generation Sequencing on Circulating Tumor DNA in Advanced Solid Cancer: Swiss Army Knife for the Molecular Tumor Board? A Review of the Literature Focused on FDA Approved Test. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 38. | Rolfo C, Manca P, Salgado R, Van Dam P, Dendooven A, Ferri Gandia J, Rutten A, Lybaert W, Vermeij J, Gevaert T, Weyn C, Lefebure A, Metsu S, Van Laere S, Peeters M, Pauwels P, Machado Coelho A. Multidisciplinary molecular tumour board: a tool to improve clinical practice and selection accrual for clinical trials in patients with cancer. ESMO Open. 2018;3:e000398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 39. | Meric-Bernstam F, Brusco L, Daniels M, Wathoo C, Bailey AM, Strong L, Shaw K, Lu K, Qi Y, Zhao H, Lara-Guerra H, Litton J, Arun B, Eterovic AK, Aytac U, Routbort M, Subbiah V, Janku F, Davies MA, Kopetz S, Mendelsohn J, Mills GB, Chen K. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 2016;27:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 40. | Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, Ni A, Thomas T, Benayed R, Ashraf A, Lincoln A, Arcila M, Stadler Z, Solit D, Hyman DM, Zhang L, Klimstra D, Ladanyi M, Offit K, Berger M, Robson M. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol. 2016;2:104-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 264] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 41. | Esagian SM, Grigoriadou GΙ, Nikas IP, Boikou V, Sadow PM, Won JK, Economopoulos KP. Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: a comprehensive systematic review. J Cancer Res Clin Oncol. 2020;146:2051-2066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 42. | Recondo G, Mahjoubi L, Maillard A, Loriot Y, Bigot L, Facchinetti F, Bahleda R, Gazzah A, Hollebecque A, Mezquita L, Planchard D, Naltet C, Lavaud P, Lacroix L, Richon C, Lovergne AA, De Baere T, Tselikas L, Deas O, Nicotra C, Ngo-Camus M, Frias RL, Solary E, Angevin E, Eggermont A, Olaussen KA, Vassal G, Michiels S, Andre F, Scoazec JY, Massard C, Soria JC, Besse B, Friboulet L. Feasibility and first reports of the MATCH-R repeated biopsy trial at Gustave Roussy. NPJ Precis Oncol. 2020;4:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Prud'homme C, Deschamps F, Allorant A, Massard C, Hollebecque A, Yevich S, Ngo-Camus M, Gravel G, Nicotra C, Michiels S, Scoazec JY, Lacroix L, Solary E, Soria JC, De Baere T, Tselikas L. Image-guided tumour biopsies in a prospective molecular triage study (MOSCATO-01): What are the real risks? Eur J Cancer. 2018;103:108-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 44. | Bayle A, Peyraud F, Belcaid L, Brunet M, Aldea M, Clodion R, Dubos P, Vasseur D, Nicotra C, Geraud A, Sakkal M, Cerbone L, Blanc-Durand F, Mosele F, Martin Romano P, Ngo Camus M, Soubeyran I, Khalifa E, Alame M, Blouin L, Dinart D, Bellera C, Hollebecque A, Ponce S, Loriot Y, Besse B, Lacroix L, Rouleau E, Barlesi F, Andre F, Italiano A. Liquid versus tissue biopsy for detecting actionable alterations according to the ESMO Scale for Clinical Actionability of molecular Targets in patients with advanced cancer: a study from the French National Center for Precision Medicine (PRISM). Ann Oncol. 2022;33:1328-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 45. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 572] [Article Influence: 143.0] [Reference Citation Analysis (0)] |
| 46. | Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, Esaki T, Komatsu Y, Kawamoto Y, Takahashi N, Ueno M, Kagawa Y, Nishina T, Kato T, Yamamoto Y, Furuse J, Denda T, Kawakami H, Oki E, Nakajima T, Nishida N, Yamaguchi K, Yasui H, Goto M, Matsuhashi N, Ohtsubo K, Yamazaki K, Tsuji A, Okamoto W, Tsuchihara K, Yamanaka T, Miki I, Sakamoto Y, Ichiki H, Hata M, Yamashita R, Ohtsu A, Odegaard JI, Yoshino T. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859-1864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 236] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 47. | Sugimori M, Sugimori K, Tsuchiya H, Suzuki Y, Tsuyuki S, Kaneta Y, Hirotani A, Sanga K, Tozuka Y, Komiyama S, Sato T, Tezuka S, Goda Y, Irie K, Miwa H, Miura Y, Ishii T, Kaneko T, Nagahama M, Shibata W, Nozaki A, Maeda S. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020;111:266-278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 48. | Bayle A, Bonastre J, Chaltiel D, Latino N, Rouleau E, Peters S, Galotti M, Bricalli G, Besse B, Giuliani R. ESMO study on the availability and accessibility of biomolecular technologies in oncology in Europe. Ann Oncol. 2023;34:934-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 52] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 49. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64104] [Article Influence: 16026.0] [Reference Citation Analysis (174)] |
| 50. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13300] [Cited by in RCA: 15454] [Article Influence: 2575.7] [Reference Citation Analysis (2)] |
| 51. | Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457-2467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1732] [Cited by in RCA: 1541] [Article Influence: 118.5] [Reference Citation Analysis (0)] |
| 52. | Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9144] [Cited by in RCA: 12711] [Article Influence: 977.8] [Reference Citation Analysis (0)] |
| 53. | AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1205] [Cited by in RCA: 1348] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 54. | Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1244] [Cited by in RCA: 1800] [Article Influence: 150.0] [Reference Citation Analysis (0)] |
| 55. | Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, Falchook GS, Price TJ, Sacher A, Denlinger CS, Bang YJ, Dy GK, Krauss JC, Kuboki Y, Kuo JC, Coveler AL, Park K, Kim TW, Barlesi F, Munster PN, Ramalingam SS, Burns TF, Meric-Bernstam F, Henary H, Ngang J, Ngarmchamnanrith G, Kim J, Houk BE, Canon J, Lipford JR, Friberg G, Lito P, Govindan R, Li BT. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020;383:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1163] [Article Influence: 232.6] [Reference Citation Analysis (0)] |
| 56. | Bekaii-Saab TS, Yaeger R, Spira AI, Pelster MS, Sabari JK, Hafez N, Barve M, Velastegui K, Yan X, Shetty A, Der-Torossian H, Pant S. Adagrasib in Advanced Solid Tumors Harboring a KRAS(G12C) Mutation. J Clin Oncol. 2023;41:4097-4106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 134] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 57. | Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 463] [Article Influence: 92.6] [Reference Citation Analysis (0)] |
| 58. | Waters AM, Der CJ. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 594] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 59. | Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2161] [Cited by in RCA: 2379] [Article Influence: 216.3] [Reference Citation Analysis (0)] |
| 60. | Luchini C, Paolino G, Mattiolo P, Piredda ML, Cavaliere A, Gaule M, Melisi D, Salvia R, Malleo G, Shin JI, Cargnin S, Terrazzino S, Lawlor RT, Milella M, Scarpa A. KRAS wild-type pancreatic ductal adenocarcinoma: molecular pathology and therapeutic opportunities. J Exp Clin Cancer Res. 2020;39:227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 61. | McIntyre CA, Lawrence SA, Richards AL, Chou JF, Wong W, Capanu M, Berger MF, Donoghue MTA, Yu KH, Varghese AM, Kelsen DP, Park W, Balachandran VP, Kingham TP, D'Angelica MI, Drebin JA, Jarnagin WR, Iacobuzio-Donahue CA, Allen PJ, O'Reilly EM. Alterations in driver genes are predictive of survival in patients with resected pancreatic ductal adenocarcinoma. Cancer. 2020;126:3939-3949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 62. | Strickler JH, Satake H, George TJ, Yaeger R, Hollebecque A, Garrido-Laguna I, Schuler M, Burns TF, Coveler AL, Falchook GS, Vincent M, Sunakawa Y, Dahan L, Bajor D, Rha SY, Lemech C, Juric D, Rehn M, Ngarmchamnanrith G, Jafarinasabian P, Tran Q, Hong DS. Sotorasib in KRAS p.G12C-Mutated Advanced Pancreatic Cancer. N Engl J Med. 2023;388:33-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 248] [Article Influence: 124.0] [Reference Citation Analysis (0)] |
| 63. | Ou SI, Jänne PA, Leal TA, Rybkin II, Sabari JK, Barve MA, Bazhenova L, Johnson ML, Velastegui KL, Cilliers C, Christensen JG, Yan X, Chao RC, Papadopoulos KP. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients With Advanced KRAS(G12C) Solid Tumors (KRYSTAL-1). J Clin Oncol. 2022;40:2530-2538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 186] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 64. | Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, Whaley A, Pinnelli M, Murciano-Goroff YR, Vakiani E, Valeri N, Liao WL, Bhalkikar A, Thyparambil S, Zhao HY, de Stanchina E, Marsoni S, Siena S, Bertotti A, Trusolino L, Li BT, Rosen N, Di Nicolantonio F, Bardelli A, Misale S. EGFR Blockade Reverts Resistance to KRAS(G12C) Inhibition in Colorectal Cancer. Cancer Discov. 2020;10:1129-1139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 327] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 65. | Ryan MB, Coker O, Sorokin A, Fella K, Barnes H, Wong E, Kanikarla P, Gao F, Zhang Y, Zhou L, Kopetz S, Corcoran RB. KRAS(G12C)-independent feedback activation of wild-type RAS constrains KRAS(G12C) inhibitor efficacy. Cell Rep. 2022;39:110993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 87] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 66. | Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3767] [Cited by in RCA: 3706] [Article Influence: 176.5] [Reference Citation Analysis (1)] |
| 67. | Schwartzberg LS, Rivera F, Karthaus M, Fasola G, Canon JL, Hecht JR, Yu H, Oliner KS, Go WY. PEAK: a randomized, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240-2247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 495] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 68. | Wang X, Allen S, Blake JF, Bowcut V, Briere DM, Calinisan A, Dahlke JR, Fell JB, Fischer JP, Gunn RJ, Hallin J, Laguer J, Lawson JD, Medwid J, Newhouse B, Nguyen P, O'Leary JM, Olson P, Pajk S, Rahbaek L, Rodriguez M, Smith CR, Tang TP, Thomas NC, Vanderpool D, Vigers GP, Christensen JG, Marx MA. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J Med Chem. 2022;65:3123-3133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 396] [Article Influence: 99.0] [Reference Citation Analysis (0)] |
| 69. | The KRAS(G12D) inhibitor MRTX1133 elucidates KRAS-mediated oncogenesis. Nat Med. 2022;28:2017-2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 70. | Qi SM, Dong J, Xu ZY, Cheng XD, Zhang WD, Qin JJ. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front Pharmacol. 2021;12:692574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 180] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 71. | Farnaby W, Koegl M, McConnell DB, Ciulli A. Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr Opin Pharmacol. 2021;57:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 72. | Valencia-Sama I, Ladumor Y, Kee L, Adderley T, Christopher G, Robinson CM, Kano Y, Ohh M, Irwin MS. NRAS Status Determines Sensitivity to SHP2 Inhibitor Combination Therapies Targeting the RAS-MAPK Pathway in Neuroblastoma. Cancer Res. 2020;80:3413-3423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 73. | Sheffels E, Kortum RL. The Role of Wild-Type RAS in Oncogenic RAS Transformation. Genes (Basel). 2021;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 74. | Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, Tzitzilonis C, Mordec K, Marquez A, Romero J, Hsieh T, Zaman A, Olivas V, McCoach C, Blakely CM, Wang Z, Kiss G, Koltun ES, Gill AL, Singh M, Goldsmith MA, Smith JAM, Bivona TG. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. 2018;20:1064-1073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 306] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 75. | Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, Sanderson MP, Kessler D, Trapani F, Arnhof H, Rumpel K, Botesteanu DA, Ettmayer P, Gerstberger T, Kofink C, Wunberg T, Zoephel A, Fu SC, Teh JL, Böttcher J, Pototschnig N, Schachinger F, Schipany K, Lieb S, Vellano CP, O'Connell JC, Mendes RL, Moll J, Petronczki M, Heffernan TP, Pearson M, McConnell DB, Kraut N. BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition. Cancer Discov. 2021;11:142-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 287] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 76. | Zhao Y, Xue JY, Lito P. Suppressing Nucleotide Exchange to Inhibit KRAS-Mutant Tumors. Cancer Discov. 2021;11:17-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 77. | Philip PA, Azar I, Xiu J, Hall MJ, Hendifar AE, Lou E, Hwang JJ, Gong J, Feldman R, Ellis M, Stafford P, Spetzler D, Khushman MM, Sohal D, Lockhart AC, Weinberg BA, El-Deiry WS, Marshall J, Shields AF, Korn WM. Molecular Characterization of KRAS Wild-type Tumors in Patients with Pancreatic Adenocarcinoma. Clin Cancer Res. 2022;28:2704-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 108] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 78. | Singh H, Keller RB, Kapner KS, Dilly J, Raghavan S, Yuan C, Cohen EF, Tolstorukov M, Andrews E, Brais LK, da Silva A, Perez K, Rubinson DA, Surana R, Giannakis M, Ng K, Clancy TE, Yurgelun MB, Schlechter BL, Clark JW, Shapiro GI, Rosenthal MH, Hornick JL, Nardi V, Li YY, Gupta H, Cherniack AD, Meyerson M, Cleary JM, Nowak JA, Wolpin BM, Aguirre AJ. Oncogenic Drivers and Therapeutic Vulnerabilities in KRAS Wild-Type Pancreatic Cancer. Clin Cancer Res. 2023;29:4627-4643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 23] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 79. | Fusco MJ, Saeed-Vafa D, Carballido EM, Boyle TA, Malafa M, Blue KL, Teer JK, Walko CM, McLeod HL, Hicks JK, Extermann M, Fleming JB, Knepper TC, Kim DW. Identification of Targetable Gene Fusions and Structural Rearrangements to Foster Precision Medicine in KRAS Wild-Type Pancreatic Cancer. JCO Precis Oncol. 2021;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 80. | Topham JT, Tsang ES, Karasinska JM, Metcalfe A, Ali H, Kalloger SE, Csizmok V, Williamson LM, Titmuss E, Nielsen K, Negri GL, Spencer Miko SE, Jang GH, Denroche RE, Wong HL, O'Kane GM, Moore RA, Mungall AJ, Loree JM, Notta F, Wilson JM, Bathe OF, Tang PA, Goodwin R, Morin GB, Knox JJ, Gallinger S, Laskin J, Marra MA, Jones SJM, Schaeffer DF, Renouf DJ. Integrative analysis of KRAS wildtype metastatic pancreatic ductal adenocarcinoma reveals mutation and expression-based similarities to cholangiocarcinoma. Nat Commun. 2022;13:5941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 81. | Jones MR, Williamson LM, Topham JT, Lee MKC, Goytain A, Ho J, Denroche RE, Jang G, Pleasance E, Shen Y, Karasinska JM, McGhie JP, Gill S, Lim HJ, Moore MJ, Wong HL, Ng T, Yip S, Zhang W, Sadeghi S, Reisle C, Mungall AJ, Mungall KL, Moore RA, Ma Y, Knox JJ, Gallinger S, Laskin J, Marra MA, Schaeffer DF, Jones SJM, Renouf DJ. NRG1 Gene Fusions Are Recurrent, Clinically Actionable Gene Rearrangements in KRAS Wild-Type Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2019;25:4674-4681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 82. | Qin S, Bai Y, Wang Z, Chen Z, Xu R, Xu J, Zhang H, Chen J, Yuan Y, Liu T, Yang L, Zhong H, Chen D, Shen L, Hao C, Fu D, Cheng Y, Yang J, Bai X hong, Li J. Nimotuzumab combined with gemcitabine versus gemcitabine in K-RAS wild-type locally advanced or metastatic pancreatic cancer: A prospective, randomized-controlled, double-blinded, multicenter, and phase III clinical trial. J Clin Oncol. 2022;40:LBA4011-LBA4011. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 83. | Nicolle R, Gayet O, Duconseil P, Vanbrugghe C, Roques J, Bigonnet M, Blum Y, Elarouci N, Armenoult L, Ayadi M, de Reyniès A, Puleo F, Augustin J, Emile JF, Svrcek M, Arsenijevic T, Hammel P, Giovannini M, Grandval P, Dahan L, Moutardier V, Gilabert M, Van Laethem JL, Bachet JB, Cros J, Iovanna J, Dusetti NJ. A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann Oncol. 2021;32:250-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 84. | Fraunhoffer N, Chanez B, Teyssedou C; Pdac Chemo Sensitivity Prediction Working Group, Iovanna JL, Mitry E, Dusetti NJ. A Transcriptomic-Based Tool to Predict Gemcitabine Sensitivity in Advanced Pancreatic Adenocarcinoma. Gastroenterology. 2023;164:476-480.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 85. | Fraunhoffer N, Teyssedou C, Pessaux P, Bigonnet M, Dusetti NJ, Iovanna J. Development and evaluation of 5FUCore, OxaliCore, and IrinoCore transcriptomic signatures for predicting response to individual drugs in the FOLFIRINOX regimen: Towards toxicity reduction and personalized chemotherapy in PDAC. J Clin Oncol. 2023;41:4141-4141. [DOI] [Full Text] |
| 86. | Reiss KA, Mick R, O'Hara MH, Teitelbaum U, Karasic TB, Schneider C, Cowden S, Southwell T, Romeo J, Izgur N, Hannan ZM, Tondon R, Nathanson K, Vonderheide RH, Wattenberg MM, Beatty G, Domchek SM. Phase II Study of Maintenance Rucaparib in Patients With Platinum-Sensitive Advanced Pancreatic Cancer and a Pathogenic Germline or Somatic Variant in BRCA1, BRCA2, or PALB2. J Clin Oncol. 2021;39:2497-2505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 140] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 87. | Shroff RT, Hendifar A, McWilliams RR, Geva R, Epelbaum R, Rolfe L, Goble S, Lin KK, Biankin AV, Giordano H, Vonderheide RH, Domchek SM. Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precis Oncol. 2018;2018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 88. | Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, Turpin B, Dowlati A, Brose MS, Mascarenhas L, Federman N, Berlin J, El-Deiry WS, Baik C, Deeken J, Boni V, Nagasubramanian R, Taylor M, Rudzinski ER, Meric-Bernstam F, Sohal DPS, Ma PC, Raez LE, Hechtman JF, Benayed R, Ladanyi M, Tuch BB, Ebata K, Cruickshank S, Ku NC, Cox MC, Hawkins DS, Hong DS, Hyman DM. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med. 2018;378:731-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1577] [Cited by in RCA: 1923] [Article Influence: 274.7] [Reference Citation Analysis (0)] |
| 89. | Subbiah V, Cassier PA, Siena S, Garralda E, Paz-Ares L, Garrido P, Nadal E, Vuky J, Lopes G, Kalemkerian GP, Bowles DW, Seetharam M, Chang J, Zhang H, Green J, Zalutskaya A, Schuler M, Fan Y, Curigliano G. Pan-cancer efficacy of pralsetinib in patients with RET fusion-positive solid tumors from the phase 1/2 ARROW trial. Nat Med. 2022;28:1640-1645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 136] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 90. | Subbiah V, Wolf J, Konda B, Kang H, Spira A, Weiss J, Takeda M, Ohe Y, Khan S, Ohashi K, Soldatenkova V, Szymczak S, Sullivan L, Wright J, Drilon A. Tumour-agnostic efficacy and safety of selpercatinib in patients with RET fusion-positive solid tumours other than lung or thyroid tumours (LIBRETTO-001): a phase 1/2, open-label, basket trial. Lancet Oncol. 2022;23:1261-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 224] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 91. | Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, Paulson AS, Borad MJ, Gallinson D, Murphy AG, Oh DY, Dotan E, Catenacci DV, Van Cutsem E, Ji T, Lihou CF, Zhen H, Féliz L, Vogel A. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 809] [Cited by in RCA: 1065] [Article Influence: 213.0] [Reference Citation Analysis (0)] |
| 92. | Javle M, Roychowdhury S, Kelley RK, Sadeghi S, Macarulla T, Weiss KH, Waldschmidt DT, Goyal L, Borbath I, El-Khoueiry A, Borad MJ, Yong WP, Philip PA, Bitzer M, Tanasanvimon S, Li A, Pande A, Soifer HS, Shepherd SP, Moran S, Zhu AX, Bekaii-Saab TS, Abou-Alfa GK. Infigratinib (BGJ398) in previously treated patients with advanced or metastatic cholangiocarcinoma with FGFR2 fusions or rearrangements: mature results from a multicentre, open-label, single-arm, phase 2 study. Lancet Gastroenterol Hepatol. 2021;6:803-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 264] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 93. | Goyal L, Meric-Bernstam F, Hollebecque A, Valle JW, Morizane C, Karasic TB, Abrams TA, Furuse J, Kelley RK, Cassier PA, Klümpen HJ, Chang HM, Chen LT, Tabernero J, Oh DY, Mahipal A, Moehler M, Mitchell EP, Komatsu Y, Masuda K, Ahn D, Epstein RS, Halim AB, Fu Y, Salimi T, Wacheck V, He Y, Liu M, Benhadji KA, Bridgewater JA; FOENIX-CCA2 Study Investigators. Futibatinib for FGFR2-Rearranged Intrahepatic Cholangiocarcinoma. N Engl J Med. 2023;388:228-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 275] [Article Influence: 137.5] [Reference Citation Analysis (0)] |
| 94. | André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, Yamashita T, Lu YS, Inoue K, Takahashi M, Pápai Z, Longin AS, Mills D, Wilke C, Hirawat S, Juric D; SOLAR-1 Study Group. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2019;380:1929-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1607] [Cited by in RCA: 1690] [Article Influence: 281.7] [Reference Citation Analysis (0)] |
| 95. | Di Leo A, Johnston S, Lee KS, Ciruelos E, Lønning PE, Janni W, O'Regan R, Mouret-Reynier MA, Kalev D, Egle D, Csőszi T, Bordonaro R, Decker T, Tjan-Heijnen VCG, Blau S, Schirone A, Weber D, El-Hashimy M, Dharan B, Sellami D, Bachelot T. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018;19:87-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 303] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 96. | Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, Garbe C, Jouary T, Hauschild A, Grob JJ, Chiarion-Sileni V, Lebbe C, Mandalà M, Millward M, Arance A, Bondarenko I, Haanen JB, Hansson J, Utikal J, Ferraresi V, Kovalenko N, Mohr P, Probachai V, Schadendorf D, Nathan P, Robert C, Ribas A, DeMarini DJ, Irani JG, Swann S, Legos JJ, Jin F, Mookerjee B, Flaherty K. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015;386:444-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 977] [Cited by in RCA: 1036] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 97. | Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, Garbe C, Schadendorf D, Krajsova I, Gutzmer R, Chiarion-Sileni V, Dutriaux C, de Groot JWB, Yamazaki N, Loquai C, Moutouh-de Parseval LA, Pickard MD, Sandor V, Robert C, Flaherty KT. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF-mutant melanoma (COLUMBUS): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018;19:603-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 711] [Article Influence: 101.6] [Reference Citation Analysis (0)] |
| 98. | Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu YL, Thomas M, O'Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Jänne PA. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385-2394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2534] [Cited by in RCA: 2683] [Article Influence: 223.6] [Reference Citation Analysis (0)] |
| 99. | Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim DW, Ou SI, Pérol M, Dziadziuszko R, Rosell R, Zeaiter A, Mitry E, Golding S, Balas B, Noe J, Morcos PN, Mok T; ALEX Trial Investigators. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2017;377:829-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1385] [Cited by in RCA: 1736] [Article Influence: 217.0] [Reference Citation Analysis (0)] |
| 100. | Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, Riely GJ, Solomon BJ, Wolf J, Thomas M, Schuler M, Liu G, Santoro A, Lau YY, Goldwasser M, Boral AL, Engelman JA. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1065] [Cited by in RCA: 1136] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 101. | von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, Wolmark N, Rastogi P, Schneeweiss A, Redondo A, Fischer HH, Jacot W, Conlin AK, Arce-Salinas C, Wapnir IL, Jackisch C, DiGiovanna MP, Fasching PA, Crown JP, Wülfing P, Shao Z, Rota Caremoli E, Wu H, Lam LH, Tesarowski D, Smitt M, Douthwaite H, Singel SM, Geyer CE Jr; KATHERINE Investigators. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N Engl J Med. 2019;380:617-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1122] [Cited by in RCA: 1668] [Article Influence: 278.0] [Reference Citation Analysis (0)] |
| 102. | Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, Ciruelos E, Ferrero JM, Schneeweiss A, Heeson S, Clark E, Ross G, Benyunes MC, Cortés J; CLEOPATRA Study Group. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1303] [Cited by in RCA: 1505] [Article Influence: 150.5] [Reference Citation Analysis (0)] |